Use of network pharmacology and molecular docking to explore the potential mechanism governing the efficacy of Jinhuaqinggan granules in the treatment of novel coronavirus-induced pneumonia
Heng Xu, Jin-Min Zhang, Yi-Zhuo Qiao, Wei Zhang, Zi-Tong Fu, Qing Zhao*
1College of Tranditional Chinese Medicine,Hebei University,Baoding 071000,China
Abstract
Background: Now that the epidemic of new coronavirus pneumonia (corona virus disease 2019) is spreading all over the world, Jinhuaqinggan granules in the Chinese treatment plan has been proved to be an effective Chinese patent medicine for the treatment of corona virus disease 2019. Methods: This study aims to clarify the possible therapeutic mechanism governing the efficacy of Jinhuaqinggan granules in the treatment of corona virus disease 2019, through using network pharmacology and molecular docking. During the analysis, 227 active components were obtained and screened by using the ADME method. Furthermore, 282 Jinhuaqinggan granule targets and 56 common targets with corona virus disease 2019 were gathered from various databases. Then the protein-protein interaction network of Jinhuaqinggan granules and corona virus disease 2019 targets were constructed and 6 core targets were selected through network topology analysis. In addition, A total of 262 biological function annotation entries (P <0.01) and 101 pathways (P < 0.01) were obtained by gene ontology functional enrichment analysis and Kyoto Encyclopedia of Genes and Genomes pathway analysis. Results: Molecular docking showed that quercetin, luteolin,kaempferol, wogonin and naringin had an affinity for SARS-CoV-2 3CL hydrolase and angiotensin-converting enzyme 2. Conclusion: corona virus disease 2019 can be prevented by the primary targets of Jinhuaqinggan granules.The most important bioactive components in Jinhuaqinggan granules—quercetin, naringenin, luteolin and wogonin—can play antiviral effect, anti-inflammatory storm, regulate immunity by regulating signal transducers and activators of transcription 1, interleukin 4, interferon-γ, heme oxygenase 1 and acting on the lipopolysaccharide response, toll-like receptor signaling pathway, mitogen-activated protein kinase signaling pathway, etc.
Key words: COVID-19, Jinhuaqinggan granules, Signaling pathway, STAT1, HMOX1, IFNG; IL4
Background
At the end of 2019, a novel coronavirus-induced pneumonia case (corona virus disease 2019 (COVID-19)) appeared in Wuhan, China, and the disease spread rapidly. As of March 23, 2020, patients with COVID-19 have appeared in various countries and regions around the world, and the epidemic situation in the world has become severe [1]. Most patients have clinical manifestations of fever, dry cough, and fatigue.A smaller number of patients have nasal congestion,sore throat, and diarrhea. Severe patients can further develop respiratory distress syndrome, acute lung injury (ALI), sepsis, coagulopathy, and multiple organ failure. Traditional Chinese medicine (TCM) therapy plays an active role in the prevention and control of epidemics in China. The New Coronavirus Pneumonia Diagnosis and Treatment Program (Trial Version 6) [2]recommends the use of Chinese patent medicines(such as Jinhuaqinggan granules (JHQG)) to prevent and control coronavirus-induced pneumonia. The diagnosis and treatment plan recommends the combination of supportive therapy and therapy of TCM for COVID-19. As of April 22, China has achieved notable results in the prevention and control of epidemics. In the clinical trial of JHQG for the treatment of new coronaviruses in the Hospital of Integrated Traditional Chinese and Western Medicine in Hubei Province, 68 patients in the treatment group and 34 patients in the control group were compared.The result is that all the differences in the incidence of adverse reactions,the disappearance of clinical symptoms (fever, fatigue, cough, expectoration),and the Hamilton anxiety scale scores were statistically significant [3].
TCM is guided by the theory of traditional Chinese medicine. Chinese materia medica treats diseases with the characteristics of multiple components, multiple targets, and various pathways [4]. JHQG is composed of 12 Chinese herbs, including Jinyinhua (Lonicerae japonicae flos),Shigao (Gypsum fibrosum), Mahuang(Ephedra herba), Kuxingren (Armeniacae semen amarum), Zhebeimu (Fritillariae thunbergii bulbus),Huangqin (Scutellariae radix), Lianqiao (Forsythiae fructus), Zhimu (Anemarrhenaerhizome), Niubangzi(Arctii fructus), Qinghao (Artemisiae annuae herba),Bohe (Menthae haplocalycis herba), andGancao(Glycyrrhizae radix et rhizoma) [5]. However, the pharmacological mechanism and material basis for the action of JHQG on COVID-19 have not been elucidated. Therefore, there is an urgent need for a comprehensive and systematic evaluation of the pharmacological mechanism of JHQG in treating COVID-19. Network pharmacology is a new discipline based on the theory of systems biology, the network analysis of biological systems, and the selection of specific signal nodes for multitarget drug molecular design. Network pharmacology is an effective tool to expound the synergistic and potential mechanisms of the networks between componenttarget-disease and protein-protein interaction (PPI),and it provides a new perspective on the therapeutic mechanisms of TCM. Molecular docking technology can be used to predict the conformation, affinity and activity between small molecules and target proteins through computational simulation, and lead compounds with higher activity and binding energy can be designed virtually. Therefore, this study used molecular docking technology to screen compounds with good affinity for SARS-CoV-2 3CL hydrolase and angiotensin converting enzyme (ACE2) and used network pharmacology to explore the potential mechanism of JHQG in the treatment of COVID-19.
Materials and methods
Collecting and screening of active ingredients
The Traditional Chinese Medicine System Pharmacology Database (TCMSP,http://lsp.nwu.edu.cn/tcmsp.php) is constructed based on scientific publications and medical literature on TCM, which contains more than 13,731 pure compounds isolated from 505 TCM herbs. TCMSP was used to collect the chemical constituents of JHQG,which captures the relationships between herbal ingredients, targets, and diseases. Ultimately, the active ingredients were screened with the recommended criteria of oral absorption availability ≥30% and drug similarity ≥ 0.18.
Prediction of chemical targets and construction of compound-target networks
The TCMSP database was used to query the target of the main components in JHQG, and the UniProt database (https://www.uniprot.org/) was used to convert the gene name of the obtained target name to standardize the target name. The relationship between TCM compounds and targets is imported into Cytoscape software (https://cytoscape.org/) to construct a TCM-compound-target network.
Filtering compounds-disease common targets
Information on COVID-19-associated target genes was collected from the GeneCards database(https://www.genecards.org/), which is a comprehensive database of such functions as genomics, proteomics, and transcriptomics. Species was set as “Homo sapiens”, and we searched the GeneCards database with the keyword “new coronavirus” to screen the targets related to COVID-19. The screened chemical targets and the targets of disease were mapped with the Venny 2.1 (https://bioinfogp.cnb.es/tools/venny/index.html) online tool,and the common targets of compound diseases were obtained as potential targets for further analysis.
PPI Network of compound-disease common targets
To further understand the interaction relationship between potential targets, the common targets that had been selected by method 2.3 were imported into the STRING database (https://string-db.org/). The species was selected as "Homo sapiens", and the confidence was set to be greater than 0.700. The obtained PPI network information was exported as a TSV file and then imported into Cytoscape. According to the results of the network topology analysis, the core targets were selected based on the criteria that the node degree value and the intermediary degree centre value both exceed the average value.
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis
To further clarify the biological function of the core target genes and the main pathway for JHQG to treat new coronary pneumonia, the common target genes for drugs and diseases were imported into the DAVID database (https://david.ncifcrf.gov/). The GO functional annotation was carried out from three aspects: biological process (BP), cell component, and molecular function (MF). GO functional annotation and KEGG pathway enrichment analyses were performed by using RStudio 3.6.3 software, and aP-value less than 0.01 was employed for further analysis.

Figure 1 Flowchart of research strategies based on network pharmacology and molecular docking technology.TCMSP, The Traditional Chinese Medicine System Pharmacology Database; OB, absorption availability; DL, drug similarity; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Composition-target molecular docking
The 2019 novel coronavirus (2019-nCoV) proteolytic enzyme (2019-nCoV Mpro) and 2019-nCoV and ACE2 protein crystal structures were obtained from the RSCB database (http://www.rcsb.org/). The enzyme crystal structure was removed from water molecules and hydrogenated, and the charge was calculated with AutodockTools. Then, the crystal structure was saved as a pdbqt format file. The Mol2 format file of the 3D structure of the five important bioactive compounds was downloaded from the ZINC database. The charge of the small molecule was balanced with AutodockTools, and the small molecule structure of the protein was processed by Pymol. Then,GirdBox was set to determine the molecular docking area, and the genetic algorithm parameters algorithm was selected for molecule docking.
Results
Active ingredients of JHQG
The TCMSP database was used to search for JHQG compounds (gypsum was not found). Based on ADME with absorption availability ≥ 30% and drug similarity ≥ 0.18, 278 active compounds were screened out. Among the screened compounds, there are 7 compounds of Zhebeimu (Fritillariae thunbergii bulbus), 23 compounds of Jinyinhua (Lonicerae japonicaeflos), 36 compounds of Huangqin(Scutellariaeradix), 92 compounds of Gancao(Glycyrrhizae radix et rhizoma), 15 compounds of Zhimu (Anemarrhenaerhizome), 8 compounds of Niubangzi (Arctii fructus), 22 compounds of Qinghao(Artemisiae annuae herba), 10 compounds of Bohe(Menthae haplocalycis herba), 23 compounds of Mahuang (Ephedraherba), 19 compounds of Kuxingren (Armeniacae semen amarum), and 23 compounds of Lianqiao (Forsythiaefructus). A total of 227 active compounds were ultimately obtained after removing the duplicate data.
Construction of compound-target network and access to important bioactive components
The 227 main compounds obtained above were used to predict the corresponding targets through the TCMSP database, and a total of 282 targets were obtained after removing the duplicate data. With the help of Cytoscape software, a TCM-compound-target network (Figure 2) was constructed, which has a total of 2025 edges and 426 nodes. The red nodes that make up the square matrix represent the relevant targets of the ingredients, and the nodes that make up the circle represent the ingredients of TCM. The node size was used to reflect the number of combined targets(degree); therefore, the targets with a larger shape represent stronger interactions in the network.According to network topology analysis, among all active ingredients, the essential bioactive components having the top five nodes of degree value include quercetin, luteolin, kaempferol, wogonin and naringenin. The top 20 compounds with degree values are shown in Table 1.
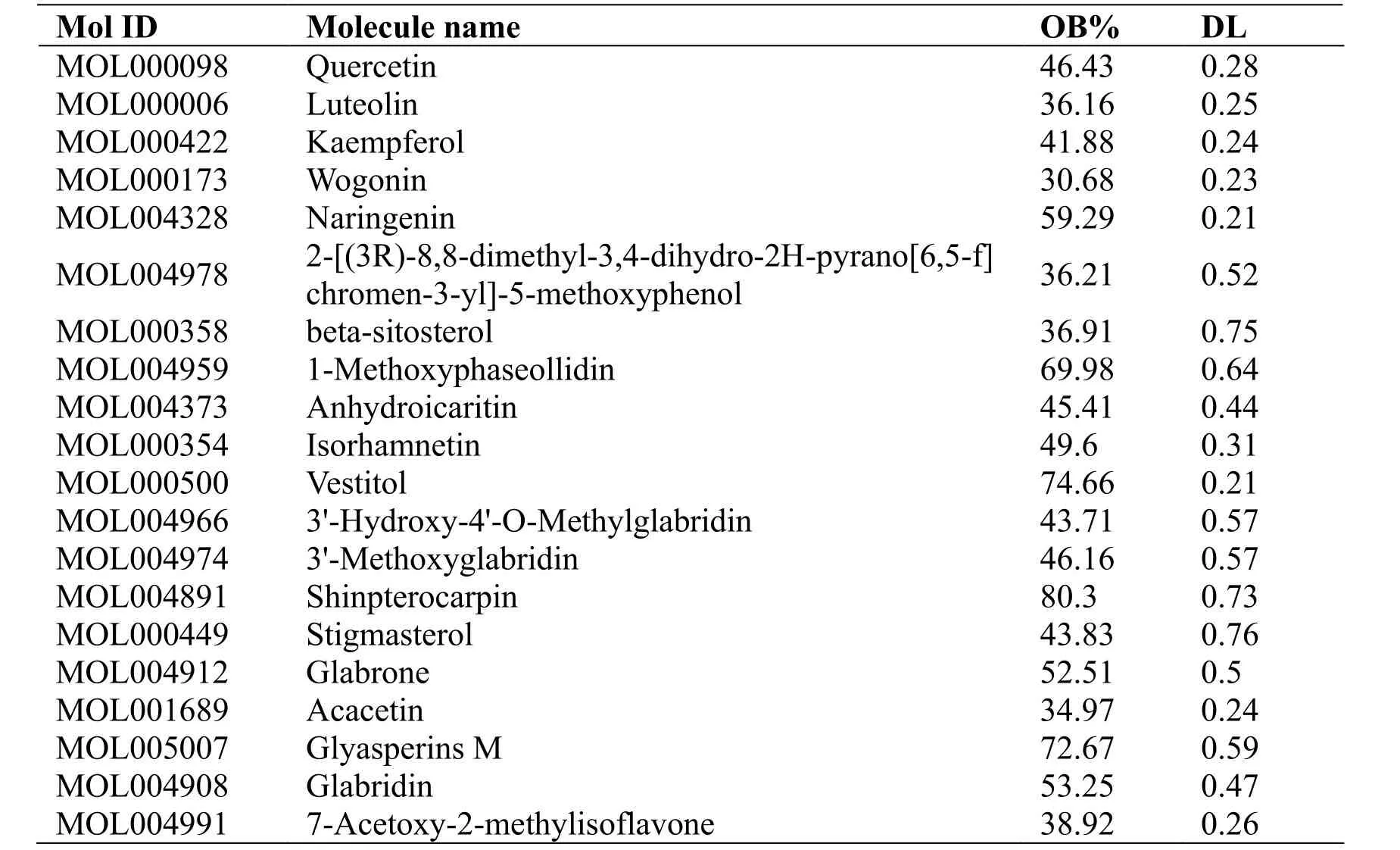
Table 1 Information on the top 20 compounds regarding degree value in a compound-target network

Figure 2 JHQG compound-target network. The red nodes represent the corresponding targets of the ingredients.JHQG, Jinhuaqinggan granules.
Collecting compounds-common disease targets(potential targets)
Through the GeneCards database, the relevant treatment targets of COVID-19 were retrieved, and a total of 350 targets were obtained. With the help of Venny 2.1 online software, the targets of the main active ingredients in JHQG were mapped to the targets of COVID-19, and 56 potential targets were obtained.The information regarding potential targets is provided in Table 2. The Venn diagram is presented in Figure 3.
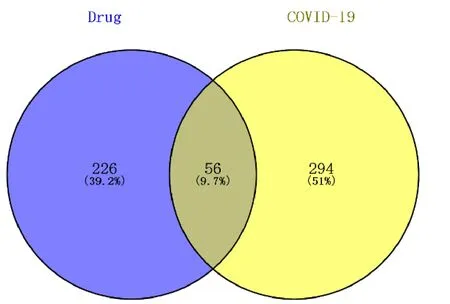
Figure 3 JHQG and COVID-19 target Wayne diagram. JHQG, Jinhuaqinggan granules; COVID-19, corona virus disease 2019.
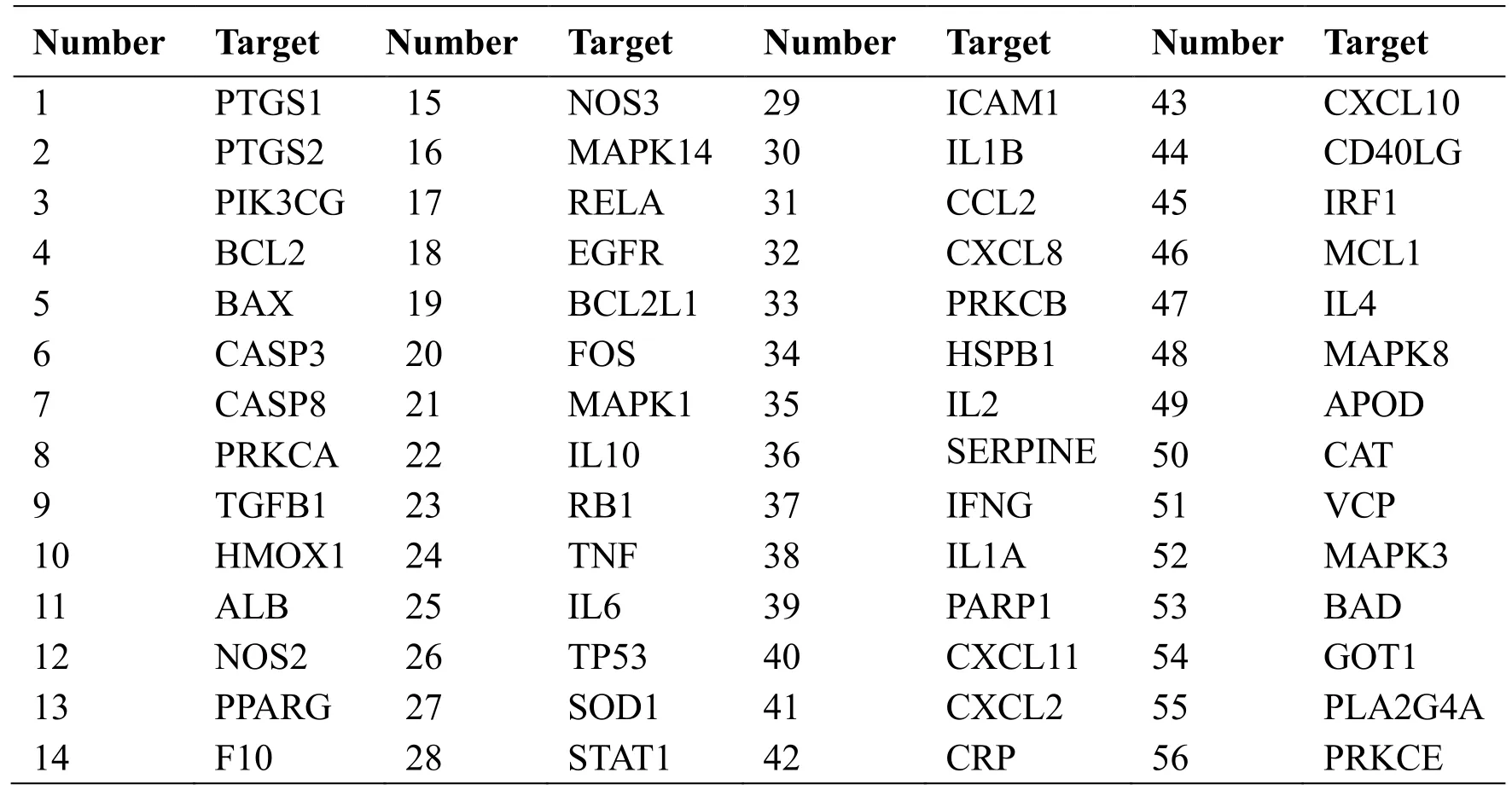
Table 2 Information on 56 potential targets of JHQG
PPI network map of compound-disease common targets.
The 56 potential genes associated with COVID-19 were imported into the STRING database for analysis with a medium confidence score of 0.700, and a network of PPIs (Figure 4) composed of 469 edges was constructed. Node degree and betweenness centrality are indicators commonly used to describe the importance of network nodes, and nodes above the average of them have stronger interactions in the PPI network. According to network topology analysis, the average node degree is 16.172, and the average betweenness centrality is 0.612. Based on the criteria that both degrees are greater than the average, 6 core targets were screened out (Table 3), namely,interleukin (IL) 10, FOS, signal transducers and activators of transcription 1 (STAT1), IL4, interferon(IFN)-γ (IFNG), and heme oxygenase 1 (HMOX1).

Figure 4 Interaction diagram of JHQG and COVID-19 protein. JHQG, Jinhuaqinggan granules; COVID-19,corona virus disease 2019.
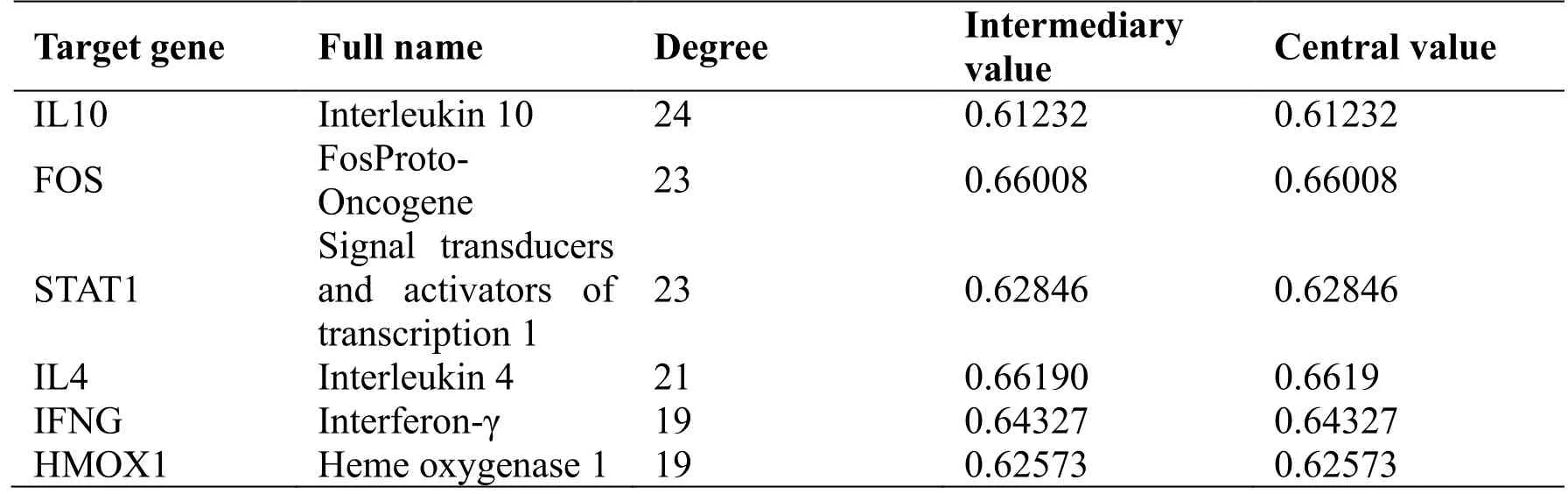
Table 3 Six core targets in the JHQG PPI network
GO enrichment analysis
The DAVID database was used to perform GO enrichment analysis on the 56 common targets.According to the screening criteria ofP< 0.01, a total of 211 BPs were enriched. The top 20 items were visualized by Rstudio software (Figure 5). These targets of BPs mainly included positive regulation of the nitric oxide (NO) BP (GO: 0045429), the lipopolysaccharide (LPS) response (GO: 0071222),the LPS-mediated signaling pathway (SP) (GO:0031663), negative regulation of the apoptosis SP(GO: 0043066), the immune response (GO: 0006955),the inflammatory response (GO: 0006954), the response to glucocorticoid (GO: 0051384), and positive regulation of the chemokine biosynthesis process (GO: 0045080).
A total of 37 cellular components GO terms were enriched, and the top 19 entries were selected based onP-values < 0.01. These targets of cellular components mainly include the cytosol, extracellular space, cytoplasm, and nucleus. In all, 60 MFs GO terms were enriched, and the top 32 entries were selected based onP-values < 0.01. The MFs of the targets mainly include protein binding, enzyme binding, cytokine activity in MFs. The top 15 items of BP and the top 15 items of MF are displayed in Figure 6.

Figure 5 Enriched GO terms for BP of potential targets of the main active ingredients of JHQG. The ordinate represents the target information involved in enrichment analysis, and the abscissa represents the number of enrichments. JHQG, Jinhuaqinggan granules; GO, gene ontology; BP, biological process.
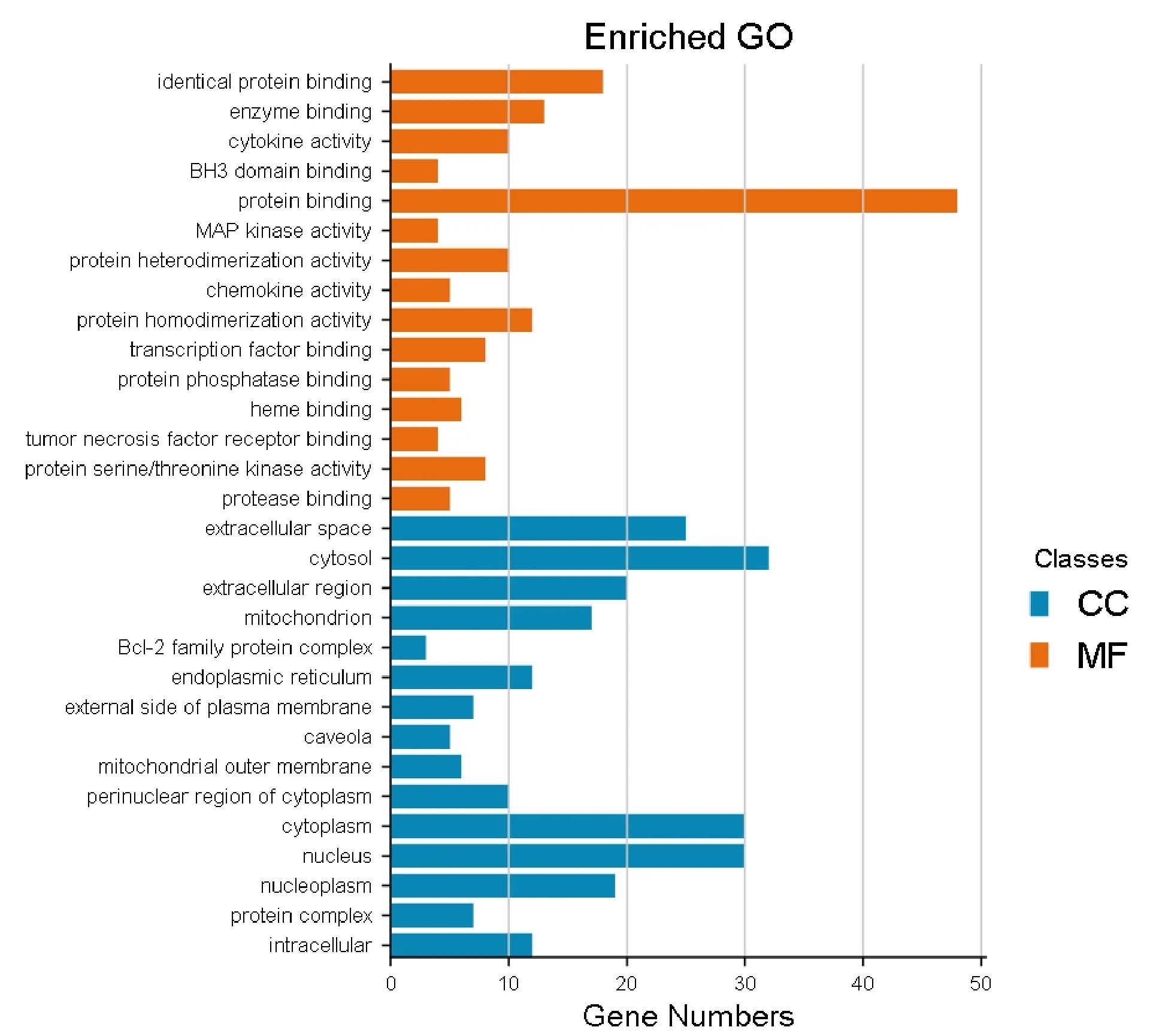
Figure 6 Enriched GO terms for MF and CC of potential targets. GO, gene ontology; MF, molecular function;CC, cell component.
KEGG pathway enrichment analysis
To further reveal the potential mechanism of JHQG on the effect of COVID-19, we conducted KEGG pathway enrichment analysis on 56 targets and screened the top 20 pathways based on the threshold ofP< 0.01. (Figure 7). Numerous pathways for potential target genes were identified, such as the tolllike receptor (TLR) SP, the tumor necrosis factor(TNF) SP, the IL17 SP, the mitogen-activated protein kinase (MAPK) SP, and the c-type lectin receptor(CLR) SP, which are associated with waterfall inflammation. The TLR SP is related to the development of ALI/acute respiratory distress syndrome (ARDS) [39]. In addition, we found other pathways, such as pathways in cancer, hepatitis B,tuberculosis, Chagas disease, influenza A, and human cytomegalovirus infection.
Target-pathway network of the top 20 KEGG terms
A target-pathway network was constructed based on the top 20 KEGG enrichment terms associated with 70 nodes and 311 edges, as shown in Figure 8, where the green rectangle represents the pathway and the purple rectangle represents the target. The details of the top 10 pathways regarding degree value are described in Table 4. The network topology analysis results show that the core targets with a degree greater than the average are MAPK3, MAPK1, RELA, MAPK8,MAPK14, TNF, IL1B, TGFB1, and FOS. It indicates that these aforementioned genes may be key to the treatment of COVID-19 with JHQG.
Molecular docking of important bioactive components in JHQG acting on SARS-CoV-2 3CL hydrolase and ACE2
SARS-CoV-2 3CL hydrolase and ACE2 are the key enzymes of COVID-19. The five bioactive compounds obtained from the compound-target network were molecularly docked with the above two key enzymes. The compound and protein can spontaneously combine when the binding energy is less than zero. Taking the binding energy ≤ -5 kcal/mol as the screening criterion, the five molecular docking modes with the smallest binding energy were screened out. The detailed results of the molecular binding energy and molecular docking model are presented in Table 5 and Table 6 and the chemical structure of five important compounds is shown in Figure 9.
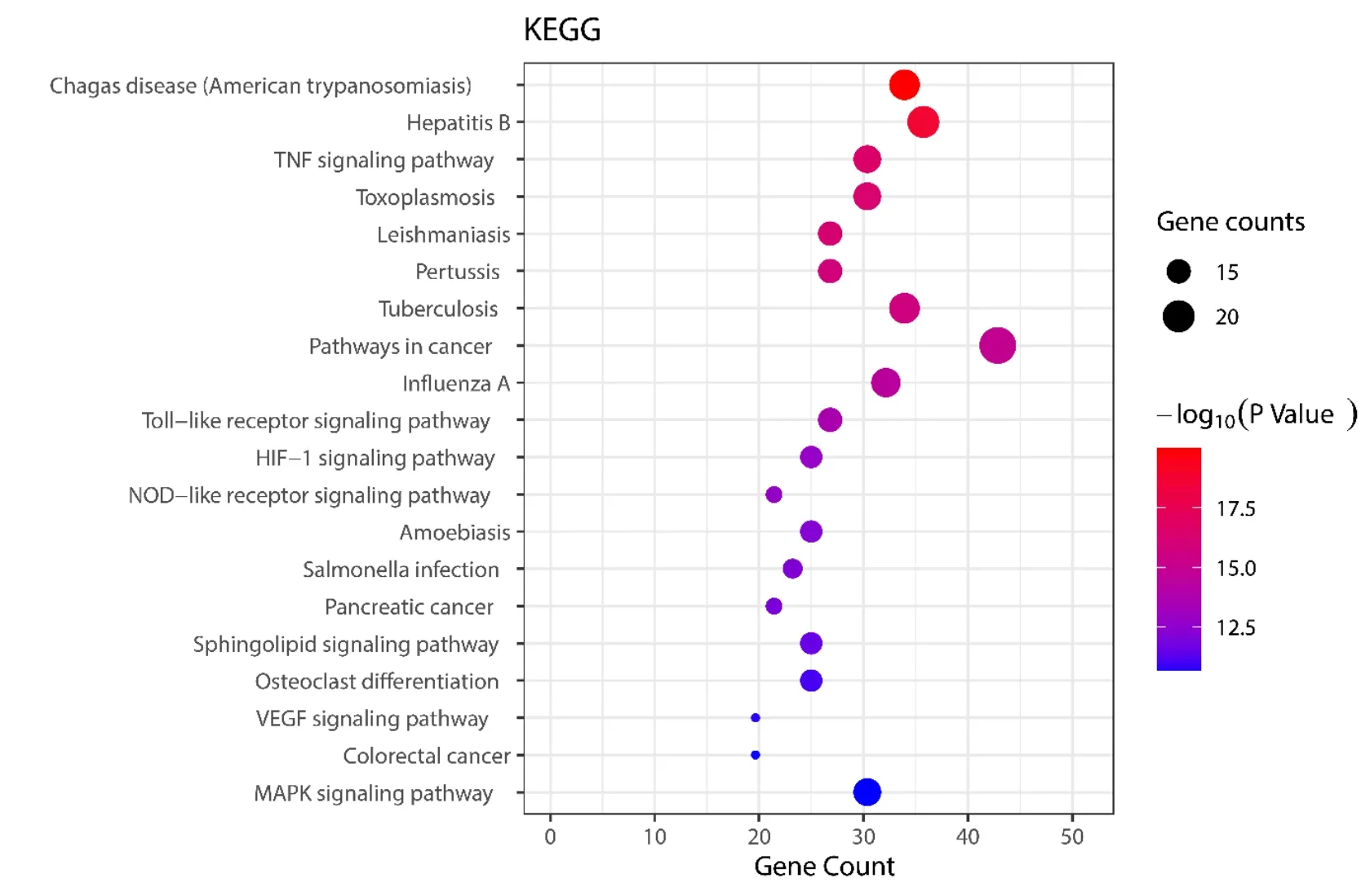
Figure 7 KEGG bubble chart of the main target of JHQG. KEGG, Kyoto Encyclopedia of Genes and Genomes;JHQG, Jinhuaqinggan granules.

Table 4 The details of top 10 pathways regarding degree value based on KEGG
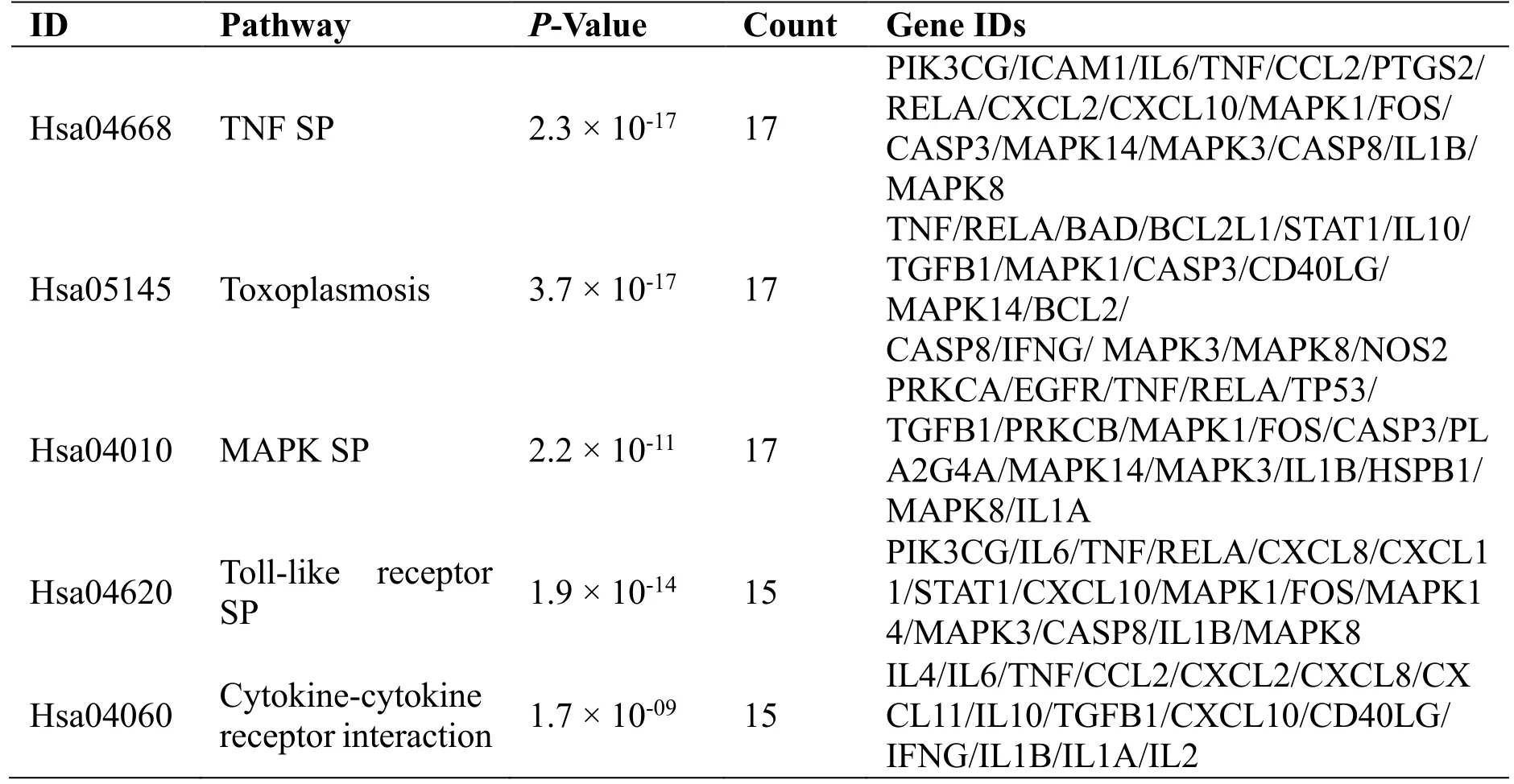
Table 4 The details of top 10 pathways regarding degree value based on KEGG (continued)

Figure 8 Target-pathway network of the top 20 KEGG terms. KEGG, Kyoto Encyclopedia of Genes and Genomes.

Table 5 Molecular docking pattern diagram of five important bioactive components with SARS-CoV-2 3CL hydrolase and ACE2

Table 6 Affinity of bioactive compounds of JHQG with SARS-CoV-2 3CL hydrolase and ACE2
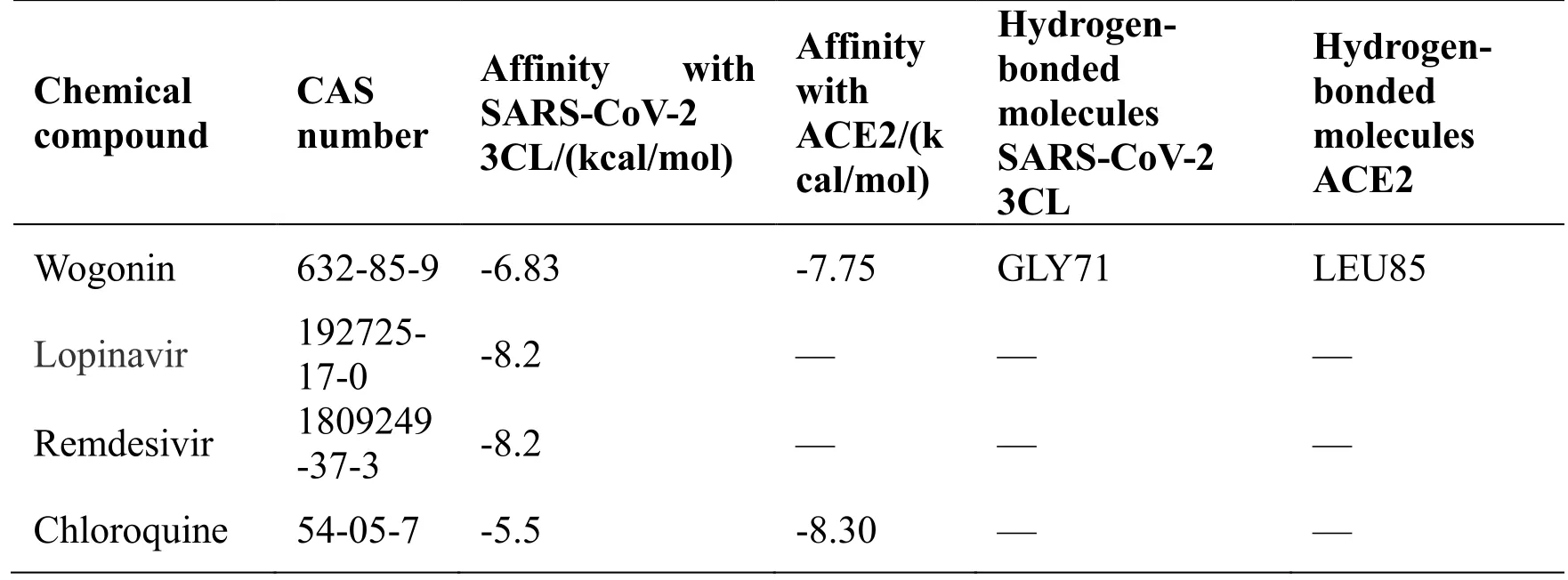
Table 6 Affinity of bioactive compounds of JHQG with SARS-CoV-2 3CL hydrolase and ACE2 (continued)

Figure 9 The chemical structure of five important compounds
Discussion
From the results of the compound-target network analysis, the top five compounds with central values were screened out: quercetin, kaempferol, luteolin,wogonin, and naringenin. These compounds can treat ALI with different mechanisms. ALI is one of the complications of COVID-19. Experimental studies have shown that [6] quercetin pre-treatment can significantly reduce bleeding, edema, congestion and expansion of capillaries and exudation of the lung interstitial and alveolar spaces in rats through significant inhibition of the release of proinflammatory cytokines, such as IL6, IL1β and TNF-α. In terms of virus suppression, quercetin [7]has both anti-infective and anti-replication activities;therefore, in monolayer cell culture, quercetin can inhibit the infection and replication of various viruses on cells by preventing viral receptor complexes from entering cells through calcium ions. Experimental studies have shown that luteolin [8] reduces airway inflammation in asthmatic rats through antioxidative stress and inhibits the COX-2 SP. Moreover, luteolin can affect the metabolism of arachidonic acid and a variety of inflammatory SPs by reducing the generation of reactive oxygen and reactive nitrogen and can ultimately inhibit the expression of inflammatory cytokines and inflammatory mediators[9-10]. Wogonin [11] has a significant immunosuppressive effect, which can significantly reduce the expression of IL1β, IL18, IL6, TNF-α,NLRP3 and caspase-1 mRNA levels, reduce NF-κB nuclear translocation, and downregulate the expression of extracellular TNF-α and intracellular ROS levels in macrophages. Naringenin has multiple pharmacological effects involving antibacterial, antiinflammatory, scavenging free radicals, antioxidant,antitussive and expectorant activities. Current research [12] shows that naringenin can improve LPS-induced ALI in rats by inhibiting the PI3K/AKT pathway, and inhibited histopathological changes,pulmonary edema and lung vascular leak. These effects are likely to be associated with the inhibition of ROS generation, neutrophil infiltration, and inflammatory cytokines levels in the serum and BALF by naringenin.
From the PPI network of the interaction between JHQG and COVID-19, six-core targets were screened,namely, IL10, proto-oncogene FOS, signal transduction and STAT1, IL4, IFNG, and HMOX1.
The continuous deterioration of new coronary pneumonia is mostly caused by inflammatory storms,also known as cytokine storms. The symptoms of COVID-19 include persistent high fever, dry cough,extreme fatigue, and even pathological changes of severe damage to organs and tissues. According to the results of prior research, cytokine storms involve ILs(IL1, IL2, IL4, IL6, IL8, IL10, and IL12), colonystimulating factors, chemokines (IFN-α, IFN-β, IFN-γ), TNF and IFN [13-14]. In the core targets of the PPI network, STAT1 can mediate the response of cells to cytokines. Regarding the JAK-STAT1 pathway,research shows that Jak1 plays an indispensable role in mediating biological responses (during airway inflammation) induced by three major cytokine receptor subfamilies: class II cytokine receptors (the receptors for IFNa/b, IFNg, and IL10), cytokine receptors that utilize the GC receptor subunit (the receptors for IL2, IL4, IL7, IL9, and IL15), and receptors that utilize the gp130 subunit (receptors for IL6, IL11, LIF, OSM, CNTF, and CT-1) [15-16]. IL4 is a cytokine secreted by type II helper T cells (Th2 cells), which can promote the proliferation and activation of B cells and T cells and plays a crucial role in regulating humoral immunity and adaptive immunity.
ARDS and ALI are critically ill cases of new coronary pneumonia. Overexpression of inflammatory mediators plays a vital role in the occurrence and development of ARDS and ALI [17].IL10 is another core target of JHQG. IL10 is a prominent inflammatory and immunosuppressive factor that can inhibit the Th1 cell response and suppress the antigen presentation of macrophages.IL10 can widely inhibit the expression of IL1, IL6,IL8, TNF-α, macrophage inflammatory protein-1α and other immune media. More importantly, IL10 can regulate IL1 receptor antagonists, soluble TNF receptors and other anti-inflammatory mediators [18-19]. The compounds of JHQG that act on the target of IL10 are quercetin, luteolin, and aloe-emodin.Through the above mechanism, it can be concluded that IL10 plays a key inhibitory role in the overexpression of inflammation.
As a member of the type III IFN family, IFN-λ plays an essential biological role in the process of antiviral and anti-respiratory infections. Pathogenic microorganisms and their metabolites stimulate the body to release large amounts of IFN-λ. IFN-λ has resistance to RNA virus infection and DNA virus infection. Studies have shown [20] that mice lacking IFN-λ are more sensitive to SARS coronavirus and influenza virus and are more susceptible to virus infection. Previous studies have also found that IFN-λ is primarily involved in the immune response of respiratory diseases induced by influenza virus and coronavirus [21]. Therefore, the active ingredients of JHQG are likely to promote the production and release of IFN-λ, thereby generating immune responses against coronavirus.
HMOX1 is the rate-limiting enzyme of haem catabolism. This enzyme is also known as heat shock protein 32, which is induced when the body is in a state of high fever. HMOX1 can confer certain heat resistance to cells; therefore, it is regarded as the main“protective enzyme” of the body. Multiple studies have shown that some drugs can induce endogenous or exogenous expression of HMOX1 to inhibit the virus. The proven virus types inhibited by HMOX1[22-24] include influenza A virus, hepatitis B virus,hepatitis C virus, and human immunodeficiency virus.The antiviral mechanism of HMOX1 [25-28] includes interfering with the virus adsorption process,inhibiting the replication of viral macromolecular nucleic acids and protein synthesis, interfering with the assembly of viruses into intact virus particles and activating IFN with haem metabolites. Among the active ingredients in JHQG, there are 5 compounds acting on HMOX1, namely, kaempferol, quercetin,luteolin, β-carotene and polyphenol.
The results of GO enrichment analysis primarily obtained such effects as LPS responses, LPS-mediated SPs, positive regulation of NO biosynthesis processes,immune responses, glucocorticoid responses, and positive regulation of chemokine biosynthesis processes. COVID-19 combined with bacterial infection is more likely to cause ALI. Clinically, the largest proportion of ALI is caused by gram-negative bacterial infections [29]. LPS is the main component of the outer wall of the gram-negative bacterial cell wall. As a common endotoxin, LPS can induce pulmonary interstitial oedema, dyspnoea and hypoxemia in rats. In humans, LPS can activate neutrophils and macrophages to release a large number of inflammatory factors. Quercetin is one of the main active ingredients of JHQG. Studies [30-31]indicate that quercetin has a protective effect on ALI induced by LPS in mice by regulating the NF-KB and Nrf-2/ARE SPs. The mechanism can be clarified as the inhibitory effect of quercetin on the release of inflammatory factors through the downregulation of the expression of NF-kB/p65 and ICAM-1. In addition,the glucocorticoid response is another important pathway of KEGG enrichment. Glycyrrhizin in liquorice has good glucocorticoid-like effects [32],such as cell membrane protection, anti-inflammatory,antiviral effects, and immune regulation. The glucocorticoid-like anti-inflammatory effects of glycyrrhizin are generated by selectively inhibiting the metabolism of arachidonic acid and the release of cytochrome C, preventing the release of prostaglandins and leukotriene inflammatory mediators [33].
The positive regulation of the NO biosynthesis process is also a major way for JHQG to treat pneumonia. Previous research has shown [34] that NO plays an important cytotoxic effect when the body is infected. NO can inhibit a variety of DNA viruses and RNA viruses by acting on the Th1/Th2 balance system,which induces IL4 and IL12 expression and inhibits IL2 and IFN-γ expression [35-39].
From the analysis of KEGG pathway enrichment,the main TLR SP, CLR SP, MAPK SP, influenza A,TNF SP, IL17 SP, and human cytomegalovirus infection were obtained. The regulation of the TLR pathway [40] is related to the development of ALI/ARDS. TLRs are innate immune protein molecules, and waterfall inflammation can be caused when the TLR 4 SP responds excessively. Studies [41]have shown that baicalin can inhibit the lung inflammation of mice caused by influenza A virus by downregulating the SP mediated by TLR7. Wogonin[42] inhibits TLR7-mediated SPs, thereby inhibiting the expression of NF-kB/p65 and alleviating influenza pneumonia.
CLRs, similar to TLRs, are important regulatory molecules in innate immunity; they haveStreptococcus pneumoniaerecognition receptors and various fungal recognition receptors [43]. There are 14 MAPK SPs in mammalian cells. Classical SPs include extracellular signal-regulated kinase, c-Jun amino-terminal kinase, and p38 MAPK, which are considered to be important to cell physiology and key regulators of the immune response [44]. After being activated, p38 MAPK can enter the nucleus to phosphorylate specific substrates and regulate the inflammatory response under the stress reflex [45].Previous experiments have suggested that [46]Zhebeimu (Fritillariae thunbergii bulbus) can reduce the airway inflammatory response by activating the MAPK SP.
Conclusion
In summary, this study used network pharmacology to retrieve the main active ingredients and potential targets in JHQG and analyze the mechanism of multitarget and multichannel treatment of COVID-19.In our research, five important bioactive ingredients were screened from the compound-target network, 56 effective targets were obtained in the Venn diagram,and 6 core targets were obtained from the PPI network.Molecular docking shows that quercetin, luteolin,kaempferol, wogonin and naringin have good affinity with SARS-CoV-2 3CL hydrolase and ACE2. The five important compounds in JHQG were quercetin,kaempferol, luteolin, wogonin, and naringenin, which can act on 6 core targets related to pneumonia,including IL10, FOS, STAT1, IL4, IFNG, and HMOX1. In keeping with these results, through enrichment analysis of GO and KEGG, JHQG was determined to regulate a variety of SPs, such as the LPS reaction pathway, the TLR SP, the CLR SP, the MAPK SP, and the TNF SP, playing an important role in the treatment of COVID-19.
 Precision Medicine Research2020年2期
Precision Medicine Research2020年2期
- Precision Medicine Research的其它文章
- Screening key target genes for pulmonary arterial hypertension based on bioinformatics
- Screening key target genes for severe acute respiratory syndrome coronavirus 2(SARS-CoV-2) based on bioinformatics and gene network
- A network pharmacology approach to investigate the mechanism of “Huangqi-Shanzhuyu” in the treatment of diabetic nephropathy
- Network pharmacological approach to explore the mechanisms of Lianhua Qingwen capsule in coronavirus disease 2019
