Expression profiles of long noncoding RNAs in retinopathy of prematurity
Yue Wang, Xue Wang, Yuan Ma, Yue-Xia Wang, Yu Di
Department of Ophthalmology, Shengjing Hospital of China Medical University, Shenyang, Liaoning Province, China
Funding: The work was supported by the National Natural Science Foundation of China, No. 81600747 (to YD) and Startup Foundation for Doctors of Liaoning Province, China, No. 201501020 (to YD).
Abstract Long noncoding RNA (lncRNA) regulates the proliferation and migration of human retinal endothelial cells, as well as retinal neovascularization in diabetic retinopathy. Based on similarities between the pathogenesis of retinopathy of prematurity (ROP) and diabetic retinopathy, lncRNA may also play a role in ROP. Seven-day-old mice were administered 75 ± 2% oxygen for 5 days and normoxic air for another 5 days to establish a ROP model. Expression of lncRNA and mRNA in the retinal tissue of mice was detected by high-throughput sequencing technology, and biological functions of the resulted differentially expressed RNAs were evaluated by Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses. The results showed that compared with the control group, 57 lncRNAs were differentially expressed, including 43 upregulated and 14 downregulated, in the retinal tissue of ROP mice. Compared with control mice, 42 mRNAs were differentially expressed in the retinal tissue of ROP mice, including 24 upregulated and 18 downregulated mRNAs. Differentially expressed genes were involved in ocular development and related metabolic pathways. The differentially expressed lncRNAs may regulate ROP in mice via microRNAs and multiple signaling pathways. Our results revealed that these differentially expressed lncRNAs may be therapeutic targets for ROP treatment. This study was approved by the Medical Ethics Committee of Shengjing Hospital of China Medical University on February 25, 2016 (approval No. 2016PS074K).
Key Words: bioinformatics; gene therapy; long noncoding RNA; microglial; neurovascular disease; optic neuropathy; retinal development; retinal neovascularization; retinopathy of prematurity; signaling pathways
Introduction
Long noncoding RNAs (lncRNAs) are involved in multiple diseases, including RNV. Increasing evidence indicates that lncRNAs are related to the development of tumors and neovascular diseases (Xu et al., 2014; Chen et al., 2017a; Long et al., 2018). In addition, lncRNAs can regulate microglial polarization and inflammation (Wang et al., 2019; Yang et al., 2019; Zhang et al., 2019c). A few studies have shown that lncRNAs exhibit tissue-specific expression in the eye (Young et al., 2005; Qiu et al., 2016). lncRNA has the function of competing with endogenous RNA or microRNA to regulate the proliferation and migration of human retinal endothelial cells, as well as RNV in diabetic retinopathy (Zhu et al., 2017;Fan et al., 2019). Based on its similarities with the pathogenesis of diabetic retinopathy, RNV in ROP may be explained by similar molecular mechanisms. As such, this study aimed to decipher the role of differential expressed lncRNA in oxygen-induced retinopathy (OIR) mice and explore how abnormal lncRNA expression affects the hypoxic process of ROP. We hope that our research on differentially expressed lncRNA can provide a theoretical basis for the identification of new clinical treatments for ROP.
Materials and Methods
Animals and OIR model
Seven-day-old C57BL/6J mice (n= 100; specific pathogen-free level) were purchased from Shenyang Changsheng Biological Technology Co, Ltd. [Shenyang, China; license No. SCXK(Liao)2015-0001]. Mice were bred at the specific pathogen-free Laboratory Animal Center of Shengjing Hospital (Benxi, China) at 23 ± 2°C with 12-hour light-dark cycles. The Medical Ethics Committee of Shengjing Hospital of China Medical University approved the animal experiments on February 25, 2016 (approval No. 2016PS074K).
Animals were randomly divided into two groups with 50 mice in each group. Mice in the control group were kept in normoxic air with their mothers until the age of 17 days.In the experimental group, OIR was induced according to Smith’s method (Smith et al., 1994). Briefly, 7-day-old mice in the OIR group and their mother were placed for 5 days in a hyperoxia chamber provided by the Laboratory Animal Center of Shengjing Hospital, which was composed of glass feeding box, oxygen detector (8F-3AW; Yuwell, Suzhou, China), and oxygen generator (OX-100A; Aipuins, Hangzhou,China) (75 ± 2%). Subsequently, these mice were maintained in normoxic air for another 5 days with their mothers (Smith et al., 1994; Heiduschka et al., 2019). Mice in both groups were sacrificed to collect retinas for total RNA extraction and pathology after deep anesthesia with ketamine at postnatal day 17 (Park et al., 2009).
Retina isolation and fulorescein isothiocyanate-dextran angiography
Seventeen-day-old mice were anesthetized with ketamine,and 500 μL of fluorescein isothiocyanate-dextran (2 × 103kDa; 50 mg/mL; Sigma-Aldrich, St. Louis, MO, USA) was injected into the left ventricle; 2 minutes later, mice were sacrificed by cervical dislocation. Next, the eyes were enucleated and fixed with 4% paraformaldehyde for 2 hours at 4°C.After removing the cornea, lens, and vitreous, retinas were peeled from the eyeball and cut into four quadrants with an ophthalmic scalpel under a stereomicroscope (SMZ18;Nikon, Tokyo, Japan). Images were captured under a fluorescence microscope (Eclipse NI, Nikon) before applying SlowFade anti-fade reagent (Sigma-Aldrich) to retinas (Banin et al., 2006; Geng et al., 2018). The lasso tool of Adobe Photoshop CS6 (Adobe, San Francisco, CA, USA) was used to manually select areas of non-perfusion and neovascular retinal tufts. ImageJ 1.37C (National Institutes of Health,Bethesda, MD, USA) was used to automatically calculate the percentage areas selected by Photoshop CS6 (Zhang et al.,2000; Shi et al., 2013). All manual work was completed by three reviewers.
Hematoxylin-eosin staining
Ten randomly selected mice from each group were fixed for 24 hours. The eyeball was cut along the sagittal plane parallel to the optic nerve and embedded in paraffin. Next, the eyeball was cooled to room temperature to prepare a 3.5-μmthick continuous slice, and 10 pieces from each eyeball were selected for hema-toxylin-eosin staining. Images were captured under a fluorescence microscope, and the average number of all neovascular nuclei present in the vitreous side were counted for each section by three independent reviewers in a double-blind manner (Zhang et al., 2017).
RNA extraction and preparation
Total RNA from each retinal tissue sample was extracted using TRIzol reagent (Takara, Kusatsu, Japan) and quantified by Nanodrop ND-2000 (Thermo Scientific, Waltham, MA,USA). RNA integrity was detected by an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). We selected samples with optical density260/280= 1.8-2.0 for subsequent experimental studies. Chip processing was subsequently performed according to Agilent’s standards, including sample labeling, microarray hybridization, and washing.
Array analysis
Analysis of lncRNA and mRNA array data was achieved by Feature extraction (version 10.7.1.1; Agilent Technologies).Normalization of raw data by quantile and quality control was performed using GeneSpring (version 13.1; Agilent Technologies). The threshold set for screening of differentially expressed genes was fold change ≥ 2 andP< 0.05. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were used to determine the effect of differentially expressed genes. Numbers and significance of differentially expressed genes included in each GO entry were counted (http://geneontology.org/). The KEGG database was used to analyze differentially expressed genes and calculate the significance of differences in gene enrichment for each pathway entry (http://www.genome.jp/kegg/).
But he did remember, and a few years later he told his younger brother, Dave, about the all-leather, NFL regulation, 1963 Chicago Bears-inscribed football that was special and kept somewhere in the garage
Hierarchical clustering was used to verify differences in gene expression patterns between samples and calculate the distance between different samples to generate a distance matrix. Finally, String was used to constructing a protein interaction network using vascular endothelial growth factor(VEGF) and other dif-ferentially expressed mRNAs (Qu et al., 2017; Liu et al., 2018).
Quantitative real-time PCR
Total RNA extraction and quality control were performed as described above. Ten lncRNAs in retinal tissues were measured by quantitative real-time PCR (qRT-PCR) using SYBR®Green assays (Takara) with the primers listed in Table 1 (Sangon Biotech Co., Ltd., Shanghai, China). β-Actin was applied as an internal control. For quantitative analysis of differential expression, data were processed by the 2-∆∆Ctmethod, and expression of each lncRNA is expressed as the relative fold change to β-actin.
Statistical analysis
All statistical data are expressed as the mean ± standard error of mean (SEM). Differences were analyzed by Mann-WhitneyUtest using SPSS 17.0 (Release 17.0; SPSS Inc., Chicago, IL,USA).P-values < 0.05 were considered statistically significant.
Results
Retinal neovascularization in OIR mice
Retinal vascular structures were examined by fluorescein is othiocyanate-dextran angiography. Expectedly, the retinal vasculature of control group mice was intact, with the main blood vessels and vessel branches arranged regularly (Figure 1A and C). In the OIR group, the retina of mice showed severe areas without perfusion, as well as neovascularization and distorted blood vessels with irregular dilation (Figure 1B and D). Percentage areas of non-perfusion and neovascularization in the OIR group were higher compared with the control group (Z= -2.882,P= 0.004;Z= -2.739,P= 0.006; Figure 1E and F).
Neovascular nuclei of the retina in OIR mice
We quantified retinal neovascular nuclei from 10 non-continuous paraffin sections of the retina, primarily by calculating the number of neovascular nuclei that broke through the inner limiting membrane of the retina. In the control group,retinal neovascular nuclei were rarely observed, with an average number of 2.70 ± 0.58 nuclei per cross-section (Figure 2A). In the OIR group, an average of 20.70 ± 2.53 nuclei was observed for each retina cross-section (Figure 2B), which is significantly larger than observed in the control group (Z=-3.686,P< 0.01; Figure 2C).

Table 1 Primers used for quantitative real-time polymerase chain reaction
Differentially expressed lncRNAs and mRNAs in the retinal tissue of OIR mice
In total, 62,028 distinct lncRNAs and 33,419 mRNAs were detected in the retinal tissue of experimental mice using high-throughput sequencing technology. Compared with the control group, 57 lncRNAs and 42 mRNAs were differentially expressed, 43 lncRNAs and 24 mRNAs were significantly upregulated, and 14 lncRNAs and 18 mRNAs were significantly downregulated in the OIR group (fold change ≥ 2,P<0.05). A volcano plot of differentially expressed genes reflects their trends (Figure 3A), while scatter plots indicate better data normalization (Figure 3B). Most genes were concentrated on chromosomes 4, 7, 11, and 14 (Figure 3C). Cluster analysis showed that lncRNA were differentially expressed in both control mice and the OIR group (Figure 3D).
We validated the top five upregulated and top five downregulated genes in the two sets of retina samples by qRT-PCR;information about these genes is listed in Table 1. For nine of the ten genes, qRT-PCR results were consistent with the sequencing results, not including AC142098.2 (Figure 3E).
Gene function analysis of differentially expressed genes in the retinal tissue of OIR mice
To analyze gene functions, GO, KEGG, and pathway cluster analyses were performed. Differentially ex-pressed genes were involved in more than 20 pathways and many processes, mainly associated with com-plement activation, the lectin pathway, nuclear factor-κB (NF-κB)-inducing kinase/NFκB signaling pathway, G protein-coupled receptor signaling pathway, and canonical Wnt signaling pathway, suggesting that lncRNAs regulate oxygen-induced retinal neovascularization by various signaling pathways (Figure 4).
Protein interaction network of RNV
Based on differentially expressed mRNAs, VEGF mRNA was selected as the main mRNA in the protein interaction network because most of the remaining differentially expressed mRNAs were associated with VEGF; thus, differentially expressed mRNAs may be related to RNV (Figure 5).
Discussion
Normal development of the retina provides nutrition for photoreceptor cells and has regenerative and repair functions(Ishikawa et al., 2015; Morken et al., 2019). ROP is a serious neurovascular disease. However, currently used drugs, which are administered be intravitreal injection, only control neovascularization and have no effect on protecting nerve function (Kang et al., 2019; Tsang et al., 2019). Previous studies have shown that nerve growth factor, progenitor cells, and bone marrow mesenchymal stem cells can contribute to the recovery of OIR in mice (Li Calzi et al., 2019; Ma et al.,2019; Troullinaki et al., 2019). Moreover, progenitor cells in combination with other treatments can normalize retinal vascular development (Li Calzi et al., 2019), and are associated with lncRNAs (Yao et al., 2019; You and You, 2019). An increasing number of studies have shown that lncRNAs exert essential functions in cancer cells, particularly during proliferation, differentiation, and malignant transformation (Shen et al., 2016; Zhang et al., 2019a). Furthermore, lncRNAs are involved in choroidal neovascularization, proliferative vitreoretinopathy, and diabetic retinopathy (Yu et al., 2019;Zhang et al., 2019b). In addition, they contribute to retinal microangiopathy by regulating cell proliferation, apoptosis,inflammatory responses, and VEGF expression (Nieminen et al., 2018; Wei et al., 2019). Thus, they may regulate target gene expression in ROP; however, the specific molecular mecha-nisms underlying retinal vasculogenesis and angiogenesis in ROP have yet to be determined. In this study, the OIR mouse model and high-throughput sequencing were used to detect lncRNAs involved in the path-ogenesis of ROP.
GO and KEGG analyses were used to gain insight into the functions of lncRNAs in OIR. Abnormally ex-pressed genes were widely distributed in retinal tissues and found to be components of signaling pathways related to retinal development and function, such as NF-κB, G protein-coupled receptor, and canonical Wnt signaling pathways. Previous studies suggested that lncRNAs competitively block the binding of mRNAs to microRNAs to exert anti-oxidative and anti-inflammatory roles in retinal endothelial cells via the NF-κB sig-naling pathway, thereby inhibiting angiogenesis (Hui and Yin, 2018; Ruan et al., 2018; Chen et al., 2019). We speculate that the mechanisms by which lncRNAs regulate retinopathy in OIR mice are similar to those involved in diabetic retinopathy. Indeed, previous studies verified abnormal expression of microRNA in the retina of OIR mice, which could regulate the formation of neovascularization (Chen et al.,2017b; Rattner et al., 2019). Thus, lncRNAs regulate retinal neovascularization via microRNA.
VEGF plays a pivotal role in retinal neovascularization and, thus, was selected as the fulcrum of our network analysis (Zhou et al., 2019). We found that differentially expressed genes were linked to VEGF expression by analysis of protein interactions and gene function. The claudin family gene claudin-4, which had the highest differential expression in the OIR group, encodes a vital tight junction protein mainly expressed in epithelial cells and endothelial cells. Claudin-4 is highly expressed in breast, prostate, and ovarian cancers,and participates in angiogenesis of ovarian cancer (Li et al.,2009; Rambabu and Jayanthi, 2018). In addition, claudin-4 is highly expressed during neovascularization of the corneal endothelium (Chng et al., 2013), whereas claudin-1, -2, and-5 are highly expressed in OIR tissues of mice, consistent with our experimental results (Luo et al., 2011). However,further studies are needed to determine the specific upstream regulatory mechanism involved and its relationship to lncRNA.
In conclusion, bioinformatics methods can provide a practical approach to screen lncRNAs associated with ROP.However, this study still has significant limitations. First, as it is difficult to obtain clinical tissue samples of ROP, the current experimental studies are based on OIR of mice. Second,the study lacked in-depth analysis of specific genes involved in ROP. As such, more investigations including analyses of larger sample sizes and clinical samples are required to determine the biological functions and regulatory mechanisms of lncRNAs associated with ROP, thus providing a theoretical framework for clinical research and development of new treatments.
Author contributions:Study design: YD; study implementation: YW,XW; data collection and analysis: YM, YXW; manuscript writing: YW.All authors approved the final version of the paper.
Conflicts of interest:The authors declare that they have no competing interests.
Financial support:The work was supported by the National Natural Science Foundation of China, No. 81600747 (to YD) and Startup Foundation for Doctors of Liaoning Province, China, No. 201501020 (to YD).The funders had no roles in the study design, conduction of experiment,data collection and analysis, decision to publish, or preparation of the manuscript.
Institutional review board statement:This study was approved by the Medical Ethics Committee of Shengjing Hospital of China Medical University on February 25, 2016 (approval No. 2016PS074K). All experimental procedures were performed in accordance with the National Institutes of Health (NIH) guidelines for the Care and Use of Laboratory Animals(NIH Publication No. 85-23, revised 1996).
Copyright license agreement:The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement:Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally peer reviewed.
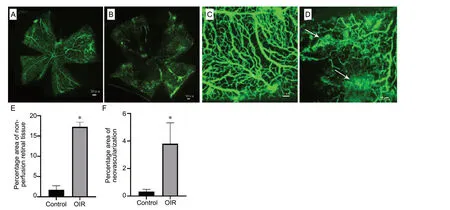
Figure 1 Retinal neovascularization in oxygen-induced retinopathy (OIR) mice.

Figure 2 Neovascular nuclei of the retina in oxygen-induced retinopathy (OIR)mice.

Figure 3 Differentially expressed lncRNAs and mRNAs in the retinal tissue of oxygen-induced retinopathy mice.

Figure 4 GO analysis and KEGG analysis of differentially expressed genes in the retinal tissue of oxygen-induced retinopathy mice.
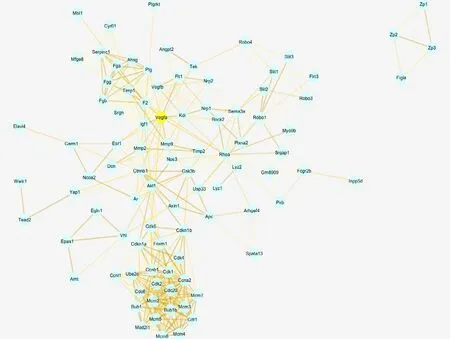
Figure 5 Differentially expressed gene-related protein network of retinal neovascularization.
Open access statement:This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-Non-Commercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
- 中国神经再生研究(英文版)的其它文章
- Transcriptional regulation of adult neural stem/progenitor cells: tales from the subventricular zone
- Targeting molecular pathways for the treatment of inherited retinal degeneration
- Green tea catechins inhibit microglial activation which prevents the development of neurological disorders
- Reversibility of visual field defects through induction of brain plasticity: vision restoration, recovery and rehabilitation using alternating current stimulation
- Role of activin receptor-like kinase 1 in vascular development and cerebrovascular diseases
- Effects of durotomy versus myelotomy in the repair of spinal cord injury

