ldentification of key genes and pathways in gastric signet ring cell carcinoma based on transcriptome analysis
Zi-Tong Zhao,Yang Li,Hong-Yu Yuan,Fu-Hai Ma,Yong-Mei Song,Yan-Tao Tian
Zi-Tong Zhao,Hong-Yu Yuan,Yong-Mei Song,State Key Laboratory of Molecular Oncology,National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital,Chinese Academy of Medical Sciences and Peking Union Medical College,Beijing 100021,China
Yang Li,Fu-Hai Ma,Yan-Tao Tian,Department of Pancreatic and Gastric Surgery,National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital,Chinese Academy of Medical Sciences and Peking Union Medical College,Beijing 100021,China
Abstract
Key words:Signet ring cell;Transcriptome sequencing;Gastric carcinoma;Bioinformatical analysis;Pathway;Gene
INTRODUCTION
Gastric cancer(GC)is the 5th most common carcinoma in the world,with a morbidity of over 1000000 new patients in 2018,and is the 3rdprincipal cause of death by cancer worldwide[1].Recently,different pathohistological standard methods have been established for GC.The massive majority of GCs are adenocarcinomas,which can be further divided into intestinal and diffuse types by Lauren classification[2,3].An alternative classification,proposed by the World Health Organization,divides gastric cancer into tubular,mucinous,papillary,and poorly cohesive carcinomas[4].Diffuse type is generated by poorly cohesive cells without gland formation and is frequently referred to as gastric signet ring cell carcinoma(GSRCC).GSRCC is a histologic diagnosis based on microscopic characteristics as the presence of signet ring cells in over 50% of the cancer cells by the World Health Organization[5].These classification methods have little clinical value.An important priority of proper classifiers should be studied to guide therapeutics.Considering the epidemiology of GSRCC,it happens more frequently in females than non-GSRCC,and occurs among patients aged between 55 to 61 years,7 years ahead of those with non-GSRCC[6,7].Since the beginning of treatment to eradicateHelicobacter pylori,the incidence of gastric adenocarcinoma has decreased.Conversely,the incidence of GSRCC is rising.The GSRCC is found in 8% to 30% of GCs[8].
The prognosis of GSRCC in entire mentioned reports has been described as uninferior than the other gastric cancer subtypes in the early stage[9,10].Contrariwise,the prognosis of GSRCC in advanced stages is quite poor,and most studies showed a significantly worse 5-year survival rate in patients with GSRCC than in those with non-GSRCC.Furthermore,Lemoineet al[11]showed that patients with advanced GSRCC appeared to benefit less from chemotherapy.Pathological response rate to chemotherapy was significantly lower in GSRCC patients(5.3%vs28.1%,P= 0.0004).Another German analysis confirmed that a worse histopathological response(16.3%vs28.9%,P< 0.001)in patients was affected by preoperative chemotherapy.An ongoing PRODIGE19 trial(NCT01717924)tends to randomize patients with resectable GCs with signet ring cells receiving perioperative chemotherapy with ECFvsan upfront surgery followed by adjuvant chemotherapy with the same regimen[12].However,the results are still uncertain.The effects of chemotherapy on GSRCC are still in controversy.According to its potential drug resistance,current studies have mainly focused on the pathogenesis study of GSRCC with regard to some diseaserelated genes.
To find out the molecular mechanisms for tumorigenesis and heterogeneity of GSRCC at the molecular level,large studies have been taken to characterize the comprehensive genomic features through transcriptome sequencing,and multiple driver alterations have been recognized.The exploration of the molecular mechanism of GSRCC is important to improve the recognition of GSRCC and find the effective therapeutics to raise the survival rate of patients.To our best knowledge,systematic and comprehensive analyses of mRNAs have not yet been conducted for GSRCC.In this study,transcriptome sequencing and comprehensive analysis were performed to identify key mRNAs and signaling pathways in GSRCC,with an aim to provide new insights into the treatment and feature of GSRCC.
MATERIALS AND METHODS
Gastric signet ring cell cancer tissue samples
A total of 60 patients who underwent surgery for GSRCC at National Cancer Center,Chinese Academy of Medical Sciences and Peking Union Medical College,Beijing,China were selected for inclusion in the study.All clinicopathological data including age,sex,histological type,and lymph node metastasis were obtained from the database.Four of samples were sent to conduct transcriptome sequencing,and 56 of samples were taken for validation.
Transcriptome sequencing
The transcriptome sequencing was used for mRNA expression profiling(CapitalBio).
KEGG and PANTHER pathway analyses
The functional enrichment of the differential genes was assessed based on the KEGG and PANTHER pathway annotations.
Protein-protein interaction network construction
To evaluate the interactive relationships among the differential genes,we mapped the differential genes to the STRING database(http://string-db.org).
RNA extraction,RT-PCR,and quantitative real-time PCR
Total RNA was extracted from frozen fresh tissues with the RNAExpress Total RNA Kit(New Cell &Molecular Biotech Co.,Ltd).cDNA was synthesized with the First-Strand cDNA Synthesis Kit(Applied Biological Materials).Quantitative real-time polymerase chain reaction(qPCR)was performed with EvaGreen 2X qPCR MasterMix(Applied Biological Materials).And the primers used are listed in Table 1.The mRNA sequence ofMAGEA2/3/6is too similar to design primers for separately verifying and validating these three genes,so we designed three pairs of primers forMAGEA2/3/6to validate the expression of these genes together.
Statistical analysis
The data are presented as the mean ± SE.The differences were assessed by the twotailed Student'st-test for group comparisons using the GraphPad Prism 5 software package.Differences were considered significant whenPvalues were less than 0.05.
RESULTS
Clinical characteristics of SRCC samples
In this study,four primary GC patients underwent transcriptome sequencing,consisting of two patients with GSRC and two with adenocarcinoma.All of them are male with a median age of 49 years old and poorly differentiated carcinomas.The positive lymph node rate was separately 26/70 and 5/26 in the GSRC group,and 0/45 and 27/34 in the adenocarcinoma group.The two GSRC samples pathologically consisted of over 90% of signet ring cells in the tumor.
Further,56 samples were chosen for validation.The analysis of the 60 samples showed that there were not significant differences between the GSRCC group and adenocarcinoma group in baseline characteristics such as age,gender,TNM stages,lymphovascular invasion,or nerve invasion,although there were significant differences in Lauren type(P< 0.001)and histology differentiation(P= 0.038,Table 2).
Identification of differential genes between GSRCC and non-GSRCC
Transcriptome sequencing was used to profile the mRNA expression in four patientswith GC(two with GSRCC and two with non-GSRCC)who underwent surgical treatment(Figure 1).There were 1162 differential genes(using a 2-fold cutoff,P<0.05),which included 682 upregulated and 480 downregulated genes in GSRCC compared with non-GSRCC.The top 10 up-regulated genes wereMAGEA2,MAGEA6,MAGEA3,MAGEA2B,ISY1-RAB43,MAGEA9,MAGEA9B,MAGEA12,KRT16P6,andNEUROD2.The top 10 down-regulated genes wereREG1B,REG1A,SPINK4,GIF,ATOH1,IGHV4-4,GUCA2A,DMBT1,SOSTDC1,andATP4B.The fold changes andPvalues of these differential genes are listed in Table 3.

Table 1 Primers used for quantitative real-time polymerase chain reaction
KEGG and PANTHER pathway enrichment analyses
KEGG pathway enrichment analyses were performed on the differential genes.The enriched KEGG pathways(Figure 2A)for the differential genes included Rap1 signaling pathway,epidermal growth factor receptor tyrosine kinase inhibitor resistance,antigen processing and presentation,HIF-1 signaling pathway,calcium signaling pathway,PI3K-Akt signaling pathway,inflammatory mediator regulation of TRP channels,phagosome,Ras signaling pathway,gastric acid secretion,metabolic pathways,chemokine signaling pathway,peroxisome proliferator-activated receptors signaling pathway,cytokine-cytokine receptor interaction,and endocytosis.For the differential genes,the enriched PANTHER pathways(Figure 2B)included angiogenesis,apoptosis signaling pathway,epidermal growth factor receptor signaling pathway,Notch signaling pathway,endothelin signaling pathway,inflammation mediated by chemokine and cytokine signaling pathway,vascular endothelial growth factor signaling pathway,glycolysis,fibroblast growth factor signaling pathway,integrin signaling pathway,insulin/insulin like growth factor pathway-protein kinase B signaling cascade,fructose galactose metabolism,CCKR signaling map,interleukin signaling pathway,and Toll receptor signaling pathway.Both KEGG and PANTHER pathways mainly focused on immune response pathways,metabolic pathways,and metastasis-associated pathways.
Construction and analysis of protein-protein interaction network
As shown in Figure 3,a regulatory network of differential genes(a 20-fold cutoff,P<0.05)was constructed.There were 141 nodes and 171 edges among these 310 genes.The top 10 hub genes wereMAGEA2,MAGEA2B,MAGEA3,MAGEA4,MAGEA6,MUC13,GUCA2A,FFAR4,REG1A,andREG1B.
Validated hub genes in GSRCC with TCGA database and tissue samples
According to the difference of gene expression and pathway and the protein-protein interaction result,we choseMAGEA2,MAGEA3,MAGEA4,MAGEA6,REG1A,andREG1Bfor validation.The data of TCGA and GTEx(GEPIA,http://gepia.cancerpku.cn/index.html)revealed that the expression ofMAGEA2,MAGEA3,MAGEA4,andMAGEA6was upregulated,but the expression ofREG1Bwas downregulated,in GC compared to the normal gastric tissue,and there was no difference inREG1Abetween GC and normal gastric tissues(Figure 4A).The correlation analysis of geneexpression and clinical data revealed a negative correlation betweenMAGEA2,MAGEA3,MAGEA4,andMAGEA6,but a positive correlation betweenREG1AandREG1B,and poor survival in patients with GC(GEPIA,http://gepia.cancerpku.cn/index.html,Figure 4B).

Table 2 Characteristics of 30 gastric signet ring cell carcinoma and 30 adenocarcinoma patients,n(%)
To investigate the clinical value of these five genes in GC,qPCR was used tomeasure their expression in 27 GSRCC and 29 non-GSRCC samples.The qPCR results revealed that the expression ofMAGEA2/3/6andMAGEA4was significantly higher,but the expression ofREG1Bwas much lower,in patients with GSRCC than in patients with non-GSRCC(Figure 4C).
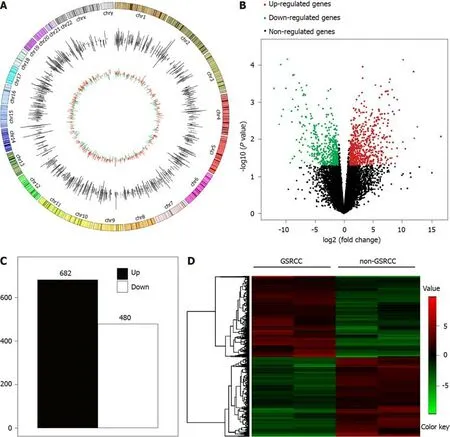
Figure 1 Summary of transcriptome sequencing results of mRNA expression in gastric carcinoma.
DISCUSSION
Advanced GSRCC is commonly considered to be more invasive,has a greater probability of lymph node metastasis than other gastric cancer types,has a worse prognosis[13,14],and is less sensitive to chemotherapy than non-SRCC[15-17].Chemotherapy remains the major treatment for advanced GC.Several studies report that chemoresistance exists in GSRCC with a poor prognosis[8].It is still unclear whether GSRCC patients can benefit from a detailed therapy.
As we know,vascular endothelial growth factor and HER-2 have been agreed as targets for molecular pathways in GC.However,SRCC cannot benefit from targeted therapy for a low expression level of HER-2[18].Some studies have mentioned that elevated expression ofPKM2andE-cadherin,and declined expression ofTMEM207in GSRCC are associated with its biological behavior of invasion and metastasis[19].
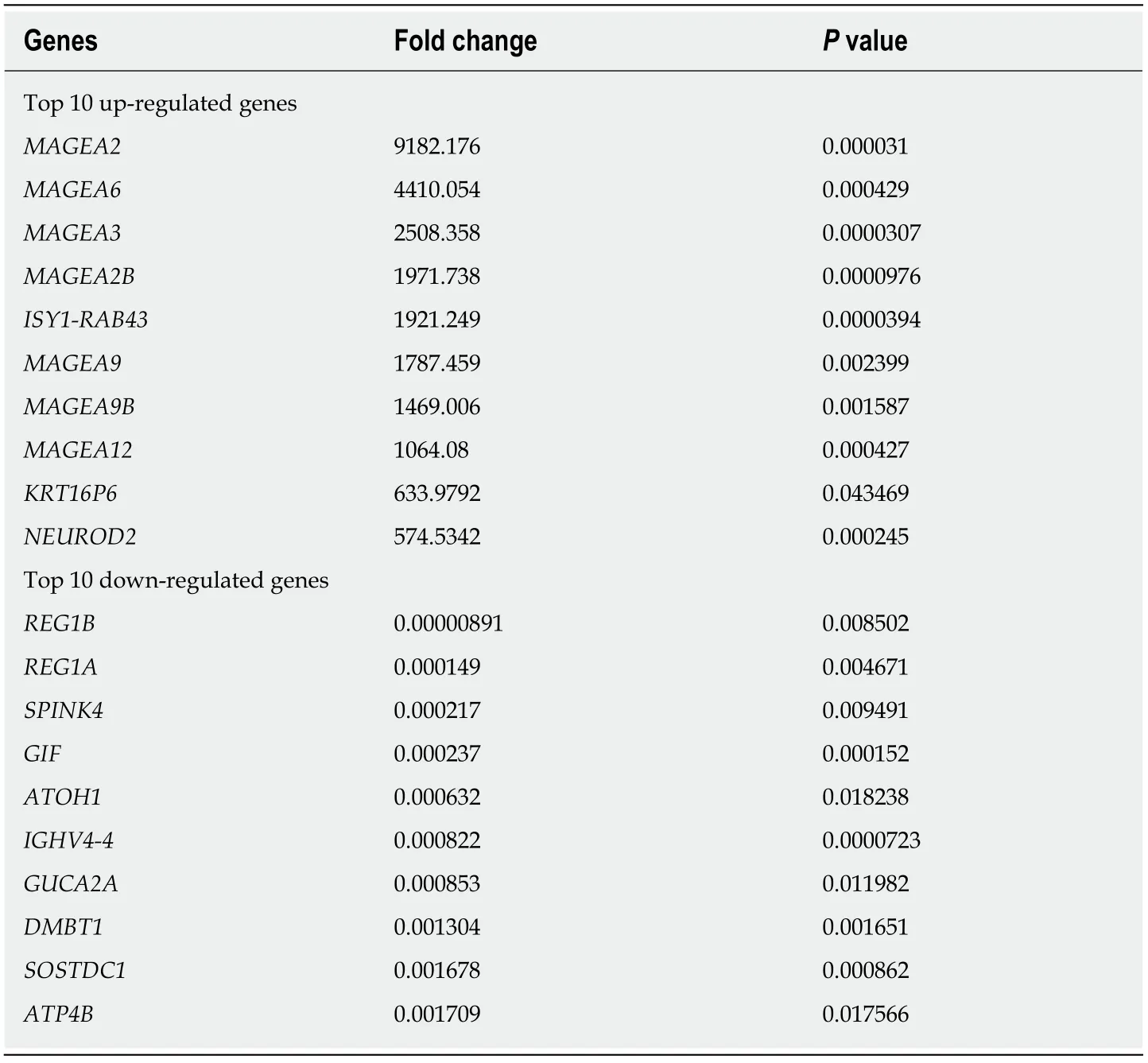
Table 3 Fold change of differential genes
Systematic analyses of mRNAs in GSRCC were rare.Chenet al[19]reported that hsamiR-665 and hsa-miR-95 were found in GSRCC compared to intestinal GC,and concluded that these two mRNAs were downregulated in GSRCC but upregulated in intestinal GC,and the relatively differential expression of the miRNAs negatively regulating their target genes could be intently associated with high invasion,metastasis,and chemoresistance of GSRCC[19].Yaoet al[20]have described the genome features of GSRCC and they found the importance of highly frequentCLDN18-ARHGAP26/6fusions in chemotherapy response in GSRCC.Kakiuchiet al[21]reported thatRHOAmutations happen specifically in diffused GCs,the majority of which were pathologically characterized by the presence of poorly differentiated components,with no available molecularly targeted drugs for this poor-prognosis subtype of GC.However,that study was not specific for GSRCC.Due to a very limited number of studies in this area,this present study aimed to identify specifically expressed genes and identify biological features in GSRCC.
By pathway analyses,we confirmed frequent alterations across various pathways in GSRCC,including immunological response pathways,metabolic pathways,and metastasis-associated pathways.Further protein-protein interaction network analyses identified the hub genes in GSRCC.Of the hub genes,MAGE-Afamily andREG1Bare immune-related genes.REG1Bhas been reported to be able to upregulate the expression of interleukin(IL)-6 mRNA and protein.
MAGE-Afamily are the best characterized cancer-testis antigens(CTA)family members,which are expressed mainly,but not exclusively,in germ cells.MAGE-Afamily are infrequently expressed in the normal human placenta,but they are widely expressed in numerous human cancers.Alviet al[22]concluded that the MSI+/CIMP+/BRAF V600E+/CD3+/PDL1+hypermethylated genotype is an ideal candidate for immune checkpoint inhibitor therapy.In addition,one-fourth of SRCC cases can potentially be targeted by KIT inhibitors.As similar as we found,MAGE-A proteins are highly immunogenic and are considered as potential targets for immunotherapy.
In conclusion,we have identified the potential key genes and pathways in GSRCC.These hub genes and pathways could be diagnostic markers and therapeutic targets for GSRCC.MAGE-Afamily as aCTAfamily member may be the potential targets for GSRCC.More research should be conducted for exploration of the mechanisms involved.

Figure 2 KEGG and PANTHER pathway analysis of differential genes.
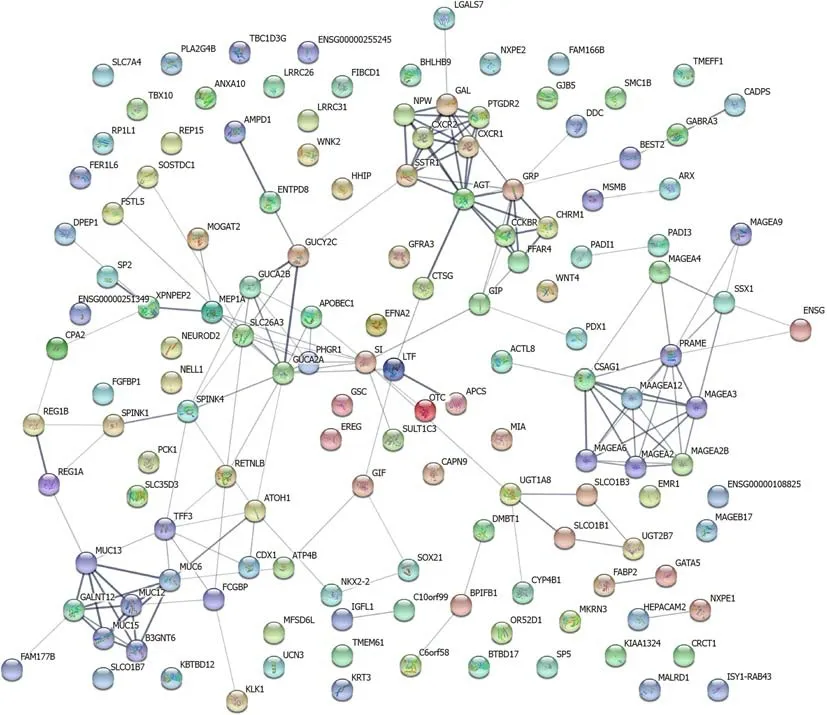
Figure 3 Protein-protein interaction network of differential genes.
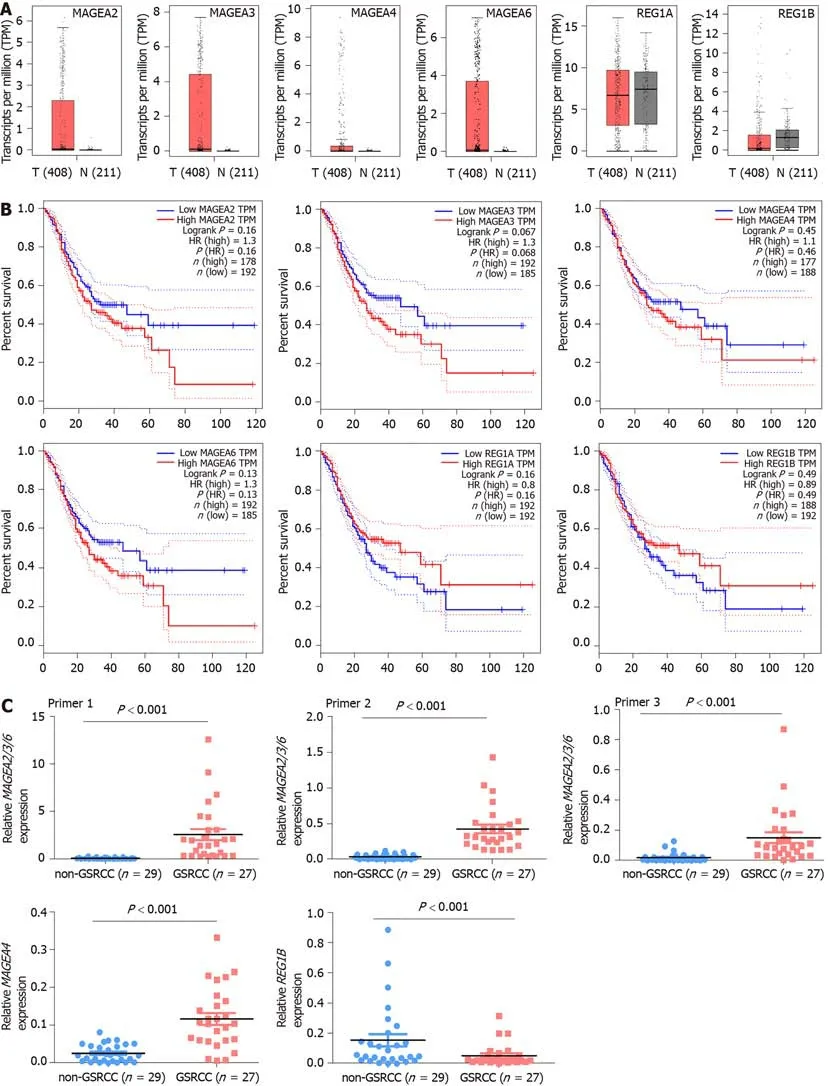
Figure 4 Expression of MAGEA2/3/6,MAGEA4,and REG1B in gastric signet ring cell carcinoma.
ARTICLE HIGHLIGHTS
Research background
Gastric signet ring cell carcinoma(GSRCC)has the features of high invasiveness,rapid progression,and resistance to chemotherapy.However,systematic analyses of mRNAs have not yet been performed.
Research motivation
The exploration of the molecular mechanism of GSRCC is important to improve the recognition of GSRCC and find the effective therapeutics to raise the survival rate of patients.
Research objectives
Transcriptome sequencing and comprehensive analysis were performed to identify key mRNAs and signaling pathways in GSRCC.
Research methods
A transcriptome analysis of two GSRCC and two non-GSRCC samples was performed.Differentially expressed mRNAs and pathways were identified.The interactive relationships among the differential genes were mapped with the STRING database.
Research results
The enriched KEGG and PANTHER pathways for the differential genes included immune response pathways,metabolic pathways,and metastasis-associated pathways.MAGEA2,MAGEA2B,MAGEA3,MAGEA4,MAGEA6,MUC13,GUCA2A,FFAR4,REG1A,andREG1Bwere identified as hub genes in the protein-protein interaction network.The expression levels ofMAGEA2,MAGEA3,MAGEA4,MAGEA6,andREG1Bshowed potential clinical value.
Research conclusions
The potential key genes and pathways of GSRCC have been identified.These hub genes and pathways could be diagnostic markers and therapeutic targets for GSRCC.
Research perspectives
MAGE-Afamily as aCTAfamily member may be the potential targets for GSRCC.More research should be conducted for exploration of the mechanisms involved.
World Journal of Clinical Cases2020年4期
- World Journal of Clinical Cases的其它文章
- Must pilots permanently quit flying career after treatment for colorectal cancer? - Medical waiver for Air Force pilots with colorectal cancer:Three case reports
- Prevalence and associated factors of suicide among hospitalized schizophrenic patients
- Lymphoepithelioma-like carcinoma of the upper urinary tract:A systematic review of case reports
- Extrapleural solitary fibrous tumor of the thyroid gland:A case report and review of literature
- Delayed right coronary ostial obstruction after J-valve deployment in transcatheter aortic valve implantation:A case report
- Diverticulum of the buccal mucosa:A case report
