miR-23a Promoting Cell Proliferation of Human Tongue Squamous Cell Carcinoma Cell through Regulating PPP2R5E
Xue LIU, Yadong TAO, Jihong SUN, Feng HUO*, Hongjie GUO
1. Chengde Medical University, Chengde 067000, China; 2. The Affiliated Hospital of Chengde Medical University, Department of Stomatology, Chengde 067000, China
Abstract [Objectives] To investigate the mechanism of miR-23a in the proliferation of human tongue squamous cell carcinoma cells. [Methods] Clinical tissue samples of tongue squamous cell carcinoma were collected and Tca8113 and CAL27 were cultured. Real-time quantitative PCR was performed to detect the expressions of miR-23a and PPP2R5E in clinical tissue samples of tongue squamous cell carcinoma. MTT assay, colony formation assay and growth curve assay were used to detect the effect of miR-23a and PPP2R5E on the proliferation of human tongue squamous carcinoma cell. Luciferase reporter assay verified the regulatory relationship between miR-23a and PPP2R5E. [Results] The expression of miR-23a in tongue squamous cell carcinoma was significantly up-regulated (P<0.01). miR-23a promoted the proliferation of tongue squamous cell carcinoma cells. Bioinformatics prediction and luciferin reporting experiments showed that PPP2R5E was a direct target gene of miR-23a. The expression of PPP2R5E was decreased in tongue squamous cell carcinoma. PPP2R5E inhibited the proliferation of tongue squamous cell carcinoma cells. Overexpression of PPP2R5E can reverse the proliferation promoting effect of miR-23a on tongue squamous cell carcinoma cells. [Conclusions] miR-23a can promote the proliferation of tongue squamous cell carcinoma cells through PPP2R5E and miR-23a plays an oncogene role in the occurrence and development of tongue squamous cell carcinoma.
Key words miR-23a, Tongue squamous cell carcinoma, PPP2R5E; Cell proliferation
1 Introduction
Oral cancer is the 11th most common malignancy in the world. Especially in developing countries, it was the ninth most common[1]. Among oral cancers, tongue squamous cell carcinoma is the most common. Tongue squamous cell carcinoma has high malignant degree, fast growth rate, strong infiltration and easy metastasis, and is the site of the highest incidence of oral cancer[2]. In recent decades, the causes of tongue squamous cell carcinoma have been gradually identified, and preventive measures and clinical diagnosis and treatment techniques have also been continuously developed, but the five-year survival rate of tongue squamous cell carcinoma has been stagnant at around 50%[3]. Multiple miRNAs are malregulated in tongue squamous cell carcinoma[4]and are closely related to the occurrence, development and prognosis of tongue squamous cell carcinoma. For example, miR-24 and miR-184 were up-regulated in tongue squamous cell carcinoma, which promoted the occurrence and development of tongue squamous cell carcinoma[5-6]. The expressions of miR-22 and miR-137 were decreased in tongue squamous cell carcinoma, which inhibited the invasion and metastasis of tongue squamous cell carcinoma[7-8]. Increased miR-23a expression in tongue squamous cell carcinoma cells can promote cisplatin induced apoptosis[9]. This project is intended to investigate the effect of miR-23a and its target genePPP2R5Ewith abnormally high expression in tongue squamous cell carcinoma tissues on the proliferation of tongue squamous cell carcinoma cells.
2 Methods and Materials
2.1SubjectandprincipalreagentTongue squamous cell carcinoma tissues were collected and provided by the Department of Stomatology, the Affiliated Hospital of Chengde Medical University. Tongue squamous cell line CAL27, rabbit anti-humanPPP2R5Eand GAPDH polyclonal antibodies were purchased from Tianjin Saierbio Technology Incorporation. RPMI1640, DMEM medium, RIPA lysate, BCA kit and various consumable materials for cell culture were purchased from Zhengzhou Senhui Bio. Fetal bovine serum from GIBCO, USA. Trizol and Lipofectamine 3000 from Invitrogen, USA. Rt-PCR reagent was purchased from Takara Biomedical Technology (Beijing) Co., Ltd.. Antisense oligonucleotides were purchased from Suzhou Genepharma Co., Ltd. The tongue squamous cell Tca8113 line and the required plasmid were donated by the life science center laboratory of Tianjin Medical University.
2.2Methods
2.2.1Cell culture and transfection. Tca8113 cells were cultured with RPMI1640 containing 10% fetal bovine serum in a cell incubator containing 5% CO2at 37 ℃. CAL27 cells were cultured with DMEM containing 10% fetal bovine serum and inoculated on a 48-well plate or 25 mL vial with appropriate cell density one day before transfection. The cells were in good condition at transfection with a cell density of approximately 85%. The plasmid and control vector were transferred into Tca8113 cells by Lipo 3000. After transfection, the 48-well plate was incubated in a cell incubator containing 5% CO2at 37 ℃ for 24 h for experiment, and the 25 mL bottle was transfected and incubated in a cell incubator containing 5% CO2at 37 ℃ for 48 h for experiment.
2.2.2Total RNA extraction. For each tissue sample, added 1 mL TRIZOL reagent and control the tissue sample to 50-100 mg.The samples were homogenized and incubated for 5 min at room temperature to completely dissociate the nucleic acid protein complex. Then added 0.2 mL of chloroform. Violently oscillated for 15 sec and incubated at room temperature for 3 min. Centrifuged at 4 ℃ for 15 min at 12 000 r/min.
The precipitated RNA transferred the supernatant to a new centrifuge tube. Added 0.5 mL isopropanol to each tube. After mixing, placed at room temperature for 10 min and centrifuged at 4 ℃ at 12 000 rpm for 10 min. Discarded the supernatant, added 1 mL to each tube of 75% ethanol refrigerated at 4 ℃, and flicked the tube wall to clean the RNA. Then centrifuged at 4 ℃, 7 500 rpm for 5 min and discarded the supernatant.
Dissolved RNA, dried the RNA precipitate in the air for about 10 min. Added 20 mL of RNA-free enzyme to each tube and dissolved for 10 min, and stored at -80 ℃.
2.2.3Synthesis of cDNA.Dispensing system; total RNA,0.8 μg; 0.5 μg/μL Oligo (dT),1 μL; dNTPs mix (2.5 mM), 1.6 μL; RNA-Free H2O, 11.1 μL; total volume 14.5 μL. Water bathed at 65 ℃ for 5 min, freezed for 2 min.
After transient separation, RT reaction liquid was added into the centrifuge tube successively: 5×First-Strand Buffer, 4 μL; 0.1 M DTT, 1μL; RNase Inhibitor, 0.3 μL; SuperScript III RT, 0.2 μL.
After mixing, 37 ℃ for 1 min, 50 ℃ for 60 min, 70 ℃ for 15 min. Placed cDNA product in an ice box for use, or stored at -20 ℃ for later use.
2.2.4Real-time quantitative PCR experiment. Increase the volume of the proportion as required.The configuration proportion is as follows: 2 × Master Mix, 5 μL; 10 μM PCR specific primer F, 0.5 μL; 10 μM PCR specific primer R, 0.5 μL; H2O, 2 μL. 8 μL mixture was added to each hole, and corresponding cDNA (2 μL/ hole) was added according to the experimental group.
Set the following reaction procedure:
95 ℃, 8 min; 95 ℃, 10 s;60 ℃, 60 s; 42 circles.
Collected the results, 2- △△CTwas used to analyze the data. Folds=2-△△Ct,△△Ct=(Ct1-Ct2)-(Ct3-Ct4).
2.2.5MTT assay. After transfection, cells were incubated for 18 h in a 37 ℃ incubator with 5% CO2and harvested for counting. At the same time point of 24, 48 and 72 h after transfection, 6 000 cells per well were seeded in 96-well plate for detection. Removed the cell culture plate, added 10 μL MTT to each hole in the corresponding time area, sterilized and put back in the incubator for 4 h. Then took out the culture plate and centrifuged for 5 min, discarded the supernatant, added 100 μL DMSO into each hole, kept in dark place and shook for 10 min, and determined the absorbance value at 570 nm of each hole with a spectrophotometer. Before processing the data, observed and removed the outliers, calculated the mean for statistical analysis.
2.2.6Cloning assay. The cells were digested and resuspended for counting after 24 h transfection. 300 cells per well were inoculated on the 12-well plate, with 3 secondary holes for each. The culture medium was changed every 3 d, and colonies containing more than 50 cells were counted after 12 d. The colony was dyed with crystal violet and the colony formation rate was used to analyze the results.
2.2.7Cell growth curve. Cells were harvested 24 h after transfection for counting, and 3 000 cells per well were inoculated on 12-well plate, with 3 secondary wells for each. The culture medium was changed every 3 d, and the cells were counted at the same time every day for 7 consecutive days. Recorded the counting results and plotted the growth curve.
2.2.8Luciferase report experiment. Bioinformatics software such as MiRbase and TargetScan predicted that miR-23a could bind toPPP2R5E3’UTR, and the binding site sequence was AATGTGA, mutating the sequence to TAAGAGT. Wild type and mutantPPP2R5E3’UTR (PPP2R5E3’UTR,PPP2R5E3’UTR mut) containing binding site length of 150bp were constructed into pCD3/EGFP vector. Tca8113 cells in 24-well plates were co-transferred with 50nmol/L ASO-NC, ASO-23a, or 200ng pcDNA3, pri-miR-23a,PPP2R5E3’UTR andPPP2R5E3’UTR mut plasmids. Lipofectamine 3000 was used to transfect, and the fluid was changed 4 h later. After 48 h transfection, RIPA lysed cells, harvested proteins, and fluorescence values were measured (Table 1).
Table 1 Primer sequence

NameSequence (5′-3′)miR-23a-RTGTCGTATCCAGTGCAGGGTCCGAGGTG-CACTGGATACGACGGAAATCC miR-23a-FGGGATCACATTGCCAGGGmiR-23a-RGTCGTATCCAGTGCGTGTCGTGGAGTCG-GCAATTGCACTGGATACGACGGAAATU6-FCTCGCTTCGGCAGCACAU6-RAACGCTTCACGAATTTGCGTPPP2R5E-FTGCAAAGTTCTGAGAAGTCCAPPP2R5E-RTGCAAAGTTCTGAGAAGTCCAGAPDH-FGGCCTCCAAGGAGTAAGACCGAPDH-RAGGGGAGATTCAGTGTGGTGASO-miR-23a GGAAATCCCTGGCAATGTGAT ASO-NCGTGGATATTGTTGCCATCA
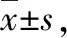
3 Results and analysis
3.1 The expression of miR-23a in tongue carcinoma tissue
Real-time quantitative PCR was used to detect the expression level of miR-23a in 10 pairs of tongue squamous cell carcinoma tissues. The results showed that the average expression level of miR-23a in tongue squamous cell carcinoma tissues was 4.617 times higher than that in the corresponding adjacent tissues. miR-23a expression in tongue squamous cell carcinoma tissues was abnormally increased (P<0.01), as shown in Fig.1.

Note:**WhenP<0.01, the statistical difference was significant.
Fig.1 Relative expression level of miR-23a in tongue squamous cell carcinoma
3.2miR-23acanpromotetheproliferationoftonguesquamouscellsIn tongue squamous cell carcinoma, we overexpressed or blocked the expression of miR-23a, and respectively used MTT assay, colony formation experiment and growth curve experiment to detect the effect of miR-23a on the activity, colony formation ability and proliferation ability of tongue squamous cell carcinoma. In Tca8113 cells, the expression level of miR-23a was changed, and the absorbance values at 24, 48, and 72 h after transfection were detected by MTT assay. The results showed that the activity of tongue squamous cell increased significantly after the overexpression of miR-23a. After blocking the expression of miR-23a, the activity of tongue squamous cell cells decreased significantly (Fig.2A). Compared with the control group the difference was statistically significant. In tongue squamous cell lines Tca8113 and CAL-27, the colony formation rate of tongue squamous cells increased significantly after overexpression of miR-23a. After blocking the expression of miR-23a, the colony formation rate of tongue squamous cell cells decreased significantly (Fig.2B and 2D). In Tca8113 cells, the expression level of miR-23a was changed, and the cell growth was counted every day for a week. After overexpression of miR-23a, cell growth was significantly accelerated. However, cell growth was significantly reduced after miR-23a was closed (Fig.2C). In conclusion, miR-23a can promote the proliferation of tongue squamous cells.
3.3PPP2R5EisthedirecttargetgeneofmiR-23aAccording to TargetScan, miRBase and other bioinformatics sites,PPP2R5Eis a potential target gene of miR-23a. miR-23a has a binding site at the 3’UTR ofPPP2R5E, as shown in Fig.3. We further confirmed by fluorescence reporter vector experiment that the seed sequence of miR-23a can directly bind to the 3’UTR ofPPP2R5EmRNA in Tca8113 cells and inhibit the expression of

Note: A. The effect of miR-23a on Tca8113 cell activity was determined by MTT assay; B. The effect of overexpression or blocking in miR-23a tongue squamous cell line Tca8113 and CAL27 on the colony formation ability was detected by colony formation experiment; C. The growth curve assay examined the effect of miR-23a on the proliferation of Tca8113 cells; D. Results of crystal violet staining from cell colonies;*whenP<0.05, the statistical difference was significant.
Fig.2 miR-23a promoted the proliferation of tongue squamous cell carcinoma cells
downstream genes. In the tongue squamous cell line, miR-23a can negatively regulate the expression of endogenousPPP2R5EmRNA.

Fig.3 Binding site information of miR-23a andPPP2R5E3’UTR
Compared with pcDNA3-EGFP-PPP2R5E3’UTR transfected with pcDNA3-EGFP alone, the fluorescence expression of EGFP was lower. Because Tca8113 cells contained endogenous miR-23a, the binding ofPPP2R5E3’UTR inhibited the expression of EGFP protein downstream. We co-transfected pcDNA3/ EGFP-PPP2R5E3’UTR with pcDNA3 and pcDNA3-pri-miR-23a respectively. The fluorescence expression intensity of pri-miR-23a group was significantly lower than that of pcDNA3 group. The binding of endogenous miR-23a andPPP2R5E3’UTR inhibited the expression of downstream EGFP protein, and the green fluorescence intensity was reduced.PPP2R5E3’UTR plasmid was co-transfected with the antisense complementary sequence ASO-23a of miR-23a and the control group ASO-NC. The result showed that the fluorescence intensity of EGFP in the ASO-23a group was significantly higher than that in the ASO-NC group. It was indicated that endogenous miR-23a could not bind toPPP2R5E3’UTR after being blocked by ASO-23a. Thereby the inhibition of downstream EGFP protein expression was released and the fluorescence intensity was increased (Fig.4A).
pcDNA3/EGFP-PPP2R5E3’UTR mut plasmid was constructed by site-specific mutation of miR-23a binding site inPPP2R5E3’UTR. In Tca8113 cellsPPP2R5E3’UTR mut plasmid was transfected with pcDNA3, pcDNA3-pri-miR-23a, ASO-23a and the control group ASO-NC, respectively. The results showed that the fluorescence expression intensity did not change significantly. It was indicated that miR-23a could not bind toPPP2R5E3’UTR after the "seed sequence" mutation ofPPP2R5E3’UTR. It could not inhibit the expression of EGFP protein, and the fluorescence intensity did not change (Fig.4B).
The expression level of miR-23a was changed in Tca8113 cells, and the expression level ofPPP2R5EmRNA was detected by real-time quantitative PCR. The results showed that after overexpression of miR-23a, the expression level ofPPP2R5EmRNA was significantly down-regulated. However, after miR-23a was blocked, the expression level ofPPP2R5EmRNA was significantly increased (Fig.4C). The difference was statistically significant. In conclusion, miR-23a can bind toPPP2R5E3’UTR and reduce the expression level of downstreamPPP2R5EmRNA. ThereforePPP2R5Eis the direct target gene of miR-23a.

Note: A. Effect of miR-23a on wild-typePPP2R5E3’UTR luciferase; B. Effect of miR-23a on luciferase of mutantPPP2R5E3’UTR; C. Effect of miR-23a onPPP2R5EmRNA level;*whenP<0.05,**P<0.01, the statistical difference was significant.
Fig.4PPP2R5E, direct target gene of miR-23a, negatively regulated by miR-23a
3.4DifferentialexpressionofPPP2R5EintonguesquamouscellcarcinomatissuesIn 10 pairs of tongue squamous cell carcinoma tissues, real-time quantitative PCR was used to detect the expression level ofPPP2R5EmRNA. The results showed that the average expression level ofPPP2R5EmRNA in tongue squamous cell carcinoma tissues was 0.582, while the expression level in corresponding para-cancer tissues was 1.119. The expression ofPPP2R5Ein tongue squamous cell carcinoma tissues decreased to 0.582,P=0.027, indicating a statistically significant difference. The expression ofPPP2R5Ein tongue squamous cell carcinoma tissues was abnormally decreased (Fig.5).

Note:**whenP<0.01, the statistical difference was significant.
Fig.5 Expression level ofPPP2R5Ein tongue squamous cell carcinoma
PPP2R5Einhibited the proliferation of tongue squamous cell. We transfected pcDNA3/ HA-PPP2R5Eand pSilencer/ sh-PPP2R5Ein the tongue squamous cell line, pcDNA3 and pSilencer respectively. Changed the expression ofPPP2R5EmRNA in tongue squamous cell carcinoma, and the effects ofPPP2R5EmRNA on the proliferation of tongue squamous cell carcinoma were detected by MTT assay, colony formation assay and growth curve assay. MTT results showed that after the overexpression ofPPP2R5E, the absorbance value of cells was significantly lower than that of the control group. Cell activity was weakened. However, after interfering with the expression ofPPP2R5E, the absorbance value of cells was significantly higher than that of the control group. Cell activity was enhanced. The difference was statistically significant (Fig.4A). The results of the colony formation experiment showed that after interfering with the expression ofPPP2R5E, the number of cell colonies was significantly higher than that of the control group. The ability of cell colony formation was enhanced. However, after the overexpression ofPPP2R5E, the number of cell colonies was significantly lower than that of the control group. The ability of cell colony formation was weakened, and the difference was statistically significant (Fig.6B and 6D). The experimental results of growth curve showed that after the overexpression ofPPP2R5E, the number of cells decreased significantly compared with the control group. After interfering with the expression ofPPP2R5E, the number of cells increased significantly compared with the control group, with a statistically significant difference (Fig.6C). In conclusion, overexpression ofPPP2R5Ecan inhibit the proliferation of tongue squamous cell, while interference in the expression ofPPP2R5Ecan promote the proliferation of tongue squamous cell.PPP2R5Egene can inhibit the proliferation of tongue squamous cell carcinoma and plays a role of tumor suppressor gene in tongue squamous cell carcinoma.

Note: A. Effect ofPPP2R5Eon the activity of Tca8113 cells; B. Effect ofPPP2R5Eon the colony formation ability of Tca8113 and CAL-27 cells; C. Effect ofPPP2R5Eon the growth ability of Tca8113 and CAL-27 cells; D. Crystal violet staining image of the colony formation of Tca8113 cells;*whenP<0.05,**P<0.01, the statistical difference was significant.
Fig.6PPP2R5Einhibited the proliferation of tongue squamous cell carcinoma cells
4 Discussions
miR-23a expression was found to be abnormally elevated in a variety of tumors, which promoted the occurrence and development of tumors[1-5]. miR-23a is highly expressed in pancreatic cancer and promotes the proliferation and metastasis of pancreatic cancer cells[10]. miR-23a expression is elevated in colon cancer tissues and cells and promotes colon cancer cell proliferation by targeting PDK4[11]. miR-23a is abnormally expressed in gastric cancer tissues and cells, and promotes proliferation and metastasis of gastric cancer cells[12]. Meanwhile, the expression of miR-23a in tongue squamous cell carcinoma tissues was also significantly increased in this study. Therefore, it is inferred that miR-23a may also play a role as an oncogene in tongue squamous cell carcinoma. This study found that overexpression of miR-23a can significantly promote the proliferation of tongue squamous cell carcinoma, while blocking the expression of miR-23a can inhibit its proliferation, which is consistent with the results obtained in the literature. miR-23a plays an oncogene role in tongue squamous cell carcinoma, and further supplements the role of miR-23a in tumor.
MiRNAs affect a series of physiological and pathological processes by regulating the expression level of their downstream target genes. miR-23a can directly target and regulate important functional genes such as NID2[13], ESRP1[14]and XIAP[15]to regulate the proliferation, apoptosis, invasion and migration of tumor cells. The expression ofPPP2R5Ein gastric cancer tissues and cells is decreased[16].
This study found that the expression ofPPP2R5Ein tongue squamous cell carcinoma tissues was significantly higher than the corresponding adjacent tissues, which was contrary to the expression trend of miR-23a. Moreover, bioinformatics predicted that miR-23a could indeed bind toPPP2R5E3’UTR. Therefore, we speculated thatPPP2R5Emight be a potential target gene of miR-23a in tongue squamous cell carcinoma tissues. The experimental results showed that miR-23a could inhibit the luciferase activity of the wild-typePPP2R5E3’UTR, but the luciferase activity of the mutatedPPP2R5E3’UTR mut was not affected, and overexpression of miR-23a could significantly inhibit the expression ofPPP2R5EmRNA. Therefore,PPP2R5Eis indeed the direct target gene of miR-23a in tongue squamous cell carcinoma. It is intended to further demonstrate thatPPP2R5Eis the functional target gene of miR-23a. In this study,PPP2R5Eoverexpression vector was used to restore the downregulation ofPPP2R5Eexpression in Tca8113 cells caused by miR-23a. The results showed thatPPP2R5Eoverexpression could restore the effect of miR-23a overexpression on Tca8113 cell proliferation. This indicates that the effect of miR-23a on tongue squamous cell carcinoma is achieved by down-regulating the expression level ofPPP2R5E.
In conclusion, this study shows that miR-23a can promote the proliferation of tongue squamous cell carcinoma by directly targeting and down-regulating the expression level ofPPP2R5E. This conclusion is expected to provide a new theoretical basis and potential targets for the early diagnosis and treatment of tongue squamous cell carcinoma.
- Medicinal Plant的其它文章
- Experimental Study on Analgesia and Hemostasis of Paris polyphylla, Scutellaria baicalensis and Their Compatibility
- Effects of Different siRNA Transfection Methods on Expression of ARID4B in HepG2 Cells
- Lignans from Seeds of Herpetospermum caudigerum Wall. Protect Against Alcoholic Liver Injury via TGF-β/Smads Pathway in Rats
- Study on the Protective Effect and Mechanism of Tibetan Medicine Ershiwuwei Songshi Pill on Acute Alcoholic Liver Injury in Rats
- Comparative Study on the Effectiveness of Two Nickel-titanium Instruments in Removing Gutta-percha
- Comparative Study on the Effect of Mongolian Medicine Euphorbia pekinensis and Pharbitidis Semen (Black)

