Private somatic mutations identified with liquid biopsy lead tumor progression in solid cancers
Maria Palmieri, Margherita Baldassarri, Nicola Iuso, Francesca Fava,, Alessandra Fabbiani,,
Francesco Cetta1, Chiara Fallerini1,2, Rossella Tita3, Maria Antonietta Mencarelli3, Alessandra Renieri1,2,3, Elisa Frullanti1,2
1Medical Genetics, University of Siena, Siena 53100, Italy.
2Department of Medical Biotechnologies, Med Biotech Hub and Competence Center, University of Siena, Siena 53100, Italy.
3Genetica Medica, Azienda Ospedaliera Universitaria Senese, Siena 53100, Italy.
Abstract Aim: Primary tumors can be divided into oncogene-addicted (e.g., lung) and non-oncogene addicted (e.g., breast). Only the former group has an Achilles-heel single gene for successful target therapy, whereas the latter has mutations of multiple causative genes. Currently, tissue biopsy used for genetic surveys do not give a complete picture of the molecular profile and clonal evolution, but only provide static information over time.Methods: A series of 133 patients with 16 different solid tumors were enrolled. Blood samples were collected and cell-free DNA (cfDNA) was extracted. cfDNA libraries were analyzed using AVENIO circulating tumor DNA (ctDNA) Expanded Kit and Illumina NextSeq 550 for sequencing was used. In order to evaluate the clinical evolution over time, a second cfDNA analysis was performed after a mean interval of 2 months.Results: Through the cfDNA liquid biopsy, we found 89 pathogenic variants in 54 genes. Breast, lung, and prostate cancers showed the largest number of mutated genes. TP53, PIK3CA, FGFR3, KRAS, and ERBB2 were the most frequently mutated genes among 16 different tumors. Gene distribution didn’t show any type of prevalence. In particular, every patient with disease progression seems to have a “private” combination of gene pair mutations, with TP53 as the most frequently mutated gene.Conclusion: We showed that the clonal evolution of tumors includes a private combination of genes, regardless of tumor type. In the future, the cancer treatment can be the targeted therapy against specific tumor mutation(s). The present approach seems promising to both identify key cancer genes and follow clonal evolution over time.
Keywords: Cell-free DNA, liquid biopsy, solid tumors, advanced tumors, private mutations, targeted-therapy
INTRODUCTION
Daily experience shows that different malignancies, such as breast or lung cancers, can be controlled using standard protocols for many years before tumor relapse. This is possible thanks to the detection of the socalled “Achilles’ heel” that represents the molecular target for different tumor groups named “oncogene addict”[1]. However, only a few tumors have a unique or a small number of mutated genes that could be susceptible to target therapy, whereas the vast majority of cancers are “non-oncogene addicted”[1]. Tissue biopsies from primary tumors usually do not show details of molecular heterogeneity and are even less informative when obtained from metastases. Additionally, biopsies from metastases are not always easy to perform[2].
Cancer is an evolving microcosm driven by selective pressures due to the environment and drug therapy. Malignant cells are in competition and/or cooperation with each other and with the surrounding environment[3]. Cancer therapy must cope with the cellular complexity of the disease and face its dynamic evolutionary aspect[4]. In order to facilitate treatment choice, it is crucial to distinguish between germline and somatic mutations. New technologies such as the liquid biopsy of cell-free DNA (cfDNA) facilitate proper detection of somatic tumor mutations, which are the key mutation of specific cancer driver genes. The technique is non-invasive, is a valuable alternative to physical biopsies, and opens new avenues for personalized medicine[5].
In this study, we show that cfDNA analysis is able to follow, over time, the clonal evolution of multiple solid tumors. In particular, patients belonging to the same tumor type exhibit different private pairs of mutations. The most frequently mutated gene isTP53that we found in combination withPIK3CA, KRAS, EGFRwith a frequency of 71%, or in combination withBRCA1,andERBB2with a frequency of 42%, or finally withPTENandMYCwhit a frequency of 28.6%.
The second liquid biopsy proves to be a powerful tool to understand which pair of mutated genes is specific for the patient and unique to him, thus leading to a fully personalized treatment.
METHODS
Patients and sampling
Our cohort has been enrolled at the Medical Genetics Unit of the Azienda Ospedaliera Universitaria Senese, Siena, Italy for diagnostic purposes. The cohort consisted of 133 patients with different solid tumors who experienced disease progression after standard therapy. Patients were previously treated in advanced/metastatic settings and most of them were not eligible for curative treatment. Written informed consent and assent for genetic analysis was obtained from all patients.
Inclusion criteria provided patients with either locally advanced or metastatic solid tumors independent from the primary tumor site. Patients were excluded if they had early-stage solid tumors and still have to experience all possible pharmacological treatments of standard guidelines. The main information collected for each patient includes oncological data, family tree, and cancer history in a genetic consultation setting.
A first peripheral blood sample for cfDNA analysis was taken during the genetic counseling visit at the stage of disease progression. Plasma was used for cfDNA extraction. A second sample for cfDNA analysis was taken after a mean time interval of 2 months (range 1-6 months).
cfDNA extraction and sequencing
Peripheral blood samples (10 mL) were collected from each patient and placed into a Cell-Free DNA BCT tube (Streck, La Vista, NE, USA). cfDNA was extracted from 4 mL of plasma using AVENIO ctDNA Expanded Kit according to the manufacturer’s instructions. cfDNA quality and quantity were verified as described in Palmieriet al.[5]. cfDNA sequencing was performed using AVENIO circulating tumor DNA (ctDNA) Expanded Kit (Roche, Basel, Switzerland) on Illumina NextSeq 550 (Illumina, San Diego, CA, USA). This technology is able to identify various types of alterations, including single nucleotide variants, insertions/deletions, gene fusions, and copy number variations present in genes linked to cancer (clinical actionable mutations) with a reportable range up to 0.05%. The sequencing analysis was performed using AVENIO Oncology Analysis Software (Roche, Basel, Switzerland).
RESULTS
Patient characteristics
From March 2018 to July 2020, a total of 133 patients with locally advanced or metastatic solid tumors were enrolled at Azienda Ospedaliera Universitaria Senese, Siena, Italy, and included in the study. The mean age of patients at the first cfDNA analysis was 56 years (range 2-83 yrs); 48% of patients were females and 52% were males. Out of 56 patients who did at least a second liquid biopsy, 22% had cancer from breast, 14% lung, 14% ovarian cancer, 5% colorectal, 6% pancreas, 6% prostate, whereas uterine cancer, retinoblastoma, cholangiocarcinoma, and gastric cancer accounted for 2% each, and soft tissue sarcoma (including right infratemporal fossa, oral, pharynx, and larynx), Wilms’ tumor, and glioblastoma accounted for 1% each of the entire series. The median follow-up time for all patients was 2 months (range 1-6 months).
Mutated gene and tumor type association
Next-generation sequencing (NGS) analysis in 133 patients at the first liquid biopsy identified 86 clinically meaningful pathogenic variants in 54 genes allowing to pick up key mutations in 67.6% (n= 90) of cases [Supplementary Table 1]. After a mean of 2 months, a second liquid biopsy was performed and 87.5% of patients remained/became positive. Table 1 summarizes all the mutated genes resulting from the second liquid biopsy associated with different tumor types. Mainly, the breast, lung, and prostate cancers were the tumors types that showed the largest number of mutated genes. The tumor type, grade, and stage as well as the drug treatments for the entire cohort of second liquid biopsy patients are summarized in the Supplementary Table 1.
Most frequently mutated genes
At the second liquid biopsy time, the mutated genes decreased from 54 to 38 in 16 different tumor types. Among these 38 mutated genes, those most frequently represented were the following:TP53(30%),PIK3CA(10%),EGFR(10%),KRAS(8%),ERBB2(8%), FGFR3(6%), andBRCA2(6%). Interestingly, these genes were mutated in 16 different tumor types without a specific prevalence among them [Figure 1].
However, clonal mutation was not confirmed in the entire cohort at the second liquid biopsy. Indeed, 9 patients resulted as negativized, 13 patients had only one mutated clone, and 18 had two clones. One patient had 5 mutated clones [Figure 2].
The most frequent mutations in our cohort were in the TP53 gene, regardless of the primary tumor type.TP53was usually mutated in association with another gene [Figure 3]. The most mutated genes in combination with TP53 were EGFR and PIK3CA in 20% of cases, KRAS and ERBB2 in 16% of cases, FGFR3 and BRCA2 in 15% of cases, or less frequently with other genes for a total number of 14 genes.

Figure 1. Distribution of the most frequently mutated genes. Among the 38 mutated genes, point mutations in TP53, PIK3CA, KRAS, and ERBB2 and copy number variation in FGFR3 were the most commonly observed alterations in all 16 tumor types

Figure 2. Second liquid biopsy: number of mutated clones. The histogram shows on the absciss axis the number of mutated clones and on the ordinate the number of patients. Most patients have two mutated clones at the time of the second liquid biopsy
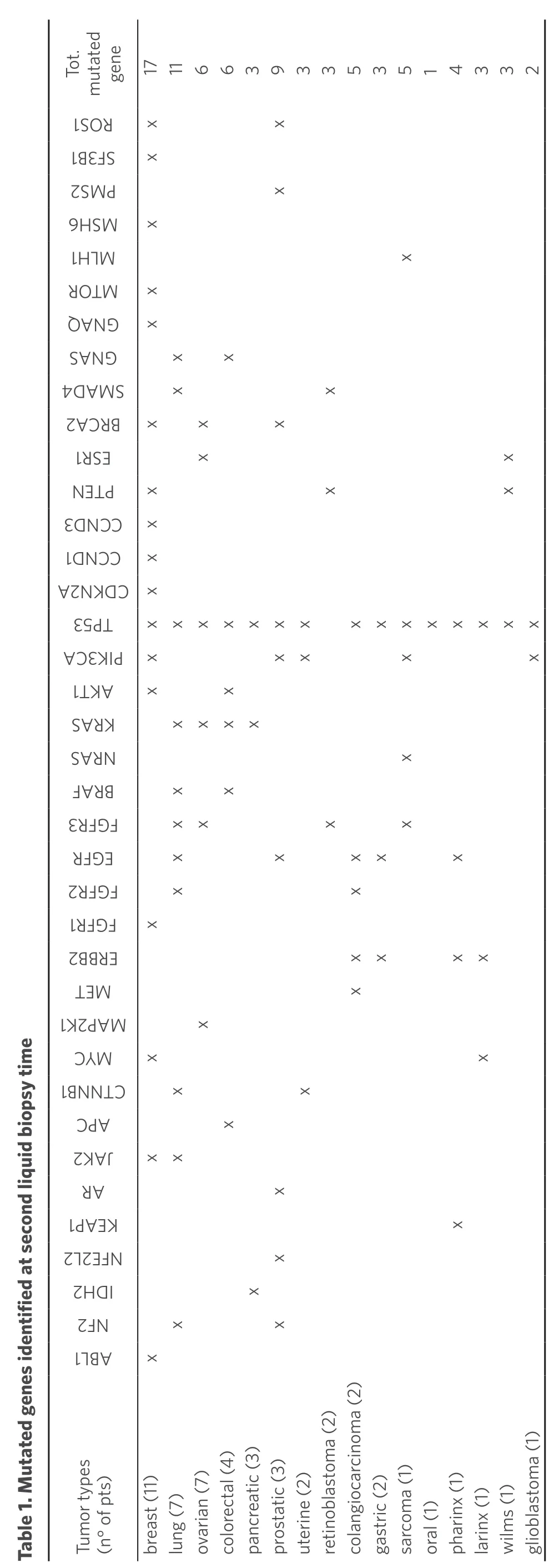
gene 171166393353514332 1 Tot. mutated SRO x x 13S BF x 2MP S x 6S HM x 1L HM x R MTO x AQ NG x SNG A x x 4AMS D x x 2ARCB x x x 1S RE x x NT EP x x x 3NCC D x 1NCC D x AND 2KC x 3P 5T xxxxxxx xxxxxxxx CA3I KP x xx x x 1K TA x x SR AK xxxx SR AN x FR AB x x 3FF RG xx x x RFEG x x xx x 2FF RG x x 1FF RG x 2BE BR xx xx TM E x KPM 12A Table 1. Mutated genes identified at second liquid biopsy time x MYC x x 1N BNCT x x CA P x 2KJA xx RA x 1AK PE x 22F LEN x 2DI H x 2N F x x 1B LA x Tumor types (n° of pts)breast (11)lung (7)ovarian (7)colorectal (4)pancreatic (3)prostatic (3)uterine (2)retinoblastoma (2)colangiocarcinoma (2)gastric (2)sarcoma (1)oral (1)pharinx (1)larinx (1)wilms (1)glioblastoma (1)
DISCUSSIONstandard. However, it is jeopardized by the wide heterogeneity that is frequently found between different regions of the same tumor (spatial heterogeneity),
Early dissemination of resistant tumor cells is the major cause of metastatic recurrence in cancer patients[6]. Currently, tissue biopsy remains the gold as well as between the primary tumor and local or distant recurrences in the same patient (temporal heterogeneity)[7]. Intratumor heterogeneity can lead to underestimation of the tumor genomics landscape portrayed from single tumor-biopsy samples and may represent a major challenge towards personalizedmedicine and biomarker development[8]. For this reason, liquid biopsy could be of primary importance to obtain a complete scenario of the molecular profile of all tumor metastases. Indeed, the cfDNA is the results of fragmented genomic DNA coming from the apoptosis or necrosis process of normal and tumoral cells. Performing the NGS-liquid biopsy on the cfDNA guarantees the total representations of the molecular information coming from the totality of metastasis.
The higher number of mutated genes occurred in breast, lung, and prostate cancer, irrespective of the number of these tumor types in the present series. Most patients had breast tumor, whereas only three patients had prostatic cancer; number of patients with lung cancer were the same as those with ovarian cancer, but only patients with lung cancer had a great number of mutated genes [Table 1].
In accordance with previous data in the literature, our study confirmed that SNVs (small nucleotide variants) inTP53,PIK3CA, andKRAS,and CNVs (copy number variations) inFGFR3andERBB2are the most commonly observed mutated genes in breast and lung cancer[9-10][Figure 1]. However, these mutations are not only confined to breast and lung cancer but are also found in other types of cancer without a specific prevalence in distribution [Figure 1].
In comparison with data from our previous study[5], showing that at the beginning of tumor expansion there was a consistent, although variable, mutational burden from tens to hundreds, disease relapse is generally characterized by a “driver” clone that is responsible for metastases. The curve in Figure 2, simulating a Gaussian bell shape (similar to the typical population curve), underlines this concept. Indeed, it shows that in metastatic patients, a negative selection occurs in the initial number of mutated clones, reducing them to a maximum of two in comparison to the considerable number of tumor clones in the early stages of the disease.

Figure 3. Eulero Venn diagram. Eulero Venn diagram shows the higher correlation between TP53 with the PIK3CA and EGFR (5 folds each), with KRAS and ERBB2 (4 folds each), or with BRCA2 and FGFR3 (3 folds each). The figure also shows correlations of other genes with each other, e.g., ERBB2 and EGFR
In particular, in relapsing patients, the second liquid biopsy showed that 37% had two mutated clones, 26% only one clone, and 18% more than two clones [Figure 2].
Notably, one of the two mutated genes was oftenTP53associated withPIK3CA(20%) orEGFR(20%),KRAS(16%) orERBB2(16%),FGFR3(15%) orBRCA2(15%), and other genes with a lower frequency [Figure 3]. We hypothesize that tumor survival does not depend only on the expansion of theTP53clone, but also requires a mutation in other genes for tumor progression. In fact, we have repeatedly found that in breast cancer, theTP53mutation is associated withPIK3CA. Indeed,PIK3CAmutations generally arise in advanced stages of breast carcinogenesis from dysplasia to carcinomain situ[11]resulting in a greater potential to migrate and invadein vitro, as well as to metastasize[12]. Moreover, in lung cancer,TP53was frequently associated withKRASmutation.
A striking finding deriving from the comparison between the first and the second liquid biopsy in the same patient has been the constant reduction in the number of mutated genes. Present data suggest that after the early phase of tumor diffusion, that is characterized by a great number of new somatic mutations, a selection takes place among the various mutated genes to determine which are the most effective clones for tumor progression.
In a small subset of patients, we were able to perform a third liquid biopsy, but numbers for the moment were too small. Data from the third biopsy, in case of further tumor relapse, will better elucidate the trend in tumor progression, showing further reduction of the number of clones, or a new increase of “random” mutations.
In conclusion, additional study in larger series is required for further confirmation of these preliminary data and suggestions. A greater use of liquid biopsy is recommended for proper detection of genetic mutations and for “the right treatment in the right patient at the right time”.
DECLARATIONS
Acknowledgments
Authors thank the support from the Regione Toscana - Istituto Toscano Tumori (ITT) (Project “Identification of genetic bases of individual predisposition to lung cancer in non-smokers”) and the ASSO (Associazione per lo Sviluppo della Scienza Oncologica) Onlus. This work was, in part, supported by the MIUR project “Dipartimenti di Eccellenza 2018-2022” to the Department of Medical Biotechnologies, University of Siena”. This work is generated within the ERN-EURACAN (European network for Rare Adult Aolid Cancer).
Authors’ contributions
Performed the experiments, analyzed the data and wrote the paper: Palmieri M Performed the experiments: Iuso N
Wrote the paper: Cetta F
Analyzed the data: Fallerini C, Tita R
Took care of the clinical part of the study: Fava F, Fabbiani A Performed genetic counseling and provided patient samples: Baldassarri M, Mencarelli MA Designed the research strategy, analyzed the data, and wrote the paper: Renieri A, Frullanti E.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Financial support and sponsorship
None.
Conflicts of interest
All authors declared that there are no conflicts of interest.
Ethical approval and consent to participate
The consent to participate was obtained from patients included in the study.
Consent for publication
The consent for publication was obtained from patients included in the study.
Copyright
© The Author(s) 2020.
 Journal of Cancer Metastasis and Treatment2020年12期
Journal of Cancer Metastasis and Treatment2020年12期
- Journal of Cancer Metastasis and Treatment的其它文章
- ROR2 regulates the survival of murine osteosarcoma cells in lung capillaries
- lsoflavone research towards healthcare applications
- Primary malignant tumors of bone surface: a review with emphasis in differential diagnosis
- Current updates in management of relapsed/refractory small cell lung cancer
- Glycogen synthase kinase 3b biology in bone and soft tissue sarcomas
- Negative effects of tumor cell nitric oxide on antiglioblastoma photodynamic therapy
