Correlation between Climatic Factors and Genetic Diversity ofPhrynocephalus forsythii
Yue QI,Wei ZHAO*,Yongjie HUANG,Xiaoning WANG and Yangyang ZHAO
1 Gansu Key Laboratory of Biomonitoring and Bioremediation for Environmental Pollution, School of Life Sciences,Lanzhou University, Lanzhou 730000, China
2 School of Nature Conservation,Beijing Forestry University, Beijing 100083,China
Abstract Global climate change is a threat to animals in nearly all biomes and ecosystems,especially for ectotherm whose life activities highly depend on environmental thermal regime.Population genetic diversity which is essential for adaptation to environmental change is a useful index for long-term species survival.In this paper,genetic diversity of eight Phrynocephalus forsythii population which distributed in Tarim Basin,China,were evaluated based on three mtDNA gene and its correlation with environment factors were investigated using RDA.Our result revealed that,the level of genetic diversity of P.forsythii populations was related to its location but there was no significant correlation between genetic distances and geographic distances in P.forsythii.However,we find that mtDNA of P.forsythii was subjected to selection pressure during evolution and population genetic diversity was significantly positively related to variation coefficient of rainfall(VCR)and altitude(AL),while significantly negatively related to longitude(N)and annual average temperature(AAT).Our result supported the previous prediction that excessive ambient heat is a threat to P.forsythii.
Keywords climatic factors,genetic diversity,selection pressure,Phrynocephalus forsythii,Tarim Basin
1.Introduction
Conservation of genetic diversity within species has been recognized as fundamentally important in striving to slow or halt the loss of biodiversity(Xuet al.,2017).The value of genetic diversity is evident from the deleterious impacts of its loss on populations through effects such as increased inbreeding and genetic drift(Oostermeijeret al.,2003).Consequently,the conservation of genetic diversity has become a renewed focus under the expectation that its loss could render populations and species less able to adapt to ongoing environmental change(Georgina,2003;Mitchellet al.,2015).Genetic diversity is an adaptation parameter of species to environmental changes in the long-term evolutionary process,thus it can be used as an indicator of environmental adaptation:the decrease of genetic diversity can reflect the weaker adaptive ability of organism to potential environmental changes(Frankhamet al.,2002).It is generally consider that genetic diversity often affected by numerous factors such as the environmental heterogeneity,habitat suitability and climate fluctuation(Caoet al.,2012;Lvet al.,2014).
Habitat environment plays an important role in the variation of biological genetic diversity(Mitchellet al.,2015).Populations that occupy different environments will be selected by different natural selection.Under the influence of natural selection of the environment,population will retain the best phenotype adapted to the habitat environment,and the genotype frequency associated with the selected phenotype of population will be changed.This process will result in the genetic differences between populations in different natural environments(Ferrari and Chi,1998).Populations will therefore show extensive geographic variation in genetics along environmental gradients when they occupy different environments(Alkon and Zhao 1993;Chen 1997).
Except for the heterogeneity of environment,natural selection during extreme events also determine the level of genetic diversity.Phenotypes of individuals are controlled by genotypes and also influenced by environment.So if the genotypes of individuals with different phenotypes differ,genotypes of the individuals which favored by natural selection will tend to increase in relative frequency,and genetic diversity of populations will thus increase.That is to say,natural selection during extreme events brings about genetic changes in population genetics(Jin and Liu,2008).
Furthermore,stability of climate is also regarded as a selective pressure,and might change the level of genetic diversity(Oliveiraet al.,2016).Continual environmental fluctuations provide a background of continuously changing selection pressures to which the species must respond.Such response caused only the individuals whose genotype pre-adapted to the environmental conditions survived during periods of environmental change,hence would influence the level of genetic diversity(Smithet al.,2014;Turchetto-Zoletet al.,2013).
Phrynocephalus forsythiiis a viviparous,agamid sand lizard endemic to the Tarim Basin with a broad altitudinal range from 800 to 3000 m(Adler,1992).A recently study which developed an eco-physiological model of extinction risk under climate change predicted that theP.forsythiiwill face a high risk of extinction due to global warming(Sinervoet al.,2018).In consideration of the crucial role of genetic diversity in future evolutionary trajectory of a specie(Leffleret al.,2012),it is important to understand the variation of population genetic diversity along environmental gradient inP.forsythii.Therefore,in this paper,mitochondrialND2,ND4andCytbgenes were used to evaluate the genetic diversity ofP.forsythii,and we predicted that the genetic diversity ofP.forsythiiwill decrease as the environment temperature increased.
2.Materials and Methods
Ethics statement Animals were treated in accordance with the guidelines of Ethics Committee of the School of Life Sciences,Lanzhou University,that specifically approved this study.
2.1.Population samplingA total of 110 individuals were sampled from eight naturalP.forsythiipopulations in Tarim Basin(Figure 1).Animals were euthanized in the field,then dissected and preserved liver and muscle tissue immediately in 100% ethanol.Longitude,latitude and altitude were recorded from the sampling localities and climatic data over 30 years were collected from the nearby weather station(National Climatic Data Center)(Table 1).Variation coefficient of temperature(VCT)was estimated through divided the standard deviation of AAT by the AAT,and variation coefficient of rainfall(VCR)through divided the standard deviation of AAR by the AAR to obtained.Humidity(H)was calculated by AAR/AAT(Kotteket al.,2006).
2.2.DNA extraction and PCR amplificationTotal genomic DNA was extracted using the TIANamp Genomic DNA Kit.Three loci of mitochondrial genes were amplified using PCR:NADH Dehydrogenase Subunit 2(ND2),NADH Dehydrogenase Subunit 4(ND4)and Cytochrome b(Cyt-b).Primers are designed with PRIMER version 6.0.All PCR products were sequenced using the Sanger sequencing method(Genewiz Biotech(Suzhou)Co.,Ltd.).All sequences were deposited in the GenBank library(Table S1).

Figure 1 Geographical distribution of sampling points of P.forsythii.The elevation gradient is represented by graduated color,from white to black(low to high).The map was downloaded from National Fundamental Geographic Information System(NFGIS)(http://nfgis.gsdi.gov.cn/).
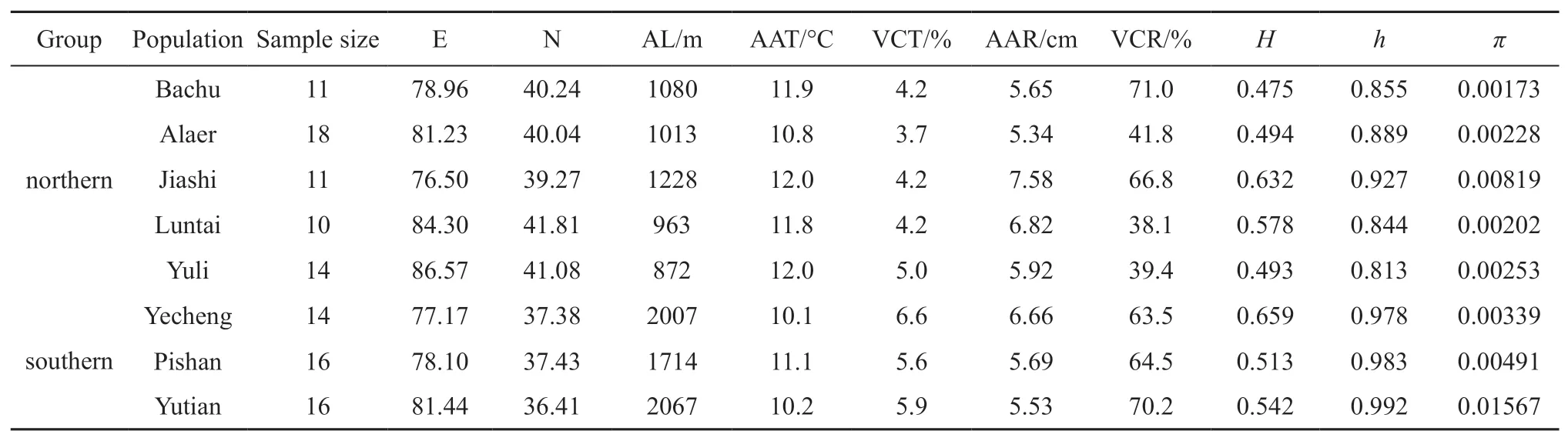
Table 1 Information of P.forsythii populations sampled in this study.
2.3.Data analysisSequences were assembled manually using SEQMANII in DNASTAR and aligned with the BioEdit version 7.0.5.2 program(Vrålstadet al.,2002).Nucleotide diversity(π)and haplotype diversity(h)calculation was run in the DnaSP version 5 program(Librado and Rozas,2009).BI analyses were performed using MrBayes version 3.2.6(Huelsenbeck &Ronquist 2001)under the AIC with the TrN+I+G model,which was selected by MrModeltest version 3.7(Posada 1998),for the BI analysis,20 million generations with Markov Chain Monte Carlo(MCMC)simulations was executed until the average standard deviation of split frequencies dropped below 0.01,and the first 25% were discarded as burn-in.Analysis of molecular variance(AMOVA)was performed to analyze amongpopulation sources of variation using ARLEQUIN version 3.5(Excoffieret al.,2005)based on the result of Bayesian inference.The genetic distances between populations obtained using a Kimura two-parameter distances(K2P)model of nucleotide substitution available in MEGA version 6.06(Wanget al.,2007).Statistical analyses of selection was used the CODEML program in PAML version 4.8(Jinet al.,2018;Yang 2007).We used the site models to detect signatures of selection.Likelihoods were obtained under the following site models:M0(one ratio),M1A(nearly neutral),M2A(positive selection),M3(discrete),M7(beta),M8(beta &ω).Then Likelihood ratio tests(LRT)were used to compare null model and experimental model:M0-M3,M1A-M2A,M7-M8.Sites were considered to have experienced positive selection when at least one codon model showed ω>1 and one LRT was significant for either the M1a-M2a or M7-M8 comparisons.The correlation between genetic distance and geographic distance was calculated by Mantel test in R version 3.5.1.Correlation analysis between genetic diversity and environmental factors(Table 1)was conducted in Canoco version 4.5(Zhanget al.,2012).
3.Result
3.1.Genetic diversityNucleotide diversity(π)and haplotype diversity(h)ranged from 0.00173-0.01567 and 0.813-0.992,respectively(Table 1).Furthermore,based on the Bayesian analysis,all total of 76 haplotypes from eight populations were divided into two major clades:the southern group(Yutian,Pishan,Yecheng)and the northern group(Jiashi,Alaer,Bachu,Luntai,Yuli),and the genetic diversity of the southern group(h=0.995,π=0.01496)was higher than the northern group(h=0.974,π=0.00858).
The AMOVA analysis also based on phylogeographic structure.Significance was detected at all three levels,and the among-group component accounted for 71.47% of the total variance(Table 2).Indicate that there is a significant genetic difference between northern and southern group.
3.2.Correlation between the genetic and geographic distancesMantel test revealed that there was no significant correlation between genetic distances and geographic distances(r=0.28,P=0.096).However,genetic distance between populations from northern and southern group was bigger than those from the same group,which indicate that there is a correlation between geographic distance and genetic distance among northern and southern group(Table 3).
3.3.Influence of climatic factors on genetic diversity of P.forsythiiIn the analyses of selection,values ofωmuch greater than 1 were detected for models M2A,M3 and M8(Table 4).The LRT that compared M0/M3 was highly significant(P<0.001),which indicated a strong signal of variable ω among sites.Furthermore,the M1a/M2a LRT and the M7/M8 LRT was significant(P<0.05;Table 4),which indicated that mtDNA ofP.forsythiiwas subjected to selection pressure during evolution.
DCA analysis for genetic diversity indicated that the gradient length of the first axis was 0.027,so the RDA model can be selected for sequencing in this study.RDA allows consideration of several environmental factors simultaneously,so it was used for analyzing relationships between environmental factors and genetic diversity.We found strong correlations between genetic diversity of mtDNA and climatic factors,the first axes explained 85.9% of the total variance(Table 5).Nucleotide diversity(π)positively correlated with AL,VCT,VCR and H,while negatively correlated with E,N,AAT and AAR,and the same correlational relationship was found between haplotype diversity(h)and environmental factors.Furthermore,Monte Carlo Permutation Tests suggested that the correlation ofhandπwithN(F=22.03,P=0.001),AL(F=15.99,P=0.001),AAT(F=5.08,P=0.024)and VCR(F=4.08,P=0.041)were statistically significant.

Table 2 Analysis of molecular variance(AMOVA)for P.forsythii populations.

Figure 2 Canonical correspondence analysis of P.forsythii and climatic factors.Direction of vectors represent climatic factors accord with genetic diversity,manifest the positive correlation of them;otherwise,they were negative correlation.Length of vectors represents the degree of correlation.

Table 3 Mantel test of genetic distance and geographic distance.

Table 4 Parameters estimation and likelihood ratio tests for the site models.

Table 5 Results of ordinations produced by canonical correspondence analysis
4.Discussion
Our study examined genetic diversity ofP.forsythiipopulations and its relationships with climatic factors at eight sites in Tarim Basin.Although correlation between genetic distances and geographical distances was no statistically significant,genetic diversity of two groups form northern and southern of Tarim Basin existed difference.We considered these differences were caused by selection pressures of different habitat environment,because mtDNA ofP.forsythiiwas subjected to selection during evolution and genetic diversity correlated strongly with N and AL through the analysis of RDA.The habitats ofP.forsythiilocated in the south of the Tarim basin are characterized by high altitude while the habitats located in the north of the Tarim basin are characterized by low altitude.In the process of adapting to the local ecological environment,population gradually differ in their genome(Yeet al.,2017).Therefore,genetic composition of northern and southern population ofP.forsythiiexist a relatively large variation.Furthermore,the genetic diversity ofP.forsythiipositively correlated with AL while while negatively correlated with N.It indicated that the environmental conditions on the southern side of Tarim basin have lower selection pressure,while the northern side has higher selection pressure forP.forsythii.Different selection pressure arising from environmental heterogeneity affects the level of the genetic diversity,and we considered that this selection pressure mainly comes from the higher ambient temperature.
According to the RDA analysis,the level of genetic diversity ofP.forsythiiwas negatively correlated with temperature but positively correlated with humidity.According to the cold-climate hypothesis(Qualls and Andrews,1999),reproductive mode of ectotherms was highly correlated with elevation,with viviparous species occurring at higher elevations than oviparous species,thus viviparity that evolved inPhrynocephaluswas adaptive to the environment of lower temperature(Wanget al.,2014).In addition,living in localities where ambient temperature is too high,ectotherms trend to retreat to thermal refugia on a daily basis and reduce foraging time,hence,constrains metabolically costly functions such as reproduction,maintenance and growth,and thus affect the level of genetic diversity(Ceia-Hasseet al.,2014).However,the positively correlation between genetic diversity and humidity was not statistic significant inP.forsythii.
Finally,it is generally believe that environmental fluctuations is considered as a selection pressure(Hancocket al.,2011;Jin and Liu,2008).However,our result of RAD run counter to this conclusion,which revealed that the level of genetic diversity ofP.forsythiipositively correlate with variation coefficients of rainfall.Considering that,VCR negatively correlated with AAT,thus the positively correlation between genetic diversity and VCR may be a byproduct of the influence produced by negative correlation between AAT and genetic diversity.
In summary,our research shows that,undergo different nature selection in heterogeneous environment,P.forsythiipopulation genetic diversity produced difference,and was mainly affected by N,AL,AAT and VCT.What's more,we find that higher temperature may be a crucial climate factor affecting population genetic diversity ofP.forsythii.
AcknowledgementsThe work was supported by the National Natural Science Foundation of China(No.31471988 and 31200287).We also thank Li Ding from Lanzhou University for the help with data analysis.
Appendix

Table S1 Sample code and GenBank Accession for mtDNA of P.forsythii.
 Asian Herpetological Research2019年4期
Asian Herpetological Research2019年4期
- Asian Herpetological Research的其它文章
- Molecular Cloning,Characterization and Sequence Analysis of KCNQ4 in Large Odorous Frog,Odorrana graminea
- Description of a New Species of Amolops(Anura:Ranidae)from Tibet,China
- Geographical Distribution and Morphological Variability of the Rapid Racerunner,Eremias velox(Pallas,1771)(Reptilia,Lacertidae)in the Eastern Periphery of Its Range
- Ecological and Geographical Reasons for the Variation of Digestive Tract Length in Anurans
- Mating Ethogram of a Video-aided Study of Mating and Parturition in Captive Chinese Crocodile Lizards(Shinisaurus crocodilurus)
- Home Range Size and Overlap of the Small Nocturnal Schlegel's Japanese Gecko(Gekko japonicus),Introduced into a City Park in Korea
