Cryogenic electron paramagnetic resonance spectroscopy of flash-frozen tissue for characterization of mitochondrial disease
Brian Bennett
Department of Physics, Marquette University, Milwaukee, WI 53233, USA.
Abstract
Keywords: Mitochondrial disease, electron paramagnetic resonance, electron paramagnetic resonance spectroscopy, mitochondria
INTRODUCTION
Mitochondria are characterized by mitochondrial respiratory chain (MRC) complexes that catalyze redox reactions and act as electron transfer conduits during energy metabolism, driving ATP synthesis while closely chaperoning potentially toxic one-electron redox equivalents.The MRC proteins contain a variety of redox-active centers, including iron-sulfur clusters, heme, copper ions, and quinones.Each of these can exist in distinct oxidation states with environmental dependencies that include (1) the redox environment of the mitochondrion and the cell; (2) the oxidative stress burden and history; (3) the integrity of the mitochondrial membrane; and (4) the functionality of the individual MRC components and the electron transfer chain overall.Other non-MRC metalloproteins, particularly aconitase and catalase, provide distinct and specific biomarkers for oxidative stress.
Mitochondrial diseases (MD) can arise where depletion of mitochondrial DNA (mtDNA)[1,2], or mutations in mtDNA and/or nuclear DNA lead to altered mitochondrial function[3-7].Altered catalytic and electron transferring activities of mitochondrial complexes I-V have been associated with MD, and physiological consequences of MRC defects include reduced metabolic capacity, reduced ATP synthesis, and increased oxidative and nitrosative stress[8-18].Symptoms are manifold and include weakness (from central nervous system, peripheral nerve, and/or skeletal muscle disease), pain, intolerance of some general anesthetics and anti-epileptic drugs, gastrointestinal disorders, ophthalmoplegia and/or visual failure, failure to thrive, cardiac and respiratory disease, liver disease, diabetes, seizures, sensorineural hearing loss, mental retardation, dementia, movement disorders, increased susceptibility to infection, and pregnancy loss[5,6,19-41].The primary manifestation of mitochondrial dysfunction is an inability to generate enough energy from metabolism to maintain the functional or structural integrity of the associated tissues.Thus, MD is particularly debilitating when dysfunctional mitochondria are present in tissues with high energy requirements, such as the cerebrum, nerves and muscle.The other major underlying pathology is oxidative stress i.e.the production of reactive oxygen species (ROS) and reactive nitrogen species (RNS).ROS and RNS are free radicals and related compounds, some of which exhibit high reactivity and can damage proteins, lipids and nucleic acids.Depolarization of the mitochondrial membrane can occur due to physical damage.Damage to individual MRC components can exacerbate oxidative stress and an oxidative stress cascade can occur.Identification of low ATP production or oxidative stress to be primarily responsible for symptoms may have profound consequences for subsequent care and therapy.
Traditional diagnosis of MD includes clinical presentation of symptoms, family history, pathology, metabolic profiling, enzyme activity levels, electrophysiology, magnetic resonance imaging of brain and magnetic resonance spectroscopy of metabolites, and mtDNA analysis[7,10,34,42-55].Diagnosis can be challenging, given that (1) mitochondrial metabolism can be affected in non-mitochondrial diseases; (2) there can be extensive variability in the distribution of abnormal mitochondria within an individual patient, resulting in “false negative” testing to occur when tissues containing the abnormal mitochondria are not tested; and (3) there are no uniform, clear cut pathological abnormalities to distinguish all MD patients from patients with other disorders, to the extent that some biopsy specimens look structurally normal.MD can also present with an extraordinary range of clinical symptoms, and laboratory testing abnormalities are common.MD is often suspected clinically as part of the differential diagnosis in patients with diseases involving the brain, muscle, or liver and MD-like symptoms are often exhibited in early childhood.Attempts to improve MD diagnosis have included the use of diagnostic algorithms to predict the likelihood of MD, DNA sequencing, and omics methods, each with associated advantages and challenges of their own[49,56-62].
Herein, the application of cryogenic electron paramagnetic resonance spectroscopy (EPR) of intact, flashfrozen tissue is described for the diagnosis and characterization of metabolic dysfunction in general, and MD in particular.The article describes (1) the principles of EPR; (2) the EPR signals exhibited by mammalian and human tissue; (3) sample preparation considerations; (4) analysis methods; and (5) the relevance of EPR to MD and integration of EPR results with other, complementary investigative methods.
ELECTRON PARAMAGNETIC RESONANCE
EPR, in the present context, is the measurement of the magnetic field-dependence of absorption of a photon by a paramagnetic substance i.e.containing unpaired electrons in one or more atomic, ionic, or molecular orbitals with a non-zero net spin-magnetic moment[63].Almost all paramagnets of biomedical importance are either free radicals, transition metal ions, or clusters.The magnetic field-dependent resonant spintransition of a paramagnetic electron occurs via its interaction with the oscillating magnetic field of an incident photon.The total “magnetic field” experienced by an electron can be due to (1) contributions from an applied laboratory magnetic field; (2) arising from spin-orbit coupling; (3) arising from zero-field splitting in individual ions or radicals containing more than one unpaired electron; (4) from nearby additional unpaired electrons (exchange- and dipolar-couplings); and (5) from nearby magnetic nuclei (electronnuclear hyperfine coupling)[64].Additional “fields” due to nuclear Zeeman and quadrupolar interactions can generally be neglected in the present context.EPR can, in principle, provide a wealth of information on the identity, chemical nature, chemical environment, electronic structure, and physical structure of the analyte from analysis of each of these interactions, typically employing computer analysis and simulations, and increasingly with quantum chemistry calculations (density functional theory, Taylor theory)[65-76].However, in the case of biological tissues, the origins and spectroscopic parameters of many of the EPR signals have been well-characterized, as described in detail below.In the studies of concern here, one is primarily interested in (1) assigning each of the signals; and (2) quantifying the species responsible.
Experimentally, the sample is placed in a resonant structure that supports a standing microwave of fixed frequency, and the applied magnetic field is scanned to search for resonant absorption across a wide field envelope[77].A typical microwave frequency is 9.5 GHz, with magnetic field scans of 0-1 T (0-10,000 G; the derived unit of magnetic flux density, the gauss G, is generally used to label the abscissae of EPR spectra).Multiple conflicting factors need to be considered when choosing a frequency for EPR.The present author’s opinion is that 18 GHz may be the best overall frequency for studies on biological tissue and commercial, high-quality 9.5 GHz instruments represent a reasonable compromise.Higher frequencies correspondingly require higher fields and superconducting magnets are generally needed for EPR at > 35 GHz.The EPR spectrum is often presented in the derivative-like mode, ∂χ”/∂B0, due to the use of magnetic field modulation and phase-sensitive detection, and resonant lines in the EPR spectrum are often labeled with “geff-values” in order to remove the frequency-dependence from the resonance position label, asgeff=hν/βB, whereνis the microwave frequency andBis the resonant field.EPR spectra recorded at different frequencies (e.g., on different instruments) can thus be compared using thegeff-values of the signals.WhereS=1/2,geffis equal tog, the Landég-factor incorporating the Zeeman and spin-orbit coupling terms; whereS>1/2,geffis related togthrough the zero-field splitting termE/D[69].Up to threegeff-values for any Kramers’ doublet may be observed, corresponding to the principal tensor orientationsxx, yy,andzz, and are often labeledgx,gy,gz, where relationships to molecular or electronic structure symmetry are obvious, org1,g2,g3, otherwise.Notably, the value ofgfor the free electron, and manyS=1/2systems, is close to 2, whereas forS>1/2the highest value,geff max≤ 4S.
EPR analysis of tissue samples for studies of MD must be carried out at low temperatures.Trivially, but nevertheless important, tissues must be maintained at a temperature sufficiently low (e.g., -80 °C freezer or liquid nitrogen at 77 K) to prevent molecular diffusion that will change the signals over time, so a low temperature is necessary for data collection to preserve sample integrity.Second, to a crude approximation, available EPR signal intensity is often inversely proportional to the absolute temperature, and signal-to-noise can be limited with raw biological material so maximizing it is important.Third, and more fundamentally, substantial population of the ground state in spin-systems with more than one unpaired electron (S>1/2) may require very low temperatures, close to liquid helium (4.2 K).Knowledge of the population of the ground state is necessary for quantitation of signals.The final and often limiting determinant of the temperature for EPR data collection is the relaxation kinetics of the EPR signals[78]; these vary considerably among the signals observed from biological tissues.Although the temperature can be tailored for specific investigation of an individual signal, there is no one optimum temperature for global EPR analysis of biological tissue analysis; however, data collection at 2 temperatures, 12 K and 40 K, is often sufficient when combined with careful analysis[79].
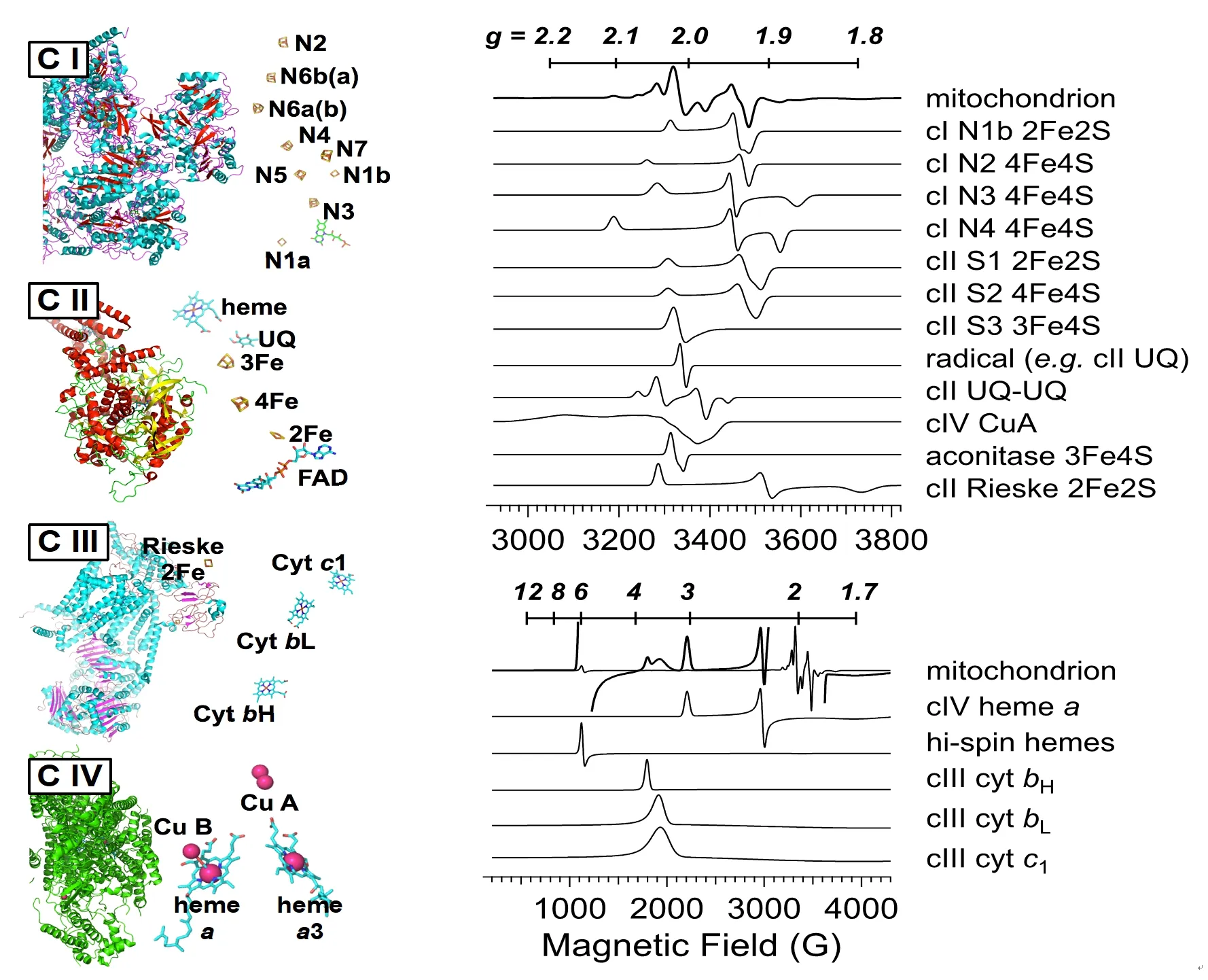
Figure 1.Redox centers in the mitochondrial respiratory chain (left) and EPR signals observed in frozen tissue with geff -values on the top scale and resonant fields at 9.5 GHz on the bottom scale (right).The labels “C I”, “C II”, “C III”, and “C IV” refer to mitochondrial respiratory chain complexes I, II, III, and IV, respectively.The spectra are computer simulations of the signals as expressed in whole, unprocessed, frozen tissue; details of the signals (precise geff values; splittings; line widths) can differ noticeably in isolated proteins or mitochondrial fragments, particularly due to differences in redox potential and spin-spin interactions between redox centers.The signal due to the whole mitochondrion in the lower right panel is shown at two different amplitudes superimposed with one at 20 × the amplitude of the other, in order to have appreciable amplitude of each of the contributory signals in one or other of the traces (this is clearest at geff = 6), while the signals shown individually have normalized peak amplitudes
EPR SIGNALS FROM FROZEN TISSUE
The low-temperature EPR signals from tissues including liver, muscle, heart, and brain, have largely been assigned from comparison of signals from tissues with those from fractionated extracts, purified proteins or protein complexes, modified or truncated proteins or protein complexes, and isolated mitochondria[80-82].The predominant signals are those from the MRC complexes I-IV, with additional signals from semiquinones, aconitase, catalase, and from ferriheme and transferrin in residual blood.The centers from MRC complexes and their computer-simulated EPR signals are shown in Figure 1.
Of the MRC complexes, Complex I (NADH: quinone oxidoreductase) provides the richest array of EPR signals due to the large number of [2Fe2S] and [4Fe4S] clusters that exhibit EPR signals in the monocationic reduced state, of which seven are proposed to be integral to electron transfer through the complex, based on the bacterial enzyme[83].There has, however, been some controversy over the assignment of EPR signals of FeS clusters in mammalian Complex I to structurally characterized ones because of the extensive overlap of signals from both Complex I and other sources in the spectra of intact tissue, and complicating weak magnetic interactions between the clusters in the intact holoproteinin situ[84-86].The most informative [FeS] clusters in Complex I are, thankfully, also the best-resolved in the EPR spectrum, N1b, N2, N3, and N4[79,83].Consideration of only these clusters reduces the number of computer-fitting parameters to avoid over-fitting to a large number of highly correlated parameters but nevertheless, provides a useful interrogation of the redox status of the mitochondrion in the low-potential (reducing) regime.Two of the clusters, the [4Fe4S]+N4 (-280 mV) and N3 (-325 mV), exhibit well-resolvedg3resonances atg= 1.88 and 1.86, respectively.These clusters exhibit the lowest midpoint potentials that can be accessed by an NADH+/NAD couple-determined overall redox potential and high intensities of signals from these centers indicate a highly reducing environment.The N1b [2Fe2S]+and N2 [4Fe4S]+clusters have higher midpoint potentials, around -205 mV to -270 mV, depending on the overall redox status of other clusters, and exhibit essentially axial EPR spectra withg⊥ ~ 1.92.
The signal from Complex II (succinate dehydrogenase) is dominated by two overlapping signals due to reduced [2Fe2S]+(S1) and [4Fe4S]+(S2) clusters.The preciseg-values for these signals are dependent on the extent of reduction and the temperature, as the influence of the spin-spin interaction between them is dependent on both.However, they both give rise to an intense derivative feature atg~1.92, adding to the contribution from Complex I signals[87-89].A third complex II signal due to an oxidized [3Fe4S]+(S3) signal overlaps atg~2.02, with that from oxidatively-deactivated cytosolic aconitase in which the labile Feaatom is lost from the active, EPR-silent [4Fe4S]2+cluster to form an [3Fe4S]+cluster[90].However, the very different temperature dependences of S3 and aconitase allow deconvolution by recording at 2 temperatures (e.g., 12 and 40 K)[80].Another signal in that region with ag~2.015 turning point and flanking resonances atg~2.03 and 1.98 has been assigned to a stable, dipolar-coupled ubisemiquinone pair in the vicinity of S3 in Complex II[91].
Although the redox cofactors of Complex III (cytochromebc1complex; CoQH2-cyctochromecreductase) have been studied extensivelyin vitro, the EPR spectra of tissue and cells do not provide much information due to their low intensities.The only signal routinely assignable to complex III is the reduced [2Fe2S]+Rieske cluster with a distinct sharpg1resonance at 2.03, a derivative feature at 1.90 (generally not resolved from the composite ‘g= 1.92’ signal), and a very broadg3feature atg~1.78 that is observable in heart and muscle samples that do not exhibit overlapping Mn(II) signals[79,92-94].The other EPR signals observable from isolated complex III are resonances that overlap in the regiongeff~3.8-3.3 and are due tog1of cytochromesc1,bL, andbH[92,95].These signals are sometimes detected but are difficult to characterize and quantify because they are weak, part of a very broad signal envelope, and their resonance positions and line widths can be sensitive to the specific environment.
The final redox-active MRC component, Complex IV (cytochromecoxidase), exhibits resonances atgeff~3.0 andgeff~2.2 due to low-spin hemea, and, under some conditions, resonances atg~2.18 and 2.0 due to the dinuclearS=1/2CuAcenter[96,97].Additional complex IV signals atgeff~12 and 2.95 can be difficult to detect in some tissues and cells due to low signal levels and overlap with the hemeageff~3.0 resonance, respectively, and are associated with the hemea3-CuBcoupled center[98-100].
Additional signals that may be observed in the spectrum arise from the [3Fe4S]+cluster of aconitase described above[90]; a rhombic high-spin ferriheme signal from catalase withgx~6.45 andgy~5.33[81,101], and a signal atg~4.2, with a characteristic splitting at the crossover, due to transferrin Fe3+[102-104].A characteristic, and sometimes intense six-lineI=5/2hyperfine-split signal due theS=5/2,MS= ±1/2manifold of55Mn2+is observed in some tissues, particularly liver, and may be distorted due to the rapid-passage relaxation effects at the low temperatures needed to observe the other signals[79].
TISSUE SAMPLE PREPARATION FOR EPR
The goals of EPR of tissue for the characterization of MD are (1) to provide a snapshot of the redox status of metabolism in actively metabolizing tissue; and (2) to report on instantaneous and chronic exposure to ROS.It is important, then, that tissue is excised and frozen before either the exhaustion of reducing equivalents or of a terminal electron acceptor (usually oxygen) alters the local and global redox potentials, and before further non-physiological ROS-mediated damage occurs.Traditional tissue mounting techniques cannot be used: EPR spectra of formalin-fixed brain tissue, for example[105], are devoid of almost all of the characteristic signals observed in freshly frozen brain[106,107].Similarly, human tissue-bank muscle samples exhibited intense free-radical signals and signals due to Fe3+but no signals ascribable to metabolic components were observed.In contrast, studies in the present author’s laboratory indicate that tissue samples frozen between 30 s and 3 min of being harvested from freshly sacrificed animals were rich in signals from MRC redox centers, and the spectra showed no time-dependent changes over that time span.Samples taken from different parts of muscle, liver and lung from the same wild-type (w/t) rat exhibited astounding reproducibility, with signal intensities within 5% for all signals.Between animals, the reproducibility was within 10% (except for the intensities of signals due to blood components, which did vary significantly).Liver samples have been found to be completely stable for at least 6 months at -80 °C.A sample of diced brain that had been stored for 1 year at -80 °C and had experienced substantial sampling handling (during which time it was likely that the temperature rose significantly above -80 °C, though it had never been thawed) showed significant change (+ 10%) in the aconitase region of the signal, indicative of oxidative conversion of the labile [4Fe4S] to [3Fe4S].Otherwise, only a very minor change (< 5%) in the composite FeS resonance atgeff= 1.92 was seen.Liver MRC signals were surprisingly tolerant of freeze-thawing, with only very small changes (< 5%) observed in the aconitase/S3/UQ2region of the spectrum.Muscle was much less tolerant of freeze-thaw cycles, with large increases in the signals from ferriheme, and aconitase and/or S3, indicating both structural damage and oxidation.Small but significant changes in other signals included an increase in CuA, a decrease in the Complex I/II composite FeS signal atgeff= 1.92, and specifically a larger decrease in N3 compared to N4 or thegeff= 1.93 feature, suggesting an increase in the overall redox potential.It appears then that medium term storage at -80 °C preserves the EPR signals and thus the redox status of the mitochondria, but sample handling must be carried out carefully to avoid warming, and thawing must be avoided.
Samples are most readily prepared by transfer of fresh tissue into an EPR tube followed by freezing in dryice/methanol, liquid nitrogen-chilled isopentane, or liquid nitrogen; the latter is a slower freezing method but is more convenient and safer in a clinical environment due to the lack of flammability.The sample is typically prepared by rapid extrusion of intact tissue from a syringe into an EPR tube that is blown from a length of quartz to leave a small hole in the bottom to prevent air spring resistance.Ideally, samples should completely fill the volume of the tube that occupies the active length of the EPR resonator for reproducible quantitation.Smaller samples can be mounted in the center of the active region of the resonator by first freezing a platform of a water-glycerol mixture at the bottom of the tube, adding the tissue, and adding more water-glycerol to the desired height before freezing.The latter step maintains a more-or-less uniform dielectric constant along the active region of the resonator, which maintains consistency ofB1, the oscillating field due to the microwave, across the sample and between samples for a given microwave power and resonator.
QUANTITATIVE ANALYSIS OF EPR SPECTRA
There are a number of challenges to data analysis.One complicating factor is that each of the contributory signals exhibits different dependencies on temperature and microwave power.Therefore, an empirical scaling factor needs to be determined for each signal for any given set of conditions.Phenomena that can reduce the observed intensity of the EPR signal include rapid-passage effects at too-low temperatures, relaxationbroadening at too-high temperatures, and power saturation at too-high microwave power.The most efficient way to arrive at scaling factors is to first determine the temperature dependence of each signal in order to (1) identify a temperature (or a small number of temperatures) at which most or all of the signals are observable; and (2) provide a temperature coefficient to account for any deficiency in signal intensity at the temperatures to be used for routine measurement compared to the maximum intensity that the temperature-dependence experiments predict.Then, a power-dependence at the preferred temperatures is carried out.Modern commercial instruments allow for two-dimensional experiments that can be programmed and run overnight to rapidly facilitate this process.
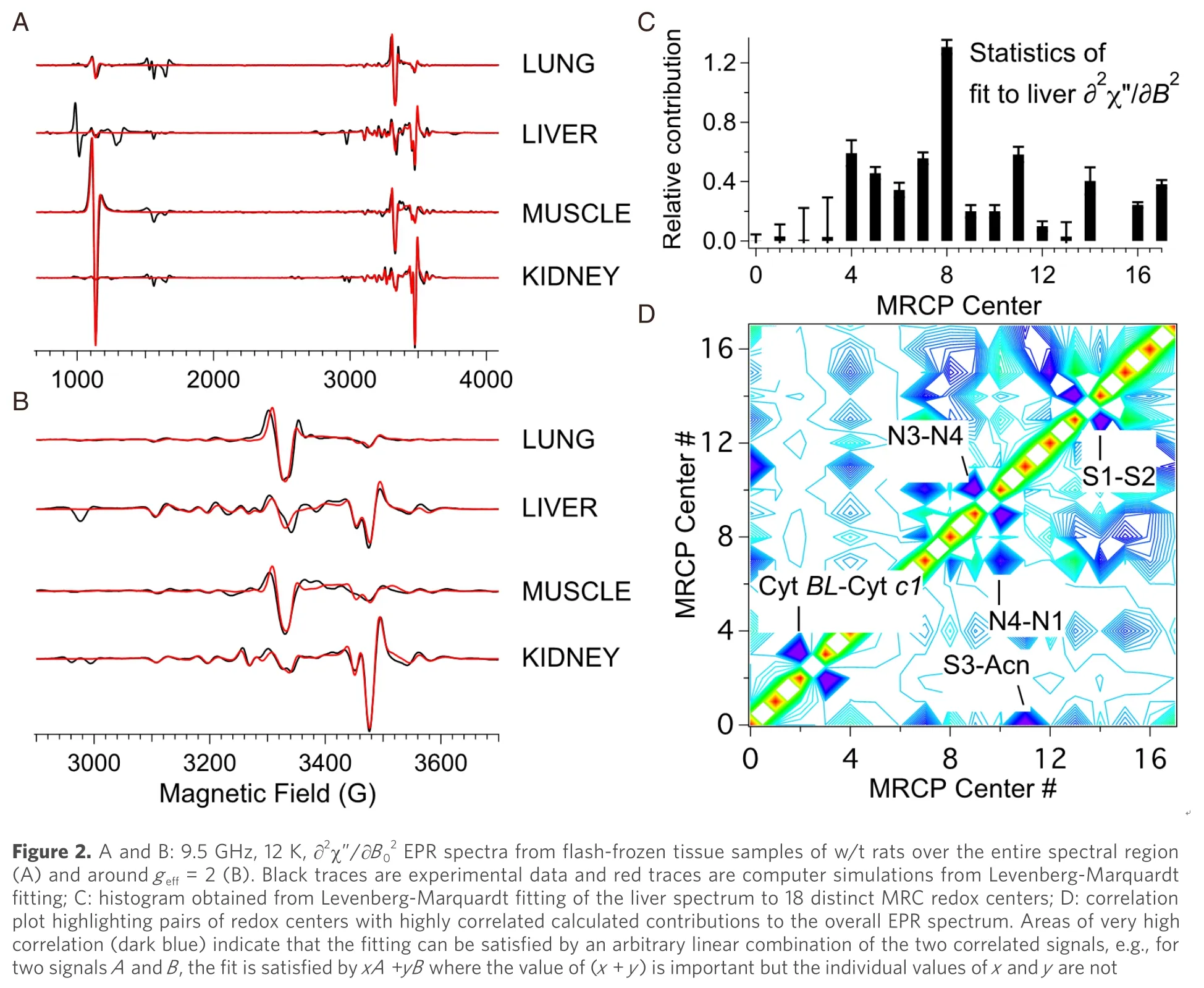
A second challenge is that many of the signals overlap, sometimes extensively.From the 24 MRC redox centers distributed among Complexes I-IV, 18 discrete EPR signals have been observed from tissues and mammalian cells under various conditions (FeS N1b, N2, N3 & N4 from Complex I; ferriheme, [UQ·]2and FeS S1, S2 & S3 from Complex II; Rieske FeS and Cyts bL, bH& c1 from Complex III; and CuA, heme a, and a spin-coupled Cu-heme a3from Complex IV), along with signals from the [3Fe4S]+of cytosolic and mitochondrial aconitases and free radical species (likely ubisemiquinone, UQ·)[79,82,108-111].Additional well-characterized tissue-specific signals include Fe(III) from transferrin (Tf), Cu(II) from ceruloplasmin (Cp), high- and low-spin hemes from catalase, and Mn(II).The overlap means that the usual method for quantifying EPR signals, that of double integration and comparison to a standard, with application of the aforementioned scaling factors, is not feasible in most cases.In any case, the wide field envelopes of many of the signals renders integration unreliable, though for some sharp and intense signals, quantitation by integration with scaling factors can be carried out.Instead, the approach taken is to iteratively fit a library of simulated EPR signals [see Figure 1, above] using a Levenberg-Marquardt damped least-squares algorithm (IGORPro, Wavemetrics) [Figure 2][112-114].The individual computed signals in the library are scaled by the aforementioned empirical scaling factor.While it is unnecessary to determine the scaling factor for each sample, it is important to determine the range of applicability of these factors across the range of samples encountered, particularly as local relaxation parameters are sensitive to the wider redox status.As with all experimentation, the space of controls must be carefully explored.
The best results have been found by fitting the ∂2χ”/∂B02“second-derivative” display [compare Figure 1 and Figure 2A and B], though this requires very high quality data that in turn requires ~300 mg of tissue, which is feasible with some animal experiments whereas only 20-50 mg of human biopsy tissue may be available, in which case the fitting is carried out on the raw experimental ∂χ”/∂B0spectrum.The Levenberg-Marquardt algorithm provides two important parameters of fitting quality.One is a standard deviation that, when compared to the actual contribution of a signal to the to the spectrum, indicates the significance of including that particular signal in the fit [Figure 2C]; a “confidence level” can also be readily calculated.The second is a correlation matrix [Figure 2D] that indicates cases where the calculated contribution of one signal is heavily dependent on the contribution of another.The highly correlated pairs that are seen in tissues are cytbL-cytc1; N3-N4; S1-S2; S3-aconitase; and, less so, N1b-N4 and N1b-N2.The N4 and N3 signals each have a small but isolated resonance corresponding to a particular principal orientation and can thus be fitted manually rather than iteratively and constrained thereafter.CytsbLandc1are rarely observed with high intensity and signals from either or both can be taken as markers of unusually elevated redox potential.[FeS] clusters S1 and S2 can be deconvoluted with care by exploiting differences in relaxation behavior but this is hardly worth the effort as they essentially provide the same information in almost all cases and are treated together as (S1 + S2); similarly, N1b and N2 can be considered together[84,85].The most important coupled pair, then, is S3-aconitase, which overlaps extensively across their narrow field envelopes and yet provide very different information that renders their deconvolution important.Fortunately, these are readily distinguished by their very different temperature dependences[80].Where disease tissue is available with a reliable control, high sensitivity to changes can be accomplished by generating difference spectra (i.e., disease minus control).One could, in principle, merely fit the difference spectrum, in which many of the signals from the individual spectra, those that are not affected by disease, would have cancelled out.A good measure of the effectiveness of the fitting procedure, however, is that the difference spectra of the two individual calculated simulations are indistinguishable from fitting to the difference spectrum directly and provide excellent reproduction of the difference of the experimental spectra[79].
APPLICATION OF EPR TO MITOCHONDRIAL DISEASE
EPR as a stand-alone technique provides three pieces of information relevant to MD and other diseases and conditions with metabolic components including cancer, neurological diseases, and cardiac dysfuncti on[79,82,106,108-111,115].The first is a measure of the redox potential across the MRC by quantitation of signals due to oxidized and reduced redox centers.This is important because the redox potential is the thermodynamic driving force for catalysis and electron transport and, ultimately, represents the potential energy available for conversion to chemical energy via ATP synthesis.The characteristic sharp distinct signal observed atg= 1.92 is largely due to thegx,yfeatures of complex I reduced FeS clusters N1b and N2, with additional contributions fromgyof N3 and N4 and fromgx,yof complex II S1 and S2.The dominant N1b and N2 clusters exhibit midpoint potentials of -205 mV to -270 mV[84], and diminution of theg= 1.92 signal compared with healthy cells or tissue therefore represents a significant departure of the redox potential from the expected -320 mV, dictated by the NAD+/NADH couple[116].More sensitive still are the N4 (g3= 1.88;Em= -280 mV) and N3 (g3= 1.86;Em= -325 mV) clusters, with the isolatedg3features upfield of theg= 1.92 signal.Care must be taken when Mn2+is present, particularly prevalent in the liver, as the N3g3feature overlaps with the often more-intense high-fieldMI= -5/2resonance of the DMS=1/2manifold ofS=5/2Mn2+[79,108].The absence of the N3 and N4 signals in spectra where theg= 1.92 is well-developed is indicative of either reduced metabolic potential (elevated redox potential), due to inefficient primary metabolism, a compromised MRC, or a membrane that allows reducing equivalents to non-productively drain from the MRC.Another possible cause, though less likely, is specific breaks in the Complex I intramolecular electron transport chain.
The second useful piece of information from EPR is the intensity of the [3Fe4S]+signal due to aconitase, that reports on the instantaneous oxidative stress burden due to ROS production as a result of MD.Upon reaction with O2·-, a non-covalently-bound iron, Fea, is lost from the active EPR-silent [4Fe4S]2+cluster and the resulting catalytically inactive but EPR-active [3Fe4S]+cluster exhibits a sharp and distinct, almostisotropic signal centered atg= 2.018[90,117-120].The magnitude of this signal has been observed to increase dramatically in tumor tissue, where excess ROS has been confirmed by other techniques, whereas in a mouse MD model the increase was moderate (125% of control)[115,79].Chronic exposure to ROS elicits an additional response.Catalase expression is known to protect against oxidative stress, and catalase overexpression protects against ROS-mediated cellular damage[121-124].Because catalase is not constitutively expressed at EPR-detectable levels in most tissues (liver is an exception)[79], the appearance of catalase ferriheme EPR signals atgeff= 6.2 and 5.7, that flank the signal atgeff= 6, provides an oxidative biomarker for sustained exposure to ROS.This signal may not be of use for monitoring MD progression, as it is for characterizing tumor growth, but could be a useful tool for evaluating therapy.
Where EPR can be much more powerful than when applied as a stand-alone tool for the characterization of MD and in subsequent therapy evaluation is when it is integrated into a comprehensive multi-technique protocol.The first and most comprehensive study of this type was the investigation of the underlying mechanism of mitochondrial dysfunction in two deoxyguanosine kinase (DGUOK; EC 2.7.1.113)-deficient rat models (“M1” and “M2”) of a genetic mitochondrial DNA depletion syndrome[79].Mitochondrial DNA assays, histology, protein immunoblot (western blot) assays, and electron transport chain activity assays were carried out in addition to EPR measurements.Both rat models were characterized by a 90% depletion of hepatic mtDNA and a 60%-80% depletion of splenetic mtDNA, similar to those observed in the human condition for which the model was developed[125].One of the models, M2, was ~50% deficient in brain mtDNA while the other, M1, was ~50% deficient in mtDNA in the quadriceps muscle overall although the deficiency appears not to be uniformly distributed throughout the muscle.Histological staining revealed numerous fibers in DGUOK-deficient rat muscle that were negative for Complexes II and IV whereas no such fibers were evident in wild-type (w/t) rat muscle.MRC protein expression assays indicated that Complexes II, IV, and V were expressed at w/t levels whereas Complexes I and III were expressed at about 50% of w/t.Activity assays of the MRC complexes, though, differed considerably from their expression levels.Complex II activities in both liver and muscle of DGUOK-deficient rats were close to w/t levels, as was that of Complex IV in muscle.Complexes I and III, however, exhibited very low activities, 10%-20% of w/t, lower than expected from protein expression assays.Interestingly, the activity of Complex IV in liver was < 20% of w/t despite being expressed at w/t levels.The EPR results were, therefore, considered in light of these observations.
EPR of liver indicated that the signals due to reduced [2Fe2S]+and [4Fe4S]+clusters of Complex I were diminished by about 50% in DGUOK-deficient rats, regardless of the cluster midpoint potential.That the same ratio of intensities of signals was observed from each of N1b, N2, N3, and N4 in w/t and MD rats indicates that the redox potential experienced by Complex I was not affected, and that the signal diminution in MD rat liver is therefore due to the proportionally diminished expression level.This result also indicates that the lower-than-expected activity of Complex I is not due to globally misfolded protein or an inability to incorporate at least the four EPR-characterized low-potential iron-sulfur clusters, but likely involves the mitochondrially-encoded ND1 subunit.The other potentially interesting results of EPR of liver were a 25% increase in the aconitase signal, and a doubling of a signal due to Mn2+.The aconitase signal suggests ongoing oxidative stress, perhaps related to the presence of the fully-reduced but partially inactive Complex I.The expression of signals due to Mn2+has been associated with oxidative stress adaptation in bacteria[126], but any role in humans is undefined.The only clear signals associated to either of Complexes III or IV were the reduced Rieske [2Fe2S]+cluster signal of Complex III, and the Complex IV hemeasignal atgeff= 3.The Rieske cluster signal from MD rats exhibited the same intensity as w/t despite lower expression and even lower activity.This observation is rationalized by the observation that Complex II is fully expressed and fully functional in MD rat liver and that electron redistribution to Complex I is thermodynamically unfavorable due to reduction of the latter.Therefore, Complex II is likely to be feeding reducing equivalents into a depleted pool of dysfunctional Complex III, ensuring complete reduction of the Rieske cluster and leading to a 50% depletion of the ferrihemeasignal.The conclusion was the inability of fully-reduced Complex I to donate its electrons into the MRC may render it the source of ROS, while a fully functional Complex II is sufficient to support reduction of Complex III which, due to the low activities of Complexes III and IV themselves, is not readily reoxidized.In liver from rat models of this MD, then, both low ATP production and ROS production are likely and both sequelae would need to be addressed in a patient.
EPR of muscle paints a different picture.The human disease being modeled is characterized by weak muscle performance[125].In rats, expression of this phenotype is far milder, possibly due to underlying metabolic corrections[79].The reduced iron-sulfur cluster signals from each of Complex I, II, and III in MD rat muscle were diminished by 50% to 75%, with the Complex I signals depleted most.In addition, the intensity of the Complex II S3 oxidized [3Fe4S]+cluster signal increased by a factor of three in MD rats.These data, in contrast to liver, suggest a global increase in redox potential (i.e., decrease in ATP-synthesizing thermodynamic driving force) in the muscle of DGUOK-depleted rats.A number of scenarios that might explain the phenotype and mitochondrial pathology were considered and rejected and, ultimately, a depletion of succinate combined with the inability of Complex I to release electrons from the mitochondrially-encoded ND1 subunit was considered the most likely explanation.
One perhaps surprising result was that the EPR of heart muscle in both MD and w/t rats was identical and indicated a fully reduced MRC and an absence of markers for oxidative stress.A reason for the observation may simply be that in any other case the rat would not be alive for study.The cause may be that the heart has large redundancy in the number of mitochondria in order to fully benefit from reducing equivalents from primary metabolism even when under stress.
CONCLUSION
EPR of biopsy tissue of a subject with suspected MD can provide some limited information, largely through the ratio of intensities of the Complex I iron-sulfur centers reporting on the ability to maintain a low redox potential, and through the observation of biomarkers for historical and ongoing ROS-mediated oxidative stress.When the EPR of such tissues can be compared with good controls, or with a yet-to-be established database of control spectra, quantitative information on the intensities of multiple MRC components can be obtained that can inform on disease mechanism.However, it is when EPR is combined with multiple complementary techniques that it becomes most useful.The primary advantages of EPR are that the sample requires no processing or fixing, and that it is relatively straightforward to determine whether the main outcome of mitochondrial dysfunction is likely to be diminished ATP synthesis, elevated ROS production, or both.One lesson from the study discussed above is that important information on an MD that is characterized in humans by poor muscle performance may be obtained from study of other tissue, e.g., liver.
DECLARATIONS
Authors’ contributions
The author contributed solely to the article.
Availability of data and materials
Not applicable.
Financial support and sponsorship
None.
Conflicts of interest
The author declared that there are no conflicts of interest.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Copyright
© The Author(s) 2020.
 Journal of Translational Genetics and Genomics2020年2期
Journal of Translational Genetics and Genomics2020年2期
- Journal of Translational Genetics and Genomics的其它文章
- Intellectual disability, the long way from genes to biological mechanisms
- Spectrum of MECP2 mutations in Indian females with Rett Syndrome - a large cohort study
- The North American mitochondrial disease registry
- Mitochondrial translation defects and human disease
- Role of transfer RNA modification and aminoacylation in the etiology of congenital intellectual disability
- Redefining infantile-onset multisystem phenotypes of coenzyme Q10-deficiency in the next-generation sequencing era
