Screening key target genes for severe acute respiratory syndrome coronavirus 2(SARS-CoV-2) based on bioinformatics and gene network
Zhi-Hua Yang, Hai-Feng Yan, Lin-Wang, Miao-Ru Han
1First Teaching Hospital of Tianjin University of Traditional Chinese Medicine, Tianjin 300381, China.
Abstract
Background: To provide a reference for the clinical development of drugs to suppress severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Methods: Retrieving genes related to SARS-CoV-2 with Genecards database and then importing the obtained gene data into the database of Database for Annotation, Visualization and Integrated Discovery (DAVID) (Version 6.8) to collect relevant information on pathways and genes. Genes enriched in the first 20 most significant pathways and genes with gene occurrence frequency ≥ 6 were respectively imported into the STRING database to construct protein-protein interaction (PPI) network diagrams, and the two network diagrams were compared. Results: In the two network graphs, RELA, MAPK1, MAPK3, PIK3CA, PIK3R1, MAPK8,JAK1, STAT1, TNF, IL6, MAPK14, and IL1B ranked higher, and the occurrence frequency of the first 20 pathways was ≥ 10. Conclusion: The pathogenesis of SARS-CoV-2 is associated with multiple pathways such as influenza A,TNF signaling pathway, chemokine signaling pathway, toll-like receptor signaling pathway, T cell receptor signaling pathway et al. RELA, MAPK1, MAPK3, PIK3CA, PIK3R1, MAPK8, JAK1, STAT1, TNF, IL6, MAPK14 and IL1B are closely related to SARS-CoV-2 and need further study. Gene interaction network and pathway analysis of diseaseassociated genes will help us to screen the key target genes of SARS-CoV-2 and provide a reference for the clinical development of effective drugs.
Keywords: Bioinformatics, Gene network, SARS-CoV-2, COVID-19, Target gene
Background
In December 2019 in Wuhan City (Hubei Province,China), multiple cases of patients with pneumonia infected by a new type of coronavirus were noted.World Health Organization (WHO) named it coronavirus disease 2019 (COVID-19) on February 11,2020. With the spread of the virus, cases in China and overseas have also been found [1, 2]. The most common clinical presentations are fever, fatigue, dry cough and some patients present with nasal congestion, runny nose and diarrhea. In severe cases, dyspnea usually occurs one week after the disease onset and some patients can rapidly progress to acute respiratory distress syndrome(ARDS), septic shock, refractory metabolic acidosis,and coagulation disorders [1]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) belongs to the coronavirus of the beta genus. The virus has enveloped and its granules are oval or round, often polygonal, and sensitive to ultraviolet and heat. It can appear in human respiratory epithelial cells for about 96 hours [3].COVID-19 was listed as a public health emergency of international concern by WHO on January 31, 2020,and it became a severe epidemic that seriously endangers people's health and public safety. SARSCoV-2 is the seventh coronavirus found to infect human beings [4, 5]. This is a new type of virus, which is highly infectious and can cause severe respiratory diseases.Clinically efficacious treatment is lacking. So far, there are no approved or verified effective drugs specific to the virus although some antiviral medicines and traditional Chinese medicine are used in the clinical treatment of SARS-CoV-2. Many therapeutic drugs are selected from the clinical treatment experience of severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS) and other infections, and the efficacy is not exact [6]. In this study, we combined bioinformatics and gene networks to analyze the genes related to SARS-CoV-2. We screened out the core target genes of SARS-CoV-2, to provided reference for clinical research and development of SARS-CoV-2 inhibition and treatment of COVID-19 effective drugs.
Research methods
Collecting SARS-CoV-2 related genes
The Genecards database (http://www.genecards.org/) is a comprehensive database of gene, transcriptome,proteome and genetic information [7]. We used the Genecards database to screen the genes related to SARS-CoV-2. We logged in to the main page of Genecards and inputted the keyword “severe acute aespiratory syndrome coronavirus 2” to obtain the virus genes related to SARS-CoV-2.
Analysis of SARS-CoV-2 related pathways with DAVID tool
The Database for Annotation, Visualization and Integrated Discovery ( DAVID) is an online database of gene and pathway function annotation. We used DAVID(https://david.ncifcrf.gov/) to analyze the pathway of SARS-CoV-2 related targets in this study. P < 0.01 was treated as the screening significance, the important signal pathways were selected for key analysis. Three hundred and forty-seven collected genes were put into the DAVID database for analysis.
Interaction network analysis of genes significantly enriched in pathways
According to the results of DAVID database analysis,genes enriched in the first 20 pathways and genes appearing on the pathway with frequency ≥ 6 were input into the STRING database (http://string-db.org/) to construct protein-protein interaction network model,and the species was set as “Homo sapiens” [8, 9]. To ensure the reliability of the data, the minimum protein interaction threshold was set to “highest confidence” ( >0.9). The protein interaction was screened, the results were saved in TSV format, and the node1, node2, and combined score data in the file were retained and imported into Cytoscape 3.5.1 software for analysis of gene interaction network diagrams. Compare the total gene network map enriched in the pathway with the gene network map with the frequency of 6 or more to find out the genes that are shared by the two and have the highest degree of value. These genes may be the key target genes to regulate the pathogenesis of SARS-CoV-2. Degrees of freedom and betweenness are two main topological parameters that measure the importance of a node in the network. They are also relevant references for determining whether a gene is a “core target” [10].The higher the degree of freedom, the stronger the biological importance, and the higher the betweenness,the more influential the node is in the network. The size of a node is determined by the degree of freedom (the greater the degree of freedom, the larger the node). The color of the node is set according to the degree of freedom, and the degree of freedom gradually increases as the color changes from blue to yellow. The thickness of the edge is determined by the betweenness, and the larger the betweenness, the thicker the edge.
Results
DAVID pathway analysis
As a result, a total of 347 genes related to SARS-CoV-2 were collected. In the DAVID database, 347 related genes were input and their Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were analyzed and 342 genes were enriched in 139 pathways. The first 20 most significant entichment pathways were selected,including influenza A, malaria, Chagas disease,hepatitis B, hepatitis C, herpes simplex infection, tolllike receptor signaling pathway, phathways in cancer, T cell receptor signaling pathway, chemokine signaling pathway, tumor necrosis factor (TNF) signaling pathway, toxoplasmosis, osteoclast differentiation,Epstein-Barr virus infection, pertussis, hypoxiainducible factor-1 (HIF-1) signaling pathway,tuberculosis, tiral carcinogenesis, Human T-cell leukemia virus-1 (HTLV-I) infection, and pancreatic cancer. The enrichment analysis results were visualized by the ImageGP software(http://www.ehbio.com/imagegp/) (Figure 1). A total of 164 gene interactions (10 genes are not in the diagram)were presented. There are 154 genes and 1,210 interaction relationships. The average value is 15.714(Figure 2). A total of 164 genes related to SARS-CoV-2 are enriched in the first 20 pathways, and the frequency of gene occurrence is ≥ 3 (Table 1).
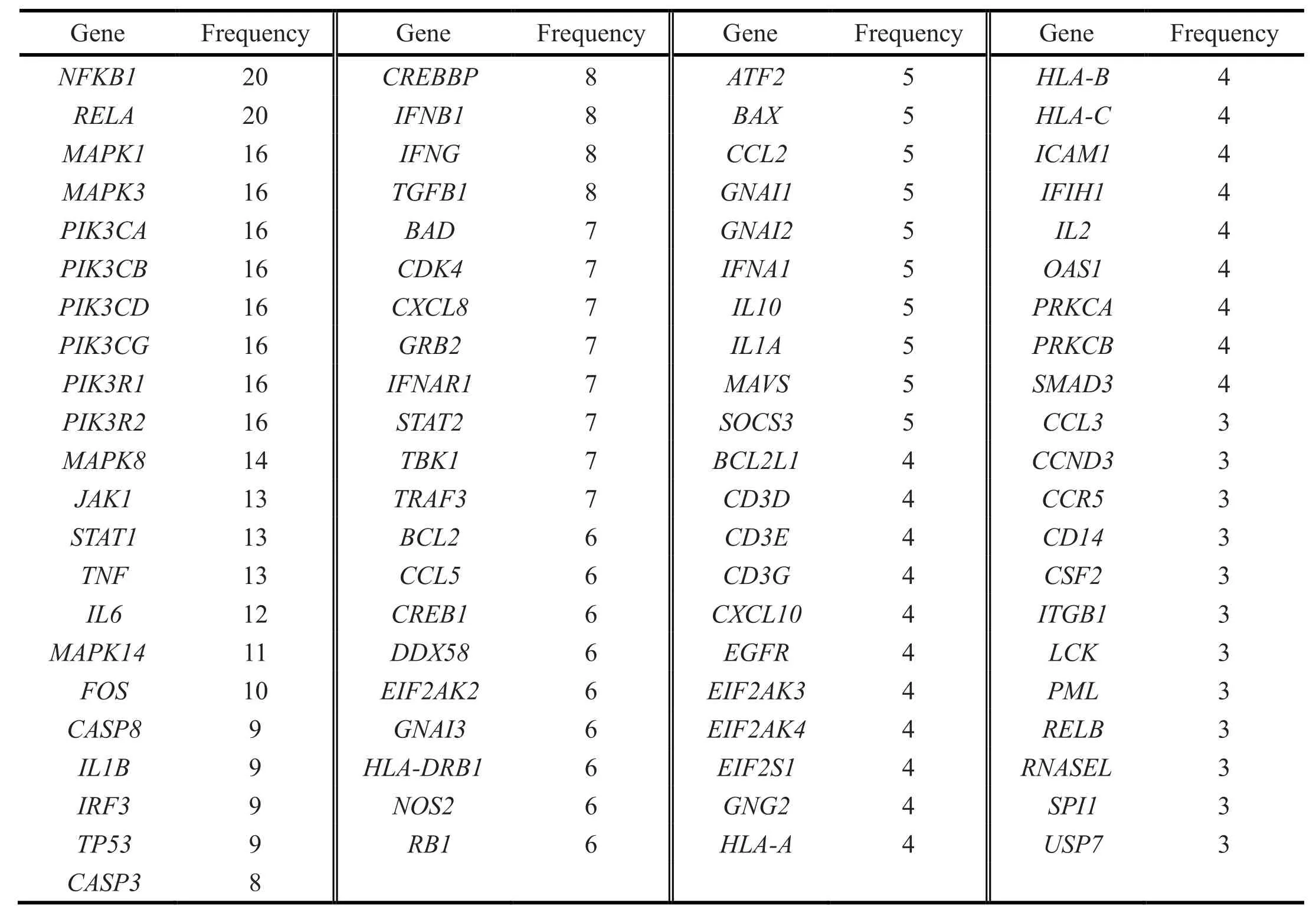
Table 1 Genes with a frequency ≥ 3 in the first 20 pathways
Network analysis of genes enriched in pathways and high-frequency genes
A total of 164 SARS-CoV-2 related genes were significantly enriched in the first 20 pathways, and 43 genes appeared in 20 pathways with a frequency of ≥ 6.These 43 genes may be closely related to the pathogenesis of SARS-CoV-2, because they are involved in the signaling of multiple pathways. The network diagram of 43 gene interactions was shown in Figure 3, with 43 nodes and 205 interaction relationships. The average value is 9.535. Twelve genes are common and highly ranked includingRELA,MAPK1,MAPK3,PIK3CA,PIK3R1,MAPK8,JAK1,STAT1,TNF,IL6,MAPK14andIL1B, indicating that these 12 genes are closely related to SARS-CoV-2 and should be our target genes for SARS-CoV-2 research.
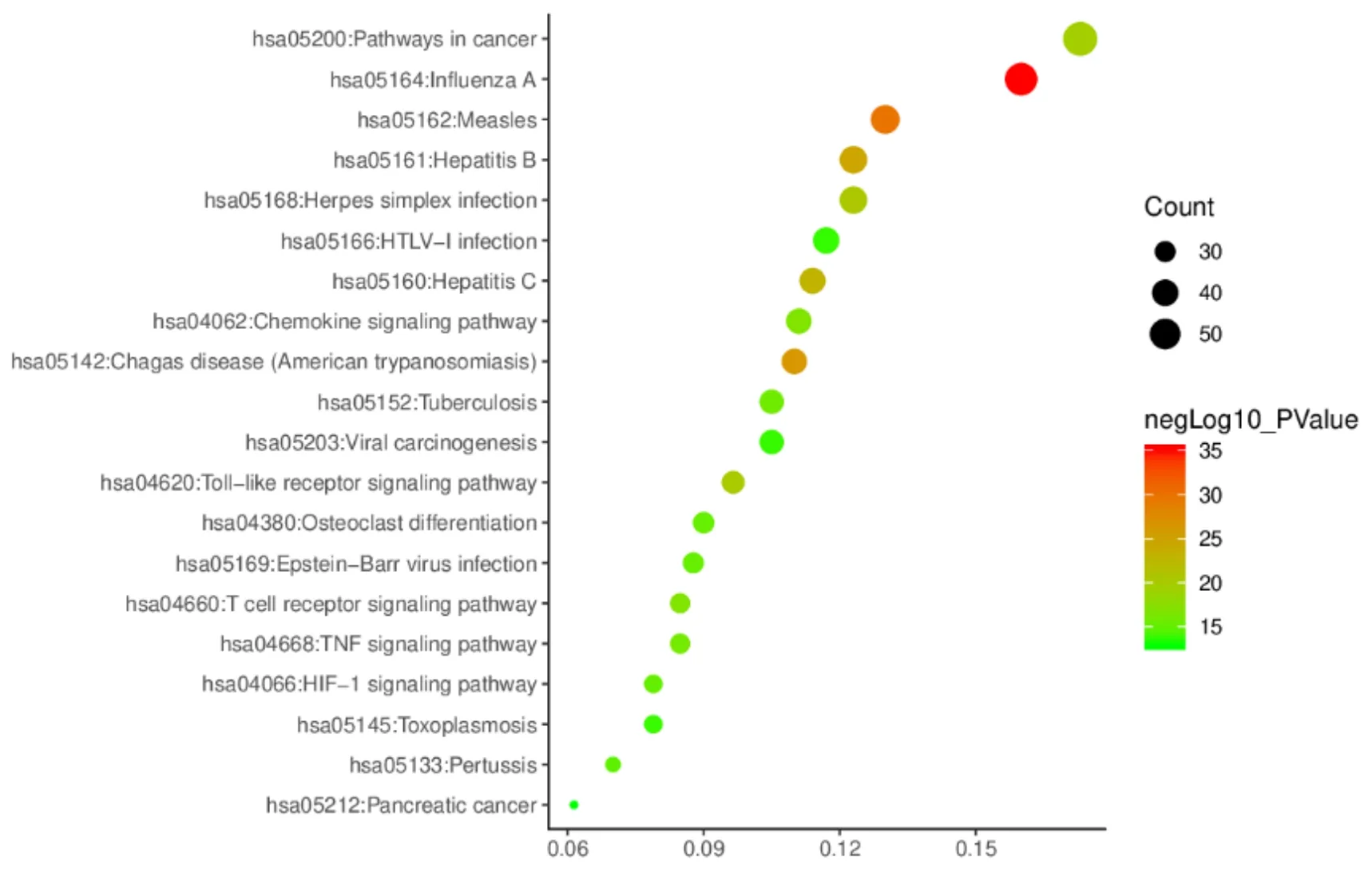
Figure 1 Enrichment analysis results of the KEGG pathway. The X-axis represents the rich factor, the Y-axis represents the pathway, and the bubble size represents the gene number of the target gene in this pathway. The larger the bubble is, the more genes are enriched in the path, and the bubble color represents the significance of enrichment.
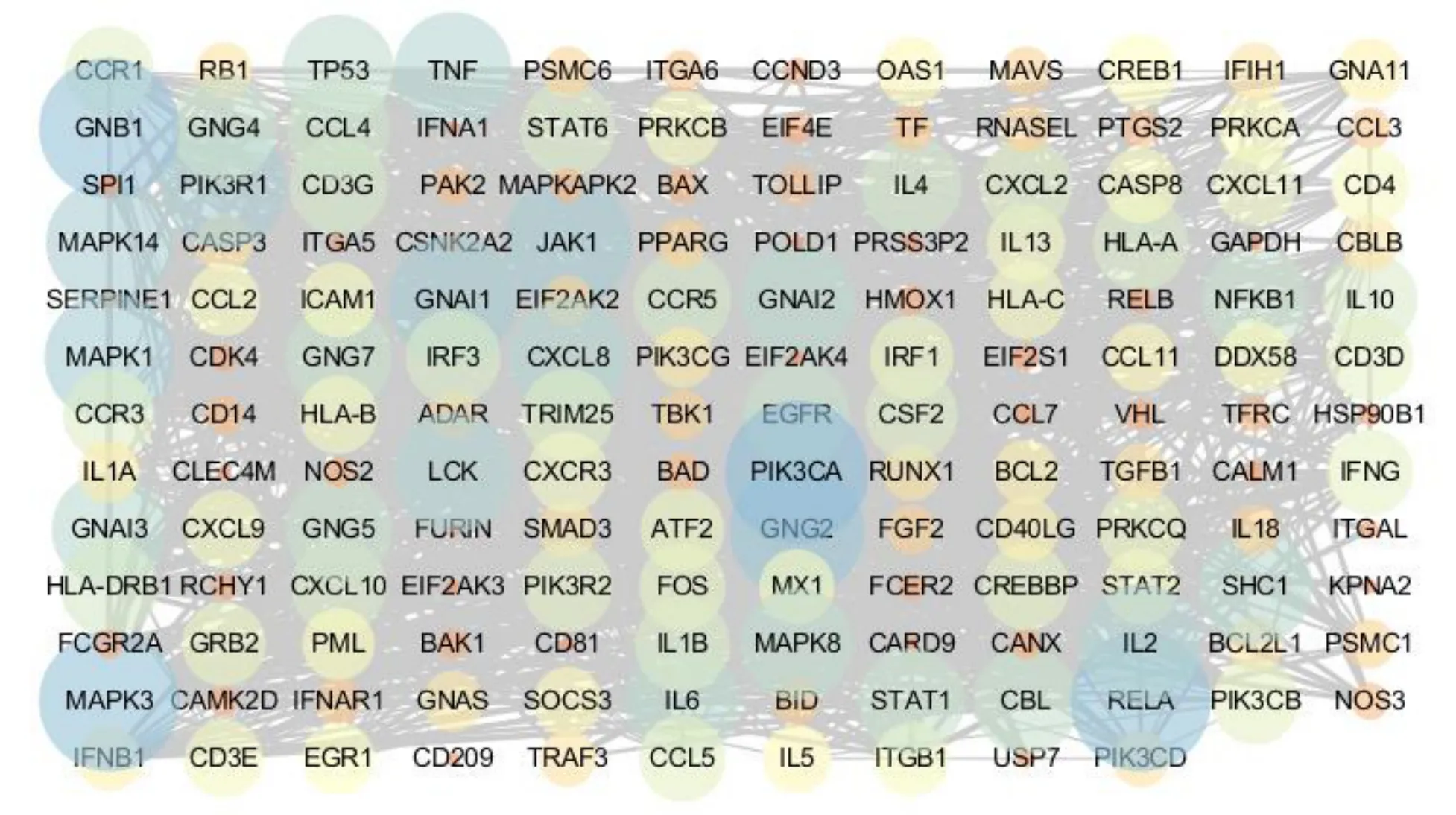
Figure 2 Interaction network of 154 genes
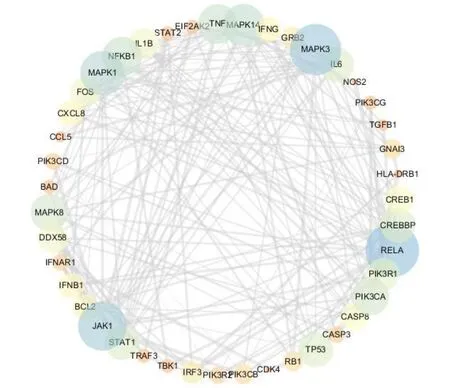
Figure 3 Interaction network of 43 genes
Discussion
At present, COVID-19 is mainly symptomatic treatment, supportive treatment and symptom improvement, and there are no clinically effective drugs to cure the disease. COVID-19 diagnosis and treatment program (trial version 6) proposed that antiviral treatment can be tried with interferon-alpha,lopinavir/ritonavir, ribavirin, chloroquine phosphate,arbidole and pointed out that these drugs in clinical application to further evaluate the efficacy of the current trial. The pathogenesis of SARS-CoV-2 is complex and involves many pathways. Therefore, we should find out the key target genes of SARS-CoV-2 and regulate the signal pathways it participates in, to prevent and treat the disease effectively. In this study, Genecards databases were used to identify 347 genes related to SARS-CoV-2. The KEGG pathway enrichment analysis of these genes was performed using the DAVID database. We found a total of 164 SARS-CoV-2 related genes were significantly enriched in the first 20 pathways and 43 genes appeared in 20 pathways with a frequency of ≥ 6. We made a network diagram of the interaction between 164 and 43 SARS-CoV-2 related genes through the STRING database. We found that the degree ofRELA,MAPK1,MAPK3,PIK3CA,PIK3R1,MAPK8,JAK1,STAT1,TNF,IL6,MAPK14andIL1Bare higher in the two network diagrams, indicating that they are closely related to SARS-CoV-2. Because these genes not only have a higher degree of freedom, but also participate in multiple pathways, they may be the key target genes of SARS-CoV-2.MAPK1,MAPK3,MAPK8andMAPK14are all mitogen-activated protein kinases, which are mainly activated under the stress of oxidative stress, DNA damage, cancer development and virus infection. They also may detect the phosphorylation of MAPK members [11].STAT3,MAPK1,PIK3CA,MAPK3,TNF,IL6may be the targets of Renshen Baidu powder in the treatment of COVID-19 by inhibiting cytokine storm [12]. Among the 20 signaling pathways, TNF signaling pathway can promote the expression of proinflammatory cytokines,chemokines, growth factors and tumor necrosis factorα (TNF-α) to amplify the inflammatory and immune responses [13]. TNF-α can activate nuclear factor kappa B (NF-κB), the latter can not only enhance the transcription level of the coding gene of TNF-α,interleukin-6 (IL-6), adhesion molecules and other coding genes, but also make the inflammatory response and body damage continue to deteriorate by the positive feedback effect between TNF-α and inflammatory factors. TNF-α is a major cytokine that mediates inflammatory response and regulates cellular and humoral immunity in the human body. Its serum level can reflect the state of the inflammatory response in the human body [14]. The severity of COVID-19 is related to the intensity of the virus and the body's inflammatory response. For example, excessive inflammatory response, may lead to serious consequences or even death [15]. In the early stage of virus infection, TNF-α inflammatory factors can be secreted rapidly and reach its peak within hours. It can induce the activation of pulmonary endothelial cells, the shedding of granulocytes and the migration of leukocytes. TNF-α is the proinflammatory factor of the cytokine storm,which may lead to the aggravation of symptoms and pathological damage after infection. Therefore, the treatment of anti-TNF-α may become a primary method of confrontation the cytokine storm [16]. Clinical studies have found that IL-6 is the main proinflammatory cytokine, and the activity of IL-6 is weakened in the secondary bacterial infection, which is conducive to virus clearance and host survival. The level of IL-6 is related to the severity of pneumonia after Influenza [16, 17]. IL1B is an important regulatory medium of inflammatory response. It is involved in cell proliferation, differentiation, apoptosis and other cell activities.IL1B can activate neutrophils, T lymphocytes and B lymphocytes, and promote the production of cytokines and antibodies. The chemokine signaling pathway is also an inflammatory signaling pathway.Inflammatory chemokines and cytokines play an important role in the immune response of viral infection.When the virus invades the organism, the organism can stimulate the autoimmune response and gather a large number of inflammatory chemokines and cytokines in the site of virus invasion, activate neutrophils,lymphocytes, and then isolate and phagocytize the virus that invades the organism. This process has a very positive sign for the inhibition of virus infection.However, the excessive and maladjusted immune response may lead to overexpression of inflammatory factors in patients' bodies, resulting in “cytokine storm”[18, 19]. Modern studies have found that the toll-like receptor signaling pathway is closely related to the pathogenesis of acute lung injury/acute respiratory distress syndrome. Toll-like receptor 4 (TLR4) is considered to be an important receptor of lipopolysaccharide. Toll-like receptor 4 (TLR4) is intracellular signal transduction, a transmembrane receptor on the target cell membrane and a major pathogen pattern recognition receptor in the natural immune system [20-22].
In this study, we explored target genes that are closely related to SARS-CoV-2 through various aspects such as the analysis of KEGG pathway enrichment, gene function, gene interaction network by using bioinformatics and gene network. Finally, we found thatRELA,MAPK1,MAPK3,PIK3CA,PIK3R1,MAPK8,JAK1,STAT1,TNF,IL6,MAPK14andIL1B are closely related to SARS-CoV-2. These 12 genes may be the key target genes of SARS-CoV-2. However, further experiments needed to verify the important role of these targets in disease treatment.
 Precision Medicine Research2020年2期
Precision Medicine Research2020年2期
- Precision Medicine Research的其它文章
- Use of network pharmacology and molecular docking to explore the potential mechanism governing the efficacy of Jinhuaqinggan granules in the treatment of novel coronavirus-induced pneumonia
- Network pharmacological approach to explore the mechanisms of Lianhua Qingwen capsule in coronavirus disease 2019
- A network pharmacology approach to investigate the mechanism of “Huangqi-Shanzhuyu” in the treatment of diabetic nephropathy
- Screening key target genes for pulmonary arterial hypertension based on bioinformatics
