Spectrum of MECP2 mutations in Indian females with Rett Syndrome - a large cohort study
Rajni Khajuria, Neerja Gupta, Kees E.P.van Roozendaal, Savita Sapra, Manju Ghosh, Sheffali Gulati, Eric E.J.Smeets, Leopold M.G.Curfs, Madhulika Kabra
1Division of Genetics, Department of Pediatrics, All India Institute of Medical Sciences, New Delhi 110029, India.
2Division of Neurology, Department of Pediatrics, All India Institute of Medical Sciences, New Delhi 110029, India.
3Department of Clinical Genetics, Maastricht University Medical Centre, P.O.Box 5800, Maastricht 6202 AZ, The Netherlands.
4Governor Kremers Center, Maastricht University Medical Centre, P.O.Box 5800, Maastricht 6202 AZ, The Netherlands.
5School for Public Health and Primary Care CAPHRI, Maastricht University, P.O.Box 616, Maastricht 6200 MD, The Netherlands.
6School for Oncology and Developmental Biology GROW, Maastricht University, P.O.Box 616, Maastricht 6200 MD, The Netherlands.
Abstract
Keywords: Rett syndrome, RTT, MECP2, DNA sequencing, genotype-phenotype correlations, mutation spectrum, India
INTRODUCTION
Rett syndrome (RTT, OMIM #312750) is a childhood neurodevelopmental disorder primarily affecting females.It is caused by mutations in the Methyl-CpG-Binding Protein 2 gene (MECP2, OMIM ★300005), an important regulator of gene expression, located at Xq28[1].
Developmental regression is a hallmark of RTT, the ongoing pathology of which is still being unraveled.Symptoms include loss of acquired skills, especially in relation to communicative and motor performance.Clinical developmental profiles, non-specific early in life, become more specific later.To support clinical diagnosis, a staging system has been developed as a framework that delineates the evolving symptoms.This includes stages of early-onset stagnation, rapid developmental regression, a pseudo-stationary stage, and late motor deterioration.We do not yet fully understand the biological pathways underlying the outward presentations of the RTT[2].The multi-functionality ofMECP2 suggests there are many downstream pathways that are interesting for understanding the pathophysiology of RTT.
Variants inMECP2 can be identified in 95%-97% of individuals with Classical RTT, using a combination of mutation detection techniques[3].Classical RTT is characterized by apparently normal early development, arrest of developmental progress at 6-18 months followed by regression of social contact, language, and hand skills.However, thereafter, improvements in social behavior and eye contact have been observed.The most recent revision of the clinical criteria for diagnosis of RTT[4]allows for a broader interpretation of regression and partial recovery than was previously acknowledged and has led to increased understanding of the disease[5].Clinicians should be aware of these criteria, for counseling of families as they seek to understand the stages their child will encounter and for the application of management strategies that may help to ameliorate or compensate for loss of skills at the different stages across the lifespan.A review of the literature of mutation analysis in large cohorts of RTT patients in Western populations indicates that the majority are sequence variations and only a small proportion of cases have large deletions/duplications[6,7].To the best of our knowledge, there are only two studies on mutation spectrum of RTT from India, including both typical and atypical RTT[8,9]and until now no study has been reported on a large cohort of classical RTT patients describing the spectrum ofMECP2sequence variations and to evaluate the genotype-phenotype correlations based on the mutation spectrum.The objectives of the present study were: (1) to study the clinical phenotype of Indian patients with classical RTT; (2) to identify the spectrum ofMECP2 sequence variations in a large cohort of Indian RTT patients and determine genotypephenotype correlation, if any; and (3) to predict the effects ofMECP2 variations on MeCP2 Protein using bioinformatics.
METHODS
Seventy-two sporadic classical RTT patients (all females) were included in this study from Pediatric OPD, Pediatric wards, Pediatric Neurology and Medical Genetics services of the All India Institute of Medical Sciences New Delhi, India.Patients were defined as classical when they showed a period of regression and fulfilled all the main inclusion criteria (partial or complete loss of acquired purposeful hand skills; partial or complete loss of acquired spoken language; gait abnormalities; impaired or absence of ability to walk; stereotypic hand movements) as per the revised diagnostic criteria for classical RTT[4].The patients not fulfilling the major criteria and having the clinical signs mentioned in exclusion criteria[4]were excluded from the study.Exclusion criteria were: evidence of brain injury secondary to perinatal or postnatal events, neuro-metabolic disorders or infection causing neurological problems.Additionally, children having grossly abnormal psychomotor development in first six months of life were also excluded.Ethical approval for the present study was taken from the Ethics Committee of the Institute.Proper information about the study was given to all families and written informed consent was obtained from the parent/guardian.Enrolled patients represented all regions of India, with the majority from northern India.Patients were evaluated by a team comprising of a clinical geneticist, a pediatric neurologist, and a child psychologist before inclusion in the study.All clinical details were recorded in a predesigned proforma.
Five-milliliter blood samples were collected from all patients in EDTA vacutainer and DNA was extracted by standard phenol-chloroform method.DNA samples were analyzed using bidirectional Sanger sequencing for sequence variations followed by quantitative analysis using Multiplex Ligationdependent Probe Amplification (MLPA) for deletion/duplication analysis ofMECP2gene.The gene nomenclature used was according to guidelines of HGNC (Hugo Genome Nomenclature Committee) and the recommended sequence variant nomenclature of HGVS (Human Genome Variation Society)[10].Any change found in DNA sequence of the RTT patients was also analyzed in their family members (except three cases) to confirm its origin.The sequence variant profile was compared with the clinical presentation to generate genotype-phenotype correlations.All novel variants identified in this study were submitted to National Centre for Biotechnology Information (NCBI) GenBank (http://www.ncbi.nlm.nih.gov/GenBank) and RettBASE: IRSFMECP2Variation Database (http://mecp2.chw.edu.au)[11].
MECP2 screening
The coding region of exons 2-4 ofMECP2gene (transcript 1, MeCP2_e2) including flanking exon/intron boundaries was amplified by PCR using seven overlapping primer sets (2.1, 2.2, 3.1, 3.2, 3.3, 4.1, and 4.2) ofMECP2gene published elsewhere[1,12].PCR amplification was performed in a final volume of 25 μL containing 10× PCR buffer with 1.5 mM MgCl2, 0.25 mM dNTPs, 0.625 U Taq polymerase, 1 pM/μL each of forward and reverse primer, and 50 ng of DNA.All samples were analyzed by direct bidirectional Sanger sequencing.The data were interpreted and compared with reference sequence ofMECP2(NM_004992.3) gene.MLPA technique was used to screen the RTT patients who were negative forMECP2sequence variations on DNA sequencing to check for gross rearrangements[6,13,14].SALSA MLPA kits P015-D2 and P015-E2 (MRC-Holland, Amsterdam, The Netherlands) were used.All cases with positive or aberrant results were rerun in a second MLPA reaction for confirmation.
Bioinformatic analysis
In the present study, various bioinformatic software packages were used for prediction of pathogenicity ofMECP2 sequence variations.Prediction tools such as PolyPhen (Polymorphism Phenotyping) (http://genetics.bwh.harvard.edu/pph/), SIFT (Sorting Tolerant from Intolerant) (http://sift.jcvi.org/), and SNPs3D (http://www.snps3d.org/) were used to predict if the non-synonymous particular variants are likely to be deleterious.Prediction of the effect of synonymous variants was done using Codon Usage Database (www.kazusa.or.jp/codon/).In-silico splice site analysis was done using Alamut (http://www.interactivebiosoftware.com/alamut/) for prediction of splice-site variants.We used MeCP2_e2 transcript of MeCP2 for pair wise alignment and the sequences were derived from NCBI HomoloGene (http://www.ncbi.nlm.nih.gov/homologene).The alignments were produced using ClustalW (http://www.ebi.ac.uk/Tools/clustalw/).Statistical analysis
We used Analysis of Variance (ANOVA) to compare the group mean score on dependent variables, covarying for age.Categorical variables were analyzed for significant associations using Pearson χ2.STATA version 9 was used for all statistical analysis.
RESULTS
Phenotypic features
All patients had apparently normal prenatal and perinatal history with normal early psychomotor development.We did not have head circumference records of all patients at birth, but all had microcephaly at the time of inclusion.The mean age of onset of symptoms in classical RTT patients was 16 ± 4.6 months (range: 6-30) and the median age was 16 months.The mean age at diagnosis of classical RTT patients was 54 ± 34.9 months (range: 18-186 months) while the median age was 42 months.All patients had partial or complete loss of acquired purposeful hand skills.Stereotypic hand movements were present in all patients and the common hand stereotypes observed in our patients were hand wringing, washing, mouthing, clenching, finger rolling, tapping, and clapping.All patients had partial or complete loss of acquired language and most patients spoke only monosyllables or babbling.All patients had gait problems, of which 40% (29/72 patients) could not walk, 39% (28/72 patients) had an impaired gait, and 21% (15/72 patients) could walk with support.
MECP2 sequence variants
Using DNA sequencing, we identifiedMECP2sequence variants in coding region with a frequency of 88.9%.Using MLPA, large deletions ofMECP2gene were identified in 9.7% of patients.One patient was identified with intronic variant and no other sequence variant was identified in this patient.In total, 38 different types ofMECP2sequence variations (25 reported and 13 novel) were identified in all 72 classical RTT patients [Table 1].
Six recurrent sequence variants (p.P152R, p.T158M, p.R168X, p.R270X, p.G269fs, and p.R306C) were identified in 36/72 RTT patients.All of these variants were identified in heterozygous form.The p.T158M and p.R270X were the most recurrent sequence variants observed, followed by p.R168X, p.G269fs, p.R306C, and p.P152R variants [Table 2].Twenty-nine patients (40.3%) were found positive with other uncommon heterozygousMECP2 sequence variants and one patient (1.38%) was identified with only one intronic variant c.378-74C>T [Table 1].
Seven patients (six negative forMECP2sequence variants on DNA sequencing and one patient who was identified with only one intronic variant c.378-74C>T) were screened using MLPA analysis for ruling out large deletion/duplications ofMECP2 gene that cannot be identified using Sanger sequencing.In six out of seven (8.33%) RTT patients, large deletions of one or more contiguous exons, especially of exon 3 and 4 ofMECP2,gene were identified.In one patient, an undefined deletion in 3’UTR ofMECP2gene was detected [Table 1], and it was confirmed that there was no sequence change at the probe hybridization site, thus it most likely was a real deletion.
Most of the mutations or sequence variants identified in patients were de novo and were not found in the family members except two cases where different mutation or variant was identified in family members.In one family, the asymptomatic mother of one RTT with p.D17fs mutation was found carrying missense mutation p.H51Q (GenBank accession No.GU812286.1) ofMECP2.In another family, the patient was carrying p.R106W mutation, whereas her asymptomatic father was a carrier of intronic c.378-74C>T variant ofMECP2gene.However, in three cases, the origin of novel large deletions could not be confirmed as the parents were not available for testing.Thirteen novelMECP2 variants were identified in this study in the RTT patients [Table 3], which included four large deletions, six frameshift [p.D17fs (GenBank accession No.FJ212168.1), p.R162fs (GenBank accession No.GU812285.1), p.E290fs (GenBank accession No.GQ203293.1), p.P384fs (GenBank accession No.FJ212171.1), p.V485fs (GenBank accession No.HQ141378), and p.S486fs (GenBank accession No.GQ203295.1)], one missense p.L138S, one synonymous p.G428G (GenBank accession No.FJ212169.1), and one nonsense (p.G92X) variant [Table 3 and Figure 1].
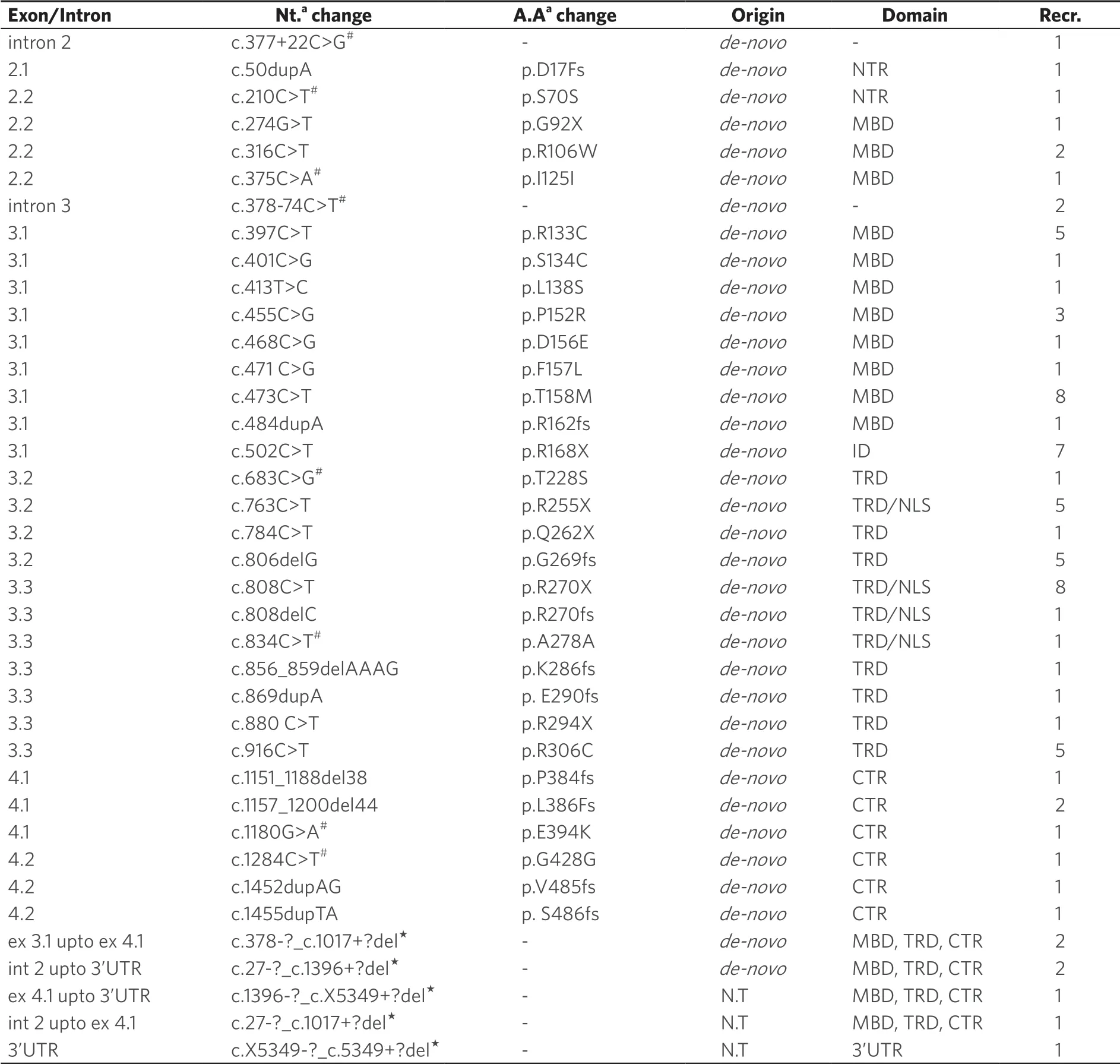
Table 1.List of MECP2 sequence variants identified in RTT patients in the present study
Seven RTT patients were found carrying more than one variant inMECP2 gene [Table 1].One of the RTT patients carrying p.L386fs mutation was also carrying a novel synonymous change p.G428G (c.1284C>T).In two patients carrying mutation p.R270X, one of them was also carrying synonymous change p.S70S (c.210C>T) and the other was carrying an intronic variation c.377+22C>G [Table 1].Another RTT patient with p.R306C (c.916C>T) mutation was also carrying an intronic variation c.378-74C>T in intron 3 ofMECP2 gene [Table 1].Another patient with mutation p.P152R was found to be carrying another missense change p.T228S inMECP2 gene.One of the patients with p.R255X mutation was found to be carrying another synonymous change p.I125I and another patient carrying p.K286fs mutation was carrying two more variants p.E394K and p.A278A.
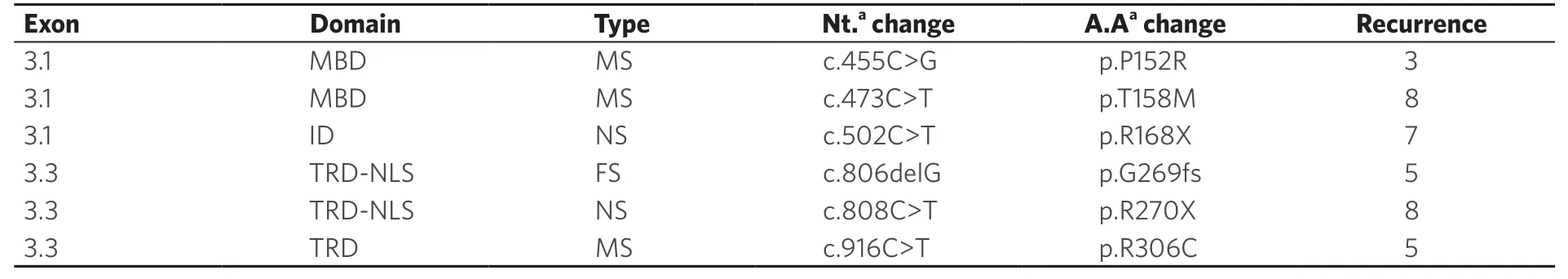
Table 2.Common MECP2 sequence variants found in classical RTT patients in present study

Table 3.List of novel MECP2 sequence variants identified in RTT patients in present study
Most of theMECP2 variants identified in present study were missense (28%), followed by frameshift (25.6%), large deletions (17.9%), nonsense (15.3%), synonymous (10.2%), and intronic (5.1%).The majority ofMECP2sequence variations were found in Methyl Binding Domain (MBD), followed by Transcription Repression Domain (TRD), C-terminal region (CTR), N-terminal region, and Interdomain (ID).Thus, they were distributed over all the domains ofMECP2[Figure 2].
Bioinformatic analysis
The majority of missense variants in the present study were predicted as deleterious, with the exception of variants such as p.T228S that were predicted as benign or non-deleterious using prediction Polyphen, SNPs3D, and SIFT; as the patient carrying this change was carrying another deleterious change p.P152R, it can be considered a neutral change.Based on the findings of codon usage database, the synonymous variant p.I125I changed the most preferred codon ATC to a least preferred codon ATA, whereas, in the sequence variations p.S70S, p.G428G, and p.A278A, the most preferred codons were changed to less preferred codons, hence were likely predicted to be associated with disease in some way, although it is difficult to explain the implication of synonymous variants in the disease process.
The sequence of human MeCP2 was aligned with MeCP2 of cattle, dog, and mouse, and it was found that most of the missense and truncating variants identified were affecting the conserved residues of the protein, thus predicted to be affecting the protein in some way.
Genotype-phenotype correlations
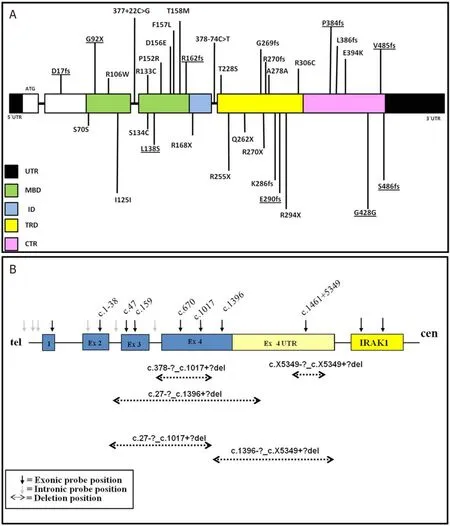
Figure 2.A: diagrammatic representation of MECP2 sequence variants identified in this study (novel variants are underlined); and B: diagrammatic representation of MECP2 deletions found in the present study
Significant correlation was seen for type of sequence variation and development of clinical features.In the present study, the recurrent nonsense sequence variations p.R270X, p.R168X, and p.R255X were found significantly associated with development of scoliosis (Pearson χ2;P= 0.002) and presence of mild facial dysmorphism (Pearsonχ2test;P= 0.006).Patients with early truncating sequence variations developed poor verbal communication as compared to patients with missense sequence variations (Pearsonχ2test;P= 0.000).The ability to walk was severely deteriorated in patients with early truncating variants as compared to patients with missense variants and large deletions (Pearsonχ2test;P= 0.011).The loss of purposeful use of hands was observed to be significantly associated with missense variants compared to truncating variants (Pearsonχ2test;P= 0.032).However, generally, no significant difference was observed with the type of sequence variation and the age of onset of symptoms; exceptions were observed in patients with p.R270X and p.R168X with earlier onset of symptoms [Table 4].The mortality was also higher in RTT patients with nonsense variants than missense variants, although we were unable to find any statistical significance due to small number of patients.Most of the missense sequence variations in our study were clustered in MBD of MeCP2 and the early truncating sequence variations were similarly clustered in TRD of MeCP2 (Pearsonχ2test;P= 0.000).

Table 4.Genotype-phenotype correlations of RTT patients based on the profile of common sequence variants
DISCUSSION
Until now, few reports on variant analysis ofMECP2 gene in classical RTT have been published from India[8,9,15], thus there are scarce data regarding mutation spectrum and genotype-phenotype correlations.The present study evaluated 72 Classical RTT females based on the revised diagnostic criteria and revealed a heterogeneous spectrum of sequence variants including 13 novel variants with a detection rate of 98.6% using a combination of Sanger sequencing and MLPA, which is higher compared to other studies on mutation spectrum of RTT.The results of the present study emphasize the need for a careful and meticulous clinical evaluation that is likely to select appropriate cases with a good yield on molecular testing, which is important in resource constrained settings.
It has been reported thatMECP2variants can be detected with a frequency of more than 95%-97% in classical RTT by screening coding region and flanking intronic regions ofMECP2gene using PCR based techniques[3,16], but these methods do not detect gross rearrangements, which could be present in a significant proportion of classical RTT patients[5].Several groups have identified gross rearrangements using quantitative analysis ofMECP2using MLPA in the patients where the cause of RTT remains unknown after sequencing[6,13,14,17].
In the present study, we were able to findMECP2 sequence variations in overall 90.3% of RTT patients using DNA sequencing.Using MLPA analysis, we were able to detect large putative deletions ofMECP2 in all the classical RTT patients, which were negative on DNA sequencing.MLPA increased the detection rate ofMECP2 sequence variants identified in RTT patients from 90.3% to 98.6%.We propose that MLPA analysis ofMECP2 is crucial and needs to be performed in classical RTT patients.Large deletions can be missed using DNA sequencing and reaffirms the view that largeMECP2 deletions are an important cause of classical RTT[6,13].In this study, the majority of the RTT patients were carrying the C>T transitions, supporting the reported literature[12].
Data from different western studies have shownMECP2sequence variation frequency between 70% and 97% in classical RTT[1,3,7,12,16,18].A literature search revealed many studies on RTT from Asia that reportedMECP2sequence variation frequency of 50%-92.5% in classical RTT patients[19-23].In the two Indian studies published thus far, the detection rates ofMECP2 variations was lower as compared to our study[8,9].The study by Lallaret al.[9], which included 19 RTT patients (14 classical and 5 atypical), reported a detection rate of 93% in classical RTT girls using a combination of Sanger sequencing followed by MLPA analysis, supporting our findings.In the other Indian study by Daset al.[8], investigating 90 individuals with suspected RTT phenotype, 19 differentMECP2 mutations and polymorphisms were identified in 27/90 (30%) patients while the rest remained uncharacterized.
The high rate ofMECP2sequence variation detected in the present study compared to the data from other Asian, Indian, and western studies can be explained by the fact that a strict inclusion/exclusion of classical RTT patients was adopted based on revised clinical diagnostic criteria of RTT[4]and involvement of a multidisciplinary team for clinical evaluation of the patients.Our study supports the previous findings that clinical stringency based on diagnostic criteria can increase the mutation detection rate in RTT patients and emphasizes the importance of diagnostic criteria in the assessment of RTT patients[24].
The worldwide reported eight hotspotMECP2 sequence variants p.R106W, p.R133C, p.T158M, p.R168X, p.R255X, p.R270X, p.R294X, and p.R306C were identified with a frequency of 57% in our study, which is similar to previously reported western and Asian studies[3,7].The hotspot variant p.R294X identified recurrently in western population was found in only one patient in the present study, whereas other hotspot variants, namely p.P152R, p.G269fs, and p.L386fs, were identified in more than one patient.In another Indian study by Daset al.[8], which included 90 patients (suspected RTT patients), showedMECP2sequence variations in only 30% of patients (27/90 patients), of which seven hotspot variants, namely p.R106W, p.R133C, p.T158M, p.R168X, p.R255X, p.R270X, and p.R306C (sequence variant p.R294X was not reported in this study), were identified with frequency of 51.8%.The other Indian study by Lallaret al.[9]reported four common hotspot variants (p.R168X, p.T158M, p.R306C, and p.R255X).The present study indicates that the variant p.R294X may not be a hotspot sequence variation in Indian RTT patients and instead p.G269fs, p.P152R, and p.L386fs, may be the hotspot sequence variations along with the seven other recurrent variants identified.Multiple recurrences of these three variant (p.G269fs, p.P152R, and p.L386fs) in the present study indicate that these variants could be the hotspot variants specific to Indian population.Larger studies from India are needed to confirm and support these findings.
The dentification of 13 novel variants, including four large deletions, was another highlight of our study, which emphasizes the genetic heterogeneity ofMECP2 variants and underlines the need for generating population specific data.In view of identification of other recurrent variants in our study along with reported hot spot mutations, sequencing theMECP2 gene (beginning with exons 3 and 4) followed by MLPA testing if sequencing results are negative is recommended rather than targeted testing.
The majority of the variants were distributed in the functional domain ofMECP2 with most missense variants clustered in MBD and truncating variants in ID and TRD ofMECP2 [Figure 2], which support the findings of previous studies[3,7,8,16].All variations identified in the patients were de novo.In one of the families, the mother was found carrying a different novel variant than her daughter and has no symptoms of the disease, as we reported previously[25].
Bioinformatic analysis revealed that most of theMECP2missense variants clustered in MBD of MeCP2 were of damaging or deleterious nature, whereas all the missense variants identified in CTR were predicted as benign or non-deleterious.These findings support previously reported studies on RTT patients[26,27].Only two missense variants were identified in TRD ofMECP2and bioinformatic analysis of the recurrent missense variant p.R306C in TRD predicted it as damaging, whereas the other non-recurrent missense variant p.T228S was predicted as benign or non-deleterious.The patient carrying this variant p.T228S was carrying another deleterious variant, p.P152R.As the number of missense variants identified in TRD in the present study was small, the effect of these variants could not be explored, but the findings of the present study strongly indicate that sequence variations inMECP2gene are the major cause of classical RTT.
There are many studies on genotype-phenotype correlation in RTT patients from 2001 to 2016 but the results are inconsistent[16,18,21,27-32].This inconsistency can be due to the use of different diagnostic criteria and severity scales for evaluation of the patients.There are currently no data on genotype-phenotype correlation in Indian patients with RTT.In this study, we tried to correlate the type and position of identified sequence variants with the phenotype of the patients.
When different types of sequence variations were compared with the phenotype, it was found that patients carrying early truncating variants showed more severe phenotype as compared to the patients with late truncating and missense sequence variants, supporting the findings from a previous study[18].
While comparing the types of sequence variations with their location inMECP2, it was found that the variants leading to severe phenotype were clustered more in functional domains of MeCP2.Only 5% of early truncating variants were present in MBD and CTR as compared to 20% present in the TRD.Only 5% of late truncating variants were observed in the CTR of MeCP2.The rare presence of missense variants in TRD or CTR of MeCP2 as compared to MBD can be explained on the basis that missense sequence variations may have mild impact on protein function compared to truncating sequence variations, resulting in a mild phenotype.These findings are in support of a previous study[26].The only recurrent missense variant in TRD was p.R306C and the other missense sequence variations found in TRD and CTR (p.T228S, p.E394K, p.E397K, and p.P430S) were observed with single occurrence, supporting the previous hypothesis that most of the missense sequence variations within the TRD might be benign variants[26].
In conclusion, this study presents the largest cohort describing the molecular genetics of classical RTT from India.To the best of our knowledge, this is the first study showing the highest detection rate ofMECP2variants in the patients with classical RTT and supports that clinical stringency based on revised diagnostic criteria can increase the variant detection rate.We propose the followingMECP2screening strategy in Indian patients with Classical RTT.Exon 3 ofMECP2should be screened first, followed by exons 2 and 4 using Sanger sequencing, and, in turn, followed by quantitative analysis using MLPA.The present study adds information on the molecular characterization of Indian patients with RTT and also reports 13 novel variants expanding the genotypic spectrum of RTT.The findings can be useful for diagnostic testing, genetic counseling, and prenatal testing.
Limitations and future research
Although the study was performed on a large cohort of patients, we were unable to prove the functional impact of novel variations on theMECP2 protein as only software prediction tools were used.It would be useful to conduct functional studies for the new variants identified.A review of the current literature indicates thatMECP2variations can cause other neurodevelopmental phenotypes such as neonatal encephalopathy and atypical RTT phenotype in both males and females.This study lacks this information as only classical cases were included.A larger study is required to provide this information.
DECLARATIONS
Acknowledgments
We gratefully thank the patients and the families for their support and participation in this study.We thank Dr.Angus Clarke, Cardiff University for providing positive controls for common mutations.We thank Dr.R.Bandts (Maastricht University Medical Center) for providing technical support related to MLPA analysis.We thank Dr.Sumita Danda (Christian Medical College and Hospital, Vellore) and Dr.Veena Kalra (Indraprastha Apollo hospitals, New Delhi) for referring their patients to our institute.
Authors’ contributions
Literature review, patient enrollment, experiment validation, planning and execution, data analysis and interpretation, bioinformatics analysis, wrote, prepared and reviewed manuscript: Khajuria R
Clinical evaluation, patient enrollment, counseling of families helped in planning the study, preparation and critical evaluation of the manuscript, reviewed the manuscript: Gupta N
Provided support with MLPA data interpretation and bioinformatics analysis using Alamut software, reviewed the manuscript: van Roozendaal KEP, Smeets EEJ, Curfs LMG
Helped in developmental assessment, behavioural evaluation and intervention for the patients, reviewed the manuscript: Sapra S
Helped in patient enrollent and counseling, reviewed the manuscript: Ghosh M
Helped in neurological evaluation and management of neurological co-morbidities and manuscript review: Gulati S
Conceptualized the study, helped in preparation of study protocol, helped in clinical evaluation, enrollment and counseling of patients, helped in preparation and critical evaluation of the study, reviewed of the manuscript, will act as the guaranteer for this study: Kabra M
Availability of data and materials
Data source is from patients enrolled for the study.For data details corresponding author may be contacted.
Financial support and sponsorship
This study was partially funded as research fellowship from Indian Council of Medical Research, New Delhi (Grant Number: 45/8/2006/Hum/BMS).
Conflicts of interest
All authors declared that there are no conflicts of interest
Ethical approval and consent to participate
Study was approved by the Institutional Ethics committee and ethical issues were considered during the study.
Consent for publication
Not applicable.
Copyright
© The Author(s) 2020.
 Journal of Translational Genetics and Genomics2020年2期
Journal of Translational Genetics and Genomics2020年2期
- Journal of Translational Genetics and Genomics的其它文章
- Redefining infantile-onset multisystem phenotypes of coenzyme Q10-deficiency in the next-generation sequencing era
- Cryogenic electron paramagnetic resonance spectroscopy of flash-frozen tissue for characterization of mitochondrial disease
- Role of transfer RNA modification and aminoacylation in the etiology of congenital intellectual disability
- Mitochondrial translation defects and human disease
- The North American mitochondrial disease registry
- Intellectual disability, the long way from genes to biological mechanisms
