染色质重塑因子在植物发育过程的功能
彭锈玲, 王剑豪, 杨松光
染色质重塑因子在植物发育过程的功能
彭锈玲1,2, 王剑豪1,2, 杨松光1*
(1. 中国科学院华南植物园, 华南农业植物分子分析与遗传改良重点实验室,广州 510650; 2. 中国科学院大学, 北京 100049)
染色质重塑复合体(chromatin remodeling complexes)通过具有ATPase活性的亚基水解ATP释放能量,通过改变核小体“构象”(包括核小体重定位、核小体滑动和核小体替换等)而改变DNA的“可及性”(accessibility),进而影响特定的生理、生化过程。染色质重塑复合体最早在酵母中发现,生化分析表明其至少含有13个亚基。目前植物染色质重塑复合体的组成还未完全解析,但通过对其酵母同源亚基(染色质重塑因子)的研究可从侧面探究植物染色质重塑复合体的功能。同时,还着重讨论了近年来在植物染色质重塑因子研究上取得的结果,以期为植物染色质重塑的作用机制提供启示。
染色质重塑因子;表观调控;功能;植物
1 染色质重塑复合体
真核生物遗传信息以核小体为基本单位,经高度包裹压缩存储于染色质中。因此真核生物在DNA复制、转录、重组和DNA修复等过程中首先要克服DNA与组蛋白之间的“紧密”结合。涉及该过程的蛋白主要包括两类,即染色质修饰酶(包括组蛋白修饰酶和DNA修饰酶)和依赖于ATP的染色质重塑因子(ATP-dependent chromatin remodelers, ATPase)。前者通过添加或者移除组蛋白和DNA上的化学基团改变DNA的“可及性”[1],而后者通过与其他蛋白组成染色质重塑复合体水解ATP释放能量,改变核小体“构象”(positioning, occupancy and composition of nucleosomes)而改变DNA的“可及性”,进而影响特定的生理过程[2–3]。
染色质重塑复合体最早从酵母()和(sucrose non-fermenting)突变体中分离鉴定。生化分析表明该复合体含有至少11个亚基(SWI1、SWI2/SNF2、SWI3、SNF5、SWP73、ARP7、ARP9、SWP82、SNF6、SNF11和TAF14),其中SWI2/SNF2蛋白具有ATPases活性[4],SWP82、SNF6、SNF11和TAF14为酵母所特有(表1)。根据ATPase亚基的结构,可将染色质重塑复合体分为4类(表1~3),即SWI/SNF、ISWI、CHD和INO80/SWR1[2], 且不同复合体可能包括相同的亚基。研究表明,这4类染色质重塑复合体ATPase亚基无论是在酵母、果蝇还是人类中均十分保守,含有保守的ATPase催化结构域(SNF2-N结构域),该催化结构域可进一步细分为DExx和HELICc两部分(图1)。除ATPase催化结构域外,不同染色质重塑复合体ATPase亚基还含有特异结构,如SWI/SNF (matingtype switching/sucrose non-fermenting)复合体ATPase亚基N端含有HSA (helicase-SANT)结构域而C端含有bromodomain结构域[5–6]。bromodomain结构域能识别组蛋白“尾巴”乙酰化的残基,使SWI/SNF复合体结合在染色质特定位点[7–8]。而ISWI (imitation switch)复合体ATPase亚基C端含有SANT和SLIDE结构域,这二者形成一个核小体识别结构与未修饰的组蛋白和DNA结合[9]。CHD (chromodomain helicase-DNA binding)复合体ATPase亚基N端有串联的chromodomain结构域[10],能识别组蛋白H3K4的甲基化位点[11–12]。与其他三类ATPase亚基相比,INO80/SWR1 (inositol requiring 80)复合体ATPase亚基结构域DExx与HELICc之间有一段较长的氨基酸残基(图1),然而这并未影响其ATPase的活性[13]。
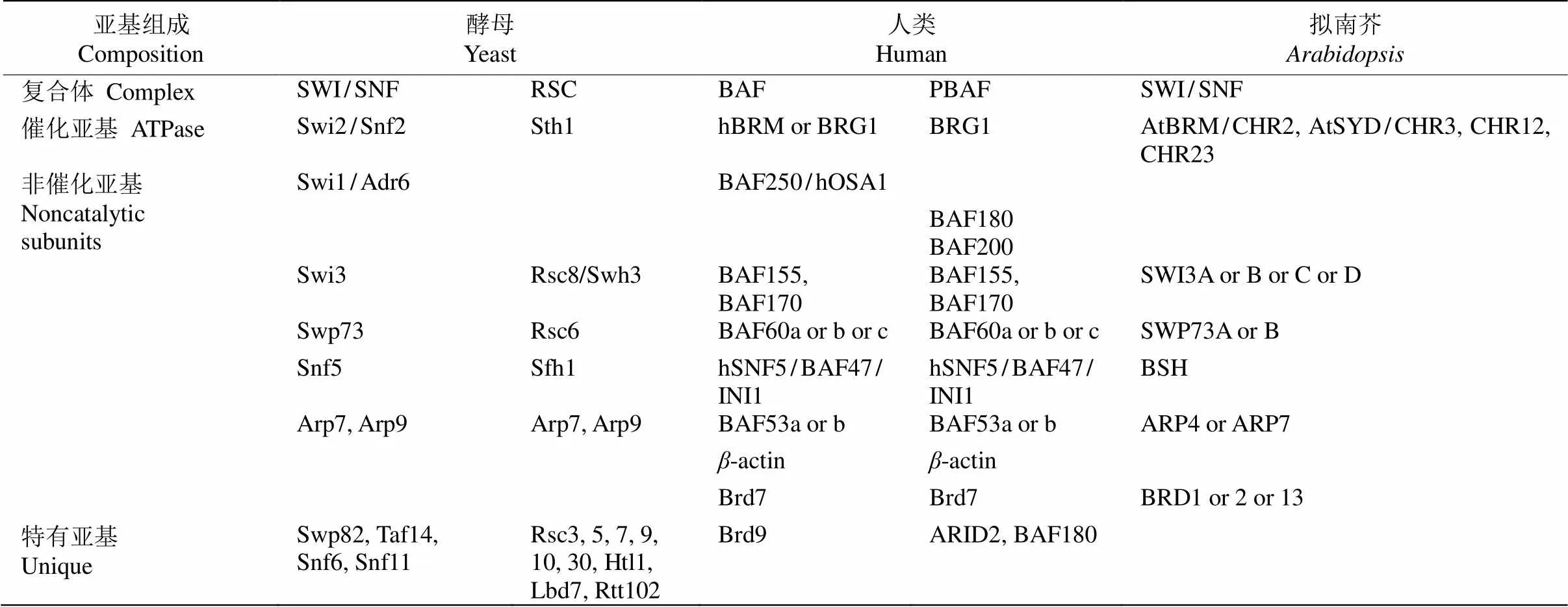
表1 不同物种的SWI/SNF复合体组成
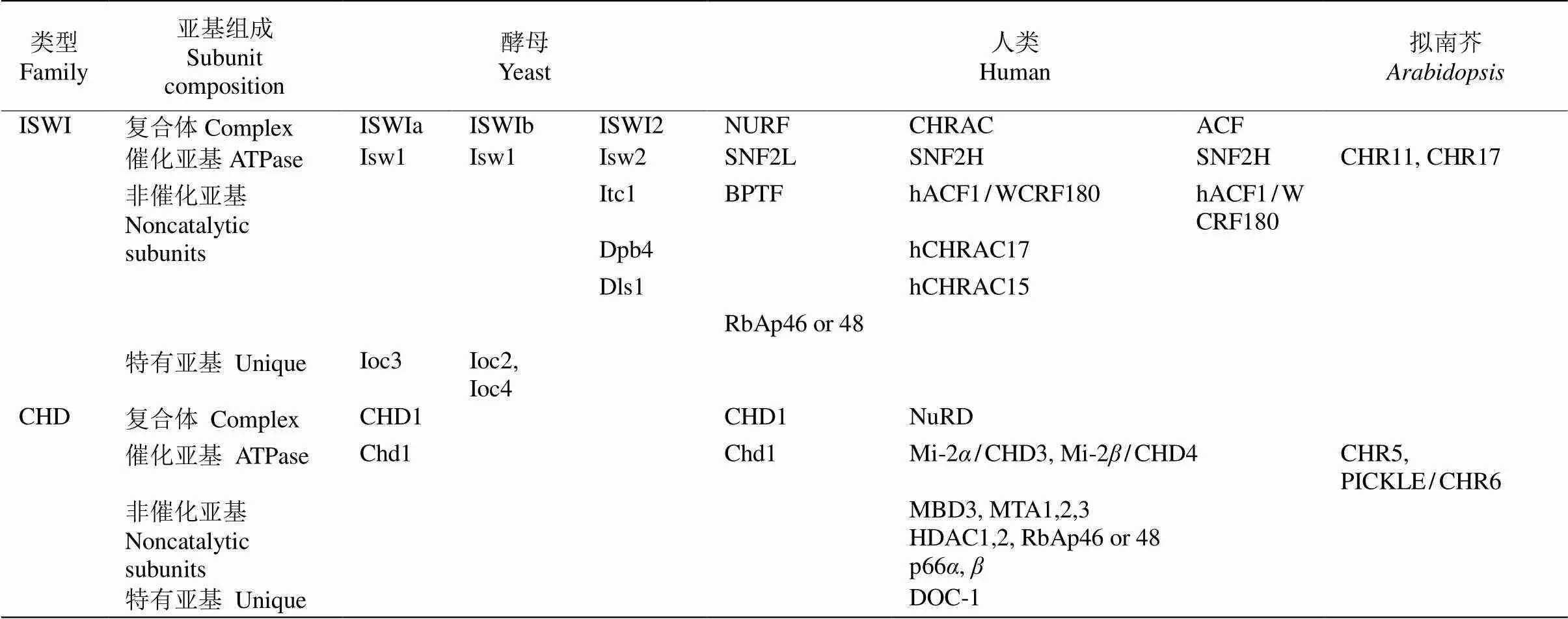
表2 不同物种的ISWI、CHD复合体组成
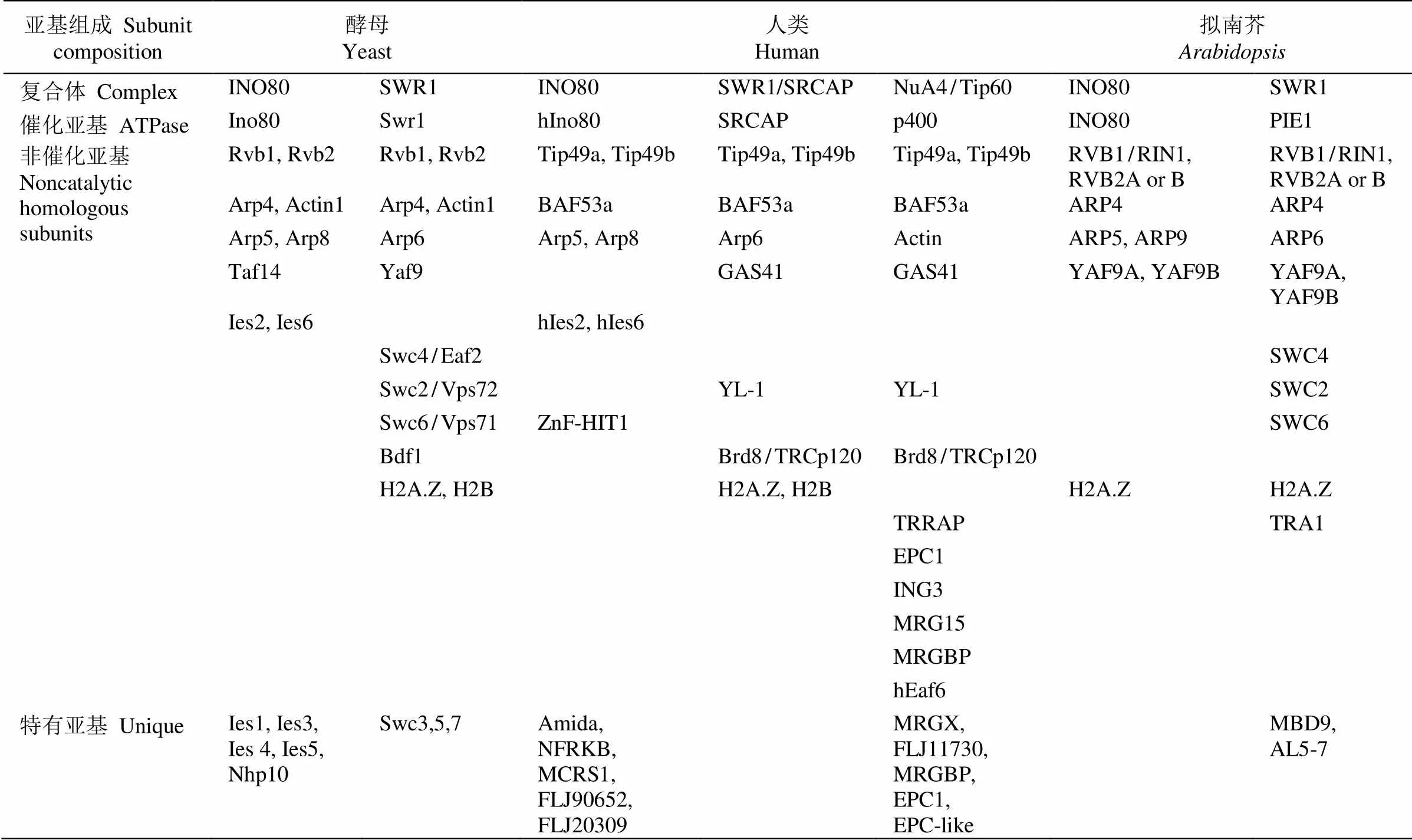
表3 不同物种INO80/SWR1复合体组成
虽然不同染色质重塑复合体ATPase亚基在结构上较为保守,但不同染色质重塑复合体具有特有的功能。如ISWI和CHD复合体主要参与DNA复制后染色质的组装[14];而SWI/SNF复合体则影响染色质的去组装和核小体稳定蛋白的替换[15–17]。INO80/SWR1复合体则介导组蛋白变体(histonevariant)的替换,其中INO80复合体介导H2A替换H2A.Z,而SWR1复合体则与之相反[18–20]。组蛋白H2A与组蛋白变体H2A.Z之间的互换对核小体结构稳定性至关重要[21–23]。
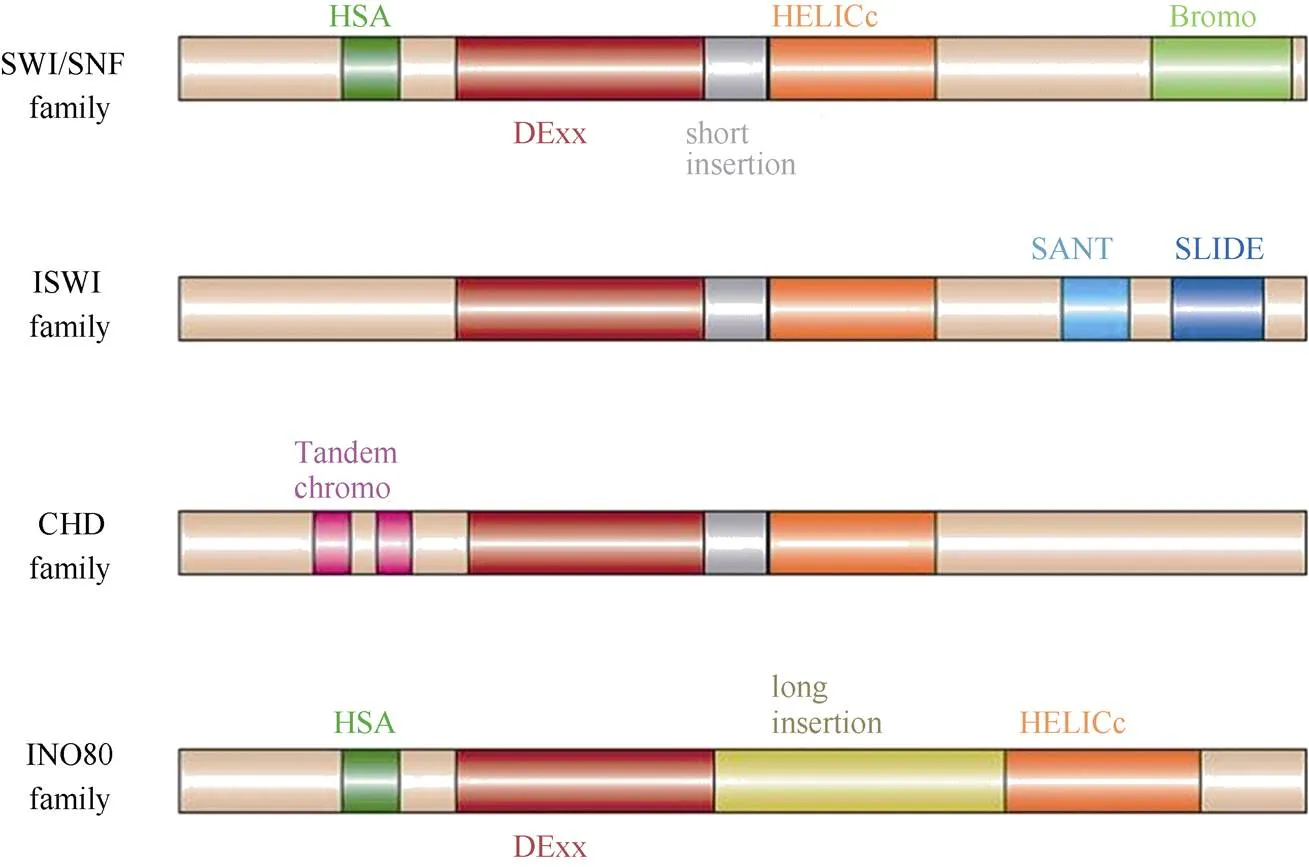
图1 不同染色质重塑复合体ATPase亚基结构示意图(引自Clapier[2])
目前对染色质重塑复合体的作用机制并不十分清楚。一般认为,染色质重塑复合体ATPase亚基与DNA易位酶(DNA translocases)具有相似之功能。当染色质重塑复合体ATPase亚基与核小体结合后,其易位酶活性将核小体之间的连接DNA (linker)推向核小体核心结构,使DNA形成一个环状结构(loop),从而使DNA与组蛋白之间结合由“紧密”状态变为“松散”状态[24–27]。该过程可能同时产生很多环状结构,这些环状结构只需改变DNA与1~2个组蛋白的结合程度即可启动核小体的滑动。然而,对形成的环状结构大小并不十分清楚, 目前的证据支持该环状结构可能由较多的碱基(约100 bp)组成[28]。而关于染色质重塑是如何被精确调节的还知之甚少,该过程可能与组蛋白的翻译后修饰有关。如H4尾巴第17~19位残基乙酰化能提高蟾蜍() ISWI复合体催化活性[29–30],而H4K16ac则抑制其活性。对酵母yISW2 (ISWI复合体催化亚基)和yChd1 (CHD复合体催化亚基)而言,H4乙酰化抑制其ATPase活性而不响应它与核小体结合,然而H4乙酰化却能提高酵母RSC复合体(SWI/SFN类)的重塑活性[31]。
2 植物染色质重塑因子及功能
目前,植物染色质重塑复合体组成尚未完全分离鉴定,然而遗传和蛋白相互作用数据提示植物中也存在多种染色质重塑复合体(表1~3)。因此通过对染色质重塑复合体同源亚基的研究可从侧面探究植物染色质重塑复合体的功能。通过与酵母、果蝇和人的染色质重塑复合体ATPase亚基的同源比对,拟南芥含41个ATPase结构域(SNF2-like)基因,可分为Snf2-like、Swr1-like、Rad54-like、Rad5/16-like、SSO1653-like和SMARCAL1-like家族,每个家族又可细分为不同亚家族[32],如Snf2-like家族可分为SWI2/SNF2、Lsh、Iswi、Chd1和Mi-2亚家族,而Swr1-like家族则可分为Ino80、Swr1和Etl1亚家族。遗传和蛋白相互作用及IP-MS研究表明,拟南芥() SWI/SNF染色质重塑复合体亚基核心组分与酵母和动物相似,但含有更多同源基因(如与酵母SWI3同源的SWI3A/B/C/D),且有植物特有的亚基(图2)。这提示植物染色质重塑复合体可能具有与动物和酵母不同的功能。围绕各组成亚基的研究表明,这些基因参与细胞分化、器官发育和激素信号转导等多种生理过程(表4)。在拟南芥所有SNF2-N蛋白中Snf2-like (11个)和Swr1-like (4个)家族成员在序列上最有可能是植物染色质重塑复合体催化亚基,围绕这些基因所取得的研究成果也最丰富。在Snf2-like家族中,SWI2/SNF2亚家族(4个)、CHD1-Mi2亚家族(4个)、Iswi-Lsh亚家族(3个)和Swr1-like家族(4个)成员分别对应于SWI/SNF、CHD、 ISWI和 INO80/SWR1复合体催化亚基(表1~3)。
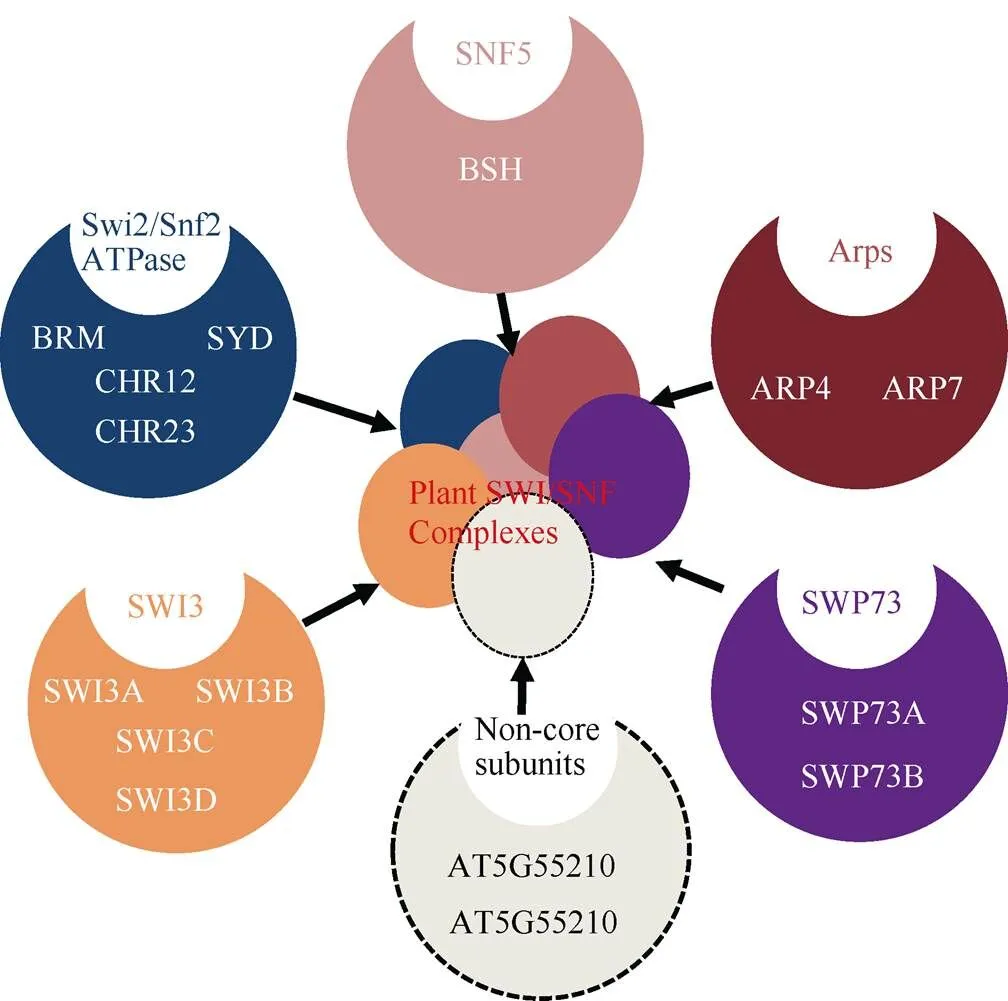
图2 植物SWI/SNF染色质重塑复合体可能组成(修改自Jerzmanowski[33])
2.1 SWI/SNF复合体
2.1.1 SWI/SNF复合体催化亚基
SWI/SNF复合体最早从酵母()中分离鉴定,随后发现该类复合体亦广泛存在于动物如果蝇()、小鼠()和人类中。目前关于植物SWI/SNF复合体的具体组成还不十分清楚,但遗传和蛋白相互作用数据显示该复合体也存在于植物中。在拟南芥41个ATPase结构域(SNF2-like)蛋白中,SWI2/SNF2亚家族成员AtBRM (BRAHMA)、AtSYD (SPLAYED)、AtCHR23 (CHROMATINREMODELL ING23)和AtCHR12 (CHROMATIN REMODELL ING 12)最有可能是植物SWI/SNF复合体催化亚基,其中AtBRM的可能性最大。首先,仅有AtBRM蛋白C-端具有与酵母SWI2/SNF2和果蝇BRAHMA蛋白一样的bromo结构域;其次,AtBRM蛋白N-端能与酵母SWI3同源蛋白AtSWI3B和AtSWI3C相互作用;最后和突变体具有相似表型[34]。

表4 拟南芥染色质重塑因子的功能分析
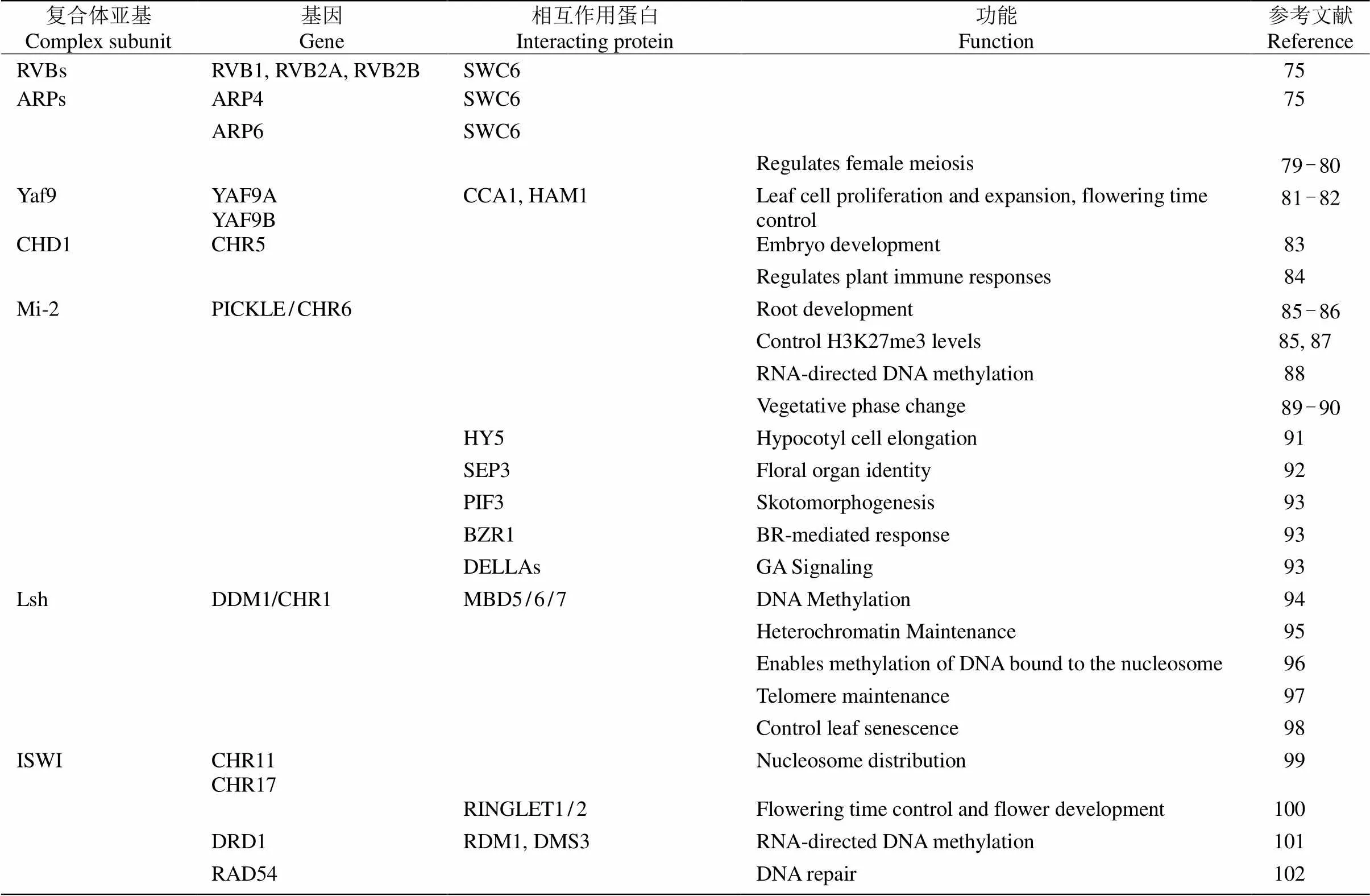
续表(Continued)
主要在分生组织和幼嫩器官中表达,其功能缺失导致2 000余基因中的一半下调而另一半上调表达[104],这表明具有双重功能。敲减的植株矮小,在长日照下叶片卷曲,花器官发育异常;在长日照和短日照下均出现早花现象[33]。缺失突变体中,有相当一部分植株在短日照下不开花,这提示在拟南芥开花过程的作用十分复杂[35]。进一步研究表明,除了影响光周期响应基因的表达外,还抑制和的表达[104–105]。在叶片中,AtBRM分别与TCP4和AN- GUSTIFOLIA3 (AN3)相互作用,共同调控叶片发育相关基因的表达[38–39]。我们的研究表明,在花序轴中AtBRM与转录因子BREVIPEDICELLUS (BP)相互作用,直接调节和的表达来调控花序轴发育[36]。在黑暗中,AtBRM与PHY-INTER- ACTING FACTOR 1 (PIF1)相互作用抑制()表达从而抑制叶绿素合成[40]。除转录因子外, AtBRM亦可与其他核蛋白相互作用。如热胁迫记忆激活因子FORGETTER1 (FGT1)与AtBRM相互作用,维持下游热胁迫相关基因处于转录激活状态[42]。而植物H3K27去甲基化酶RELATIVE OF EARLY FLO- WERING 6 (REF6)通过招募AtBRM结合于下游基因CTCTGYTY基序降低其H3K27me3水平激活转录[41]。这与动物中BRM拮抗PcG蛋白(polycomb group proteins)的作用一致。PcG蛋白作为表观遗传抑制因子维持细胞内非活化基因的抑制状态,其分别通过与Polycomb Repressive Complex 1 (PRC1) 和PRC2复合体相互作用建立和维持染色质抑制状态。PRC2复合体与目标基因结合后,催化这些基因组蛋白H3K27me3修饰,从而抑制基因表达。这与我们在拟南芥主根发育过程观察到AtBRM拮抗PcG蛋白影响生长素运输蛋白基因表达,从而影响主根根冠干细胞微环境维持的结果一致[43]。最近研究表明,翻译后修饰对染色质重塑过程也起着重要作用,AtBRM作为ABA信号途径核心组分SnRK (蔗糖非依赖1蛋白激酶)和PP2C (蛋白磷酸酶2C)的底物来调控ABA反应[46];我们亦观察到,METHYL METHANE SULFONATE SENSITIVITY 21 (MMS21)通过SUMO化修饰AtBRM调节其蛋白稳定性参与主根发育[43]。有趣的是,新的研究结果表明microRNA前体(pri-miRNAs)也能与AtBRM相互作用,AtBRM作为microRNA前体加工复合体SE (MICROPRO CESSOR COMPONENT SERRATE)组分改变microRNA前体二级结构以便后续通过DCL1和HYL1进一步加工[47]。
对拟南芥SWI2/SNF2亚家族其他成员的研究表明,参与顶端分生组织(SAM)活性的维持。缺失突变体植株矮小,生长缓慢、叶片极性和SAM缺失。其作用机理是,通过途径影响SAM的维持,因为AtSYD可与BARD1相互作用结合于启动子直接结合而调节表达, 后者促进SAM中干细胞活性[48–49]。进一步研究还表明,AtSYD通过调控JA和ET信号相关基因参与植物的生物胁迫响应[106],而这种胁迫大部分是通过抑制(SUPPRESSOR OF NPR1, CONSTITU- TIVE 1)实现的[107]。对和在植物发育中的功能还不十分清楚。过表达和均抑制植物种子萌发[50],在其他发育过程过表达显著抑制植物生长,而过表达表型则不明显[51];但在胁迫方面, 二者表型相似[52–53]。
2.1.2 SWI/SNF复合体非催化亚基-SWI3类蛋白
拟南芥基因组编码4个SWI3同源蛋白,分别为AtSWI3A、AtSWI3B、AtSWI3C和AtSWI3D, 在结构上他们均含有SWIRM、SANT和Leucine Zipper 结构域。进化分析表明植物SWI3类蛋白可明显分为两簇,即SWI3A/B和SWI3C/D,这也与AtSWI3A、AtSWI3B、AtSWI3C和AtSWI3D的生物学功能有所差异相符。与突变导致植物胚在早期发育过程异常,而和突变使得植物叶片和花器官发育异常[53]。有意思的是突变还导致植物主根发育异常,而其他3个突变体则未观察到相应表型[53]。酵母双杂交结果表明,AtSWI3A可分别与AtSWI3A、AtSWI3B、AtSWI3C、BSH、AtSYD和FCA相互作用[53–54], 提示AtSWI3A、BSH和AtSYD可能形成1个复合体。然而关于AtSWI3A与其他蛋白相互作用的生物学意义目前并不清楚。对AtSWI3B而言,除分别可与AtSWI3A、AtSWI3B、AtSWI3C、AtSWI3D、BSH、AtSYD、AtBRM相互作用外, 还分别与type 2C类磷酸酶(phosphatase type 2C) HAB1 (HYPERSENSI- TIVE TO ABA1)和长链非编码RNA结合蛋白IDN2相互作用,参与ABA信号和长链非编码RNA形成[55–56]。进一步研究还表明,AtSWI3B (包括At- SWI3C和AtSWI3D)与MORC6 (microrchidia 6)、SUVH9 [SU(VAR)3-9 homolog]和IDN2形成复合体, 通过RNA指导的DNA甲基化(RdDM, RNA-directedDNA methylation)介导的途径调控DNA的甲基化[57]。在叶片发育过程,通过调控生长素代谢酶基因(IAA carboxyl methyltransferase 1)的染色质“构象”调节其表达,从而参与叶片发育[58]。
AtSWI3C也分别与AtSWI3A、AtSWI3B、AtSYD和AtBRM相互作用,进一步研究表明其还可与转录因子AN3和酵母SWI/SNF复合体同源蛋白SWP73B以及ARP4/7 (actin-related protein4/7)相互作用调控叶片发育[39]。同时,AtSWI3C通过与DELLA蛋白RGL2和RGL3相互作用促进(GIBBERELLIN-INSENSITIVE DWARF1)和(GIBBERELLIN 3-OXIDASE)表达,参与GA信号转导[59]。相对于其他3个AtSWI3蛋白,AtSWI3D的功能还知之甚少,其缺失植株的表型与和缺失突变体相似,出现叶片卷曲,花器官发育异常和育性降低等表型[53]。
2.1.3 SWI/SNF复合体非催化亚基-SNF5类蛋白
4.4.2 强化服务监督,完善管理制度。进一步完善和优化项目立项制、项目公示制、项目审计制、项目检查验收制、后续管护等制度,加强项目事中、事后监督。
在酵母中,SNF5蛋白对染色质的装配和基因启动子与SWI/SNF复合体的结合至关重要[108],其C端保守的200个氨基酸残基形成两个重复结构负责与SWI/SNF复合体其他亚基和其他因子如cyclin E/CDK2之间的相互作用。动物发生突变往往会导致癌症。在拟南芥中,SNF5同源蛋白BSH(BUSHY GROWTH)仅由1个基因编码,其可与AtSWI3A和AtSWI3B相互作用,且在酵母中异源表达,可互补酵母突变体表型[61]。利用反义RNA技术降低表达导致植物顶端分生组织减小且出现不育表型[61]。T-DNA插入突变体虽然导致种子贮存基因在幼苗中异位表达,然而植株却未出现可以看到的表型[60]。这可能是由于T-DNA插入位置在BSH的C端,仅破坏BSH蛋白C端结构使得BSH还保留部分功能。
2.1.4 SWI/SNF复合体非催化亚基SWP73类蛋白
酵母SWI/SNF复合体亚基SWP73对SWI/SNF复合体在转录过程的作用至关重要。在植物中,拟南芥基因组编码两个SWP73蛋白:SWP73A和SWP73B,二者氨基酸序列相似度高达83.7%。蛋白互作分析表明SWP73A仅可与AtSWI3C相互作用,而SWP73B不仅可以与AtSWI3C和AtSWI3D相互作用,还能与AtBRM和ARP4/7以及转录因子AN3相互作用[39]。生物学功能分析表明,和功能亦有所差异,突变植物未出现可见表型,而突变导致植物根[63]、叶片和花发育异常[62]和开花时间推迟[64]。进一步研究表明,通过促进根中细胞分裂素的合成促进根中分生组织的维持[63];同时通过改变染色质组蛋白修饰水平和H2A.Z的替换影响表达从而参与植物成花控制[64]。最新的研究表明,SWP73B通过直接结合于下游基因的G-box区域调节其表达抑制下胚轴伸长,但与SWP73B直接结合的基因与PIF4结合的基因大部分是不同的[65]。在胁迫方面,还参与UV-B介导的DNA损伤修复,然而其作用机制还不清楚[66]。
2.1.5 SWI/SNF复合体非催化亚基ARP类蛋白
酵母和动物SWI/SNF类复合体均含有一类ARPs (actin-related proteins)蛋白。在酵母中,ARPs一共有10个(ARP1~10),其序列与酵母actin相似性按编号递减。在酵母所有ARPs中,ARP7和ARP9是SWI/SNF类(包括RSC)复合体组分,而ARP4、ARP5和ARP8为INO80/SWR1复合体组分。动物SWI/SNF类仅含有一个ARP (Baf53/BAP55),与酵母ARP4同源。虽然在结构上ARPs与actin相似均含有ATP/ADP-binding pocket (actin fold)结构,但除ARP4外,其他所有ARPs均没有像actin那样的ATPase活性。生物界所有ARPs可分为11类,其中ARP4~ARP9家族成员大多定位于细胞核[107]。拟南芥基因组编码9个ARPs (ARP2~ARP9,其中ARP4包含2个同源基因和),其中ARP4~ ARP9定位于细胞核[111]。CoIP-MS分析表明在拟南芥所有ARPs中, ARP4和ARP7为SWI/SNF复合体组分[39],而后续的研究表明ARP4也是INO80/ SWR1复合体的组分。
虽然在正常情况下,ARP4和ARP7定位于细胞核,但在有丝分裂过程,它们与染色质组装无关,且也可以定位于细胞质[109]。与相似,缺失突变导致植物不育,敲减突变体使植株生长发育受阻并出现早花、花衰老推迟和花器官发育异常等表型[67–68]。
2.2 INO80/SWR1复合体
基因最早从筛选调控酵母磷脂生物合成的突变体中分离,后续生化分析表明其与其他14个亚基组成复合体(表3)。随后,同源基因亦在酵母中发现,其主要催化组蛋白变体Htz1与H2A之间的交换。进一步研究表明,也与其他蛋白形成复合体,其中Rvb1、Rvb2、Arp4和actin亚基与INO80复合体共有(表3)。拟南芥中编码4个(包括PIE1、INO80、CHR19和CHR10)与INO80和Swr1类复合体催化亚基同源蛋白,目前仅对CHR19、INO80和PIE1的功能有所了解。
目前的研究认为拟南芥SWR1复合体最少由10个亚基组成(PIE1、SWC2、SWC4、SWC6、YAF9A、RVB1、RVB2A、RVB2B、ARP4和ARP6),其中PIE1是催化亚基[73],首先,PIE1可与SWC2、SWC6和ARP6以及组蛋白H2A相互作用调控植物开花和发育[73–74];其次,与PIE1形成的复合体在植物调节基因表达过程中也负责H2A.Z变体的交换[75–77]。然而有意思的是,在植物免疫过程中,和突变导致植物基本抗性降低,而突变则增加抗性[76],这提示在不同的生理过程中植物SWR1复合体亚基的功能可能是不一样的。进一步研究表明,通过促进和表达,抑制二者目标基因表达参与植物发育[77]。
对植物INO80/SWR1复合体非催化亚基RVB1、RVB2A和RVB2B的功能还知之甚少,但质谱鉴定表明,其能与SWC6形成复合体[75]。对SWC4的研究表明其参与植物雄配子和胚发育,且对叶片细胞的分化和伸长至关重要[75],同时质谱鉴定表明其与SWC6相互作用。利用SWC6-MYC融合蛋白进行CoIP结合质谱分析,SWC6与PIE1、SWC2、SWC4、YAF9A、RVB1、RVB2A、RVB2B、ARP4和ARP6形成复合体[75],其pre-messenger RNA通过Ski- interacting protein (SKIP)介导剪切调控、和表达参与植物开花时间决定[78]。ARP6除与PIE1和SWC6相互作用参与PIE1和SWC6相似功能外,还通过促进(DISRUPTED MEIOTIC cDNA1)表达促进雌配子的减数分裂[79], 进一步研究表明该过程是细胞色素P450基因(KLUH/ CYP78A5)通过促进表达而实现[80]。
拟南芥中有2个与酵母INO80/SWR1复合体亚基Yaf9同源的基因:和。和在功能上有部分冗余,其中能与SWC6相互作用,且通过提高组蛋白H4乙酰化水平促进其表达,从而降低和表达, 抑制开花[81]。最新的研究表明,和通过调节细胞伸长和分化来影响植株发育, 且其调控开花还存在一条独立于的途径[82], 虽然YAF9s可通过维持(并不促进) H2A.Z变体与基因区的结合和组蛋白H4乙酰化水平直接促进表达[82]。
2.3 CHD复合体
基因家族成员在结构上除含有DEAD/H- related ATP酶结构域外,其N端还含有一段串联chromodomains结构域。所有CHD蛋白分为3类:CHD1和CHD2在C端含有DNA结合区;CHD3和CHD4的C端缺少DNA结合区,其N端有一对PHD结构;CHD5~CHD9的C端含有多余结构。酵母基因组仅编码1个CHD蛋白CHD1,其可与组蛋白乙酰转移酶复合体的(SAGA and SLIK complexes)亚基相互作用,并通过其PHD结构域识别H3K4me3位点并与转录激活区结合促进下游基因转录延伸和剪切。
除催化亚基外,对拟南芥CHD复合体的其他亚基我们还知之甚少。拟南芥基因组编码4个CHD类催化亚基,分别为CHR5、PICKLE/CHR6、CHR4和CHR7。其中CHR5通过改变核小体“构象”正调控表达参与植物抗病过程[84]。在种子发育过程CHR5通过结合于和启动子促进二者表达,调控胚的发育[83]。有意思的是,在这个过程PICKLE拮抗CHR5的功能[83],这也与PICKLE在萌发阶段抑制种子胚性功能一致[112]。与AtBRM相似,PICKLE通过与拮抗PcG蛋白CLF (CURLY LEAF)的功能促进主根中分生组织的活性[83],而通过IAA14介导抑制侧根起始基因和表达抑制侧根的起始[86]。除在根中外, PICKLE还在叶片和花器官的发育过程拮抗CLF, 使其调控基因水平降低[113]。然而在14 d的幼苗中, PICKLE却促进其调控基因表达水平[87],这提示在植物不同的发育阶段PICKLE的功能不一样。进一步研究表明,PICKLE可与MADS-Box转录因子SEP3相互作用,提示在PICKLE影响花器官发育可能还依赖于SEP3[90]。PICKLE通过影响孢子体和配子体发育,调控植物的生殖生长[114],而HY5通过招募PICKLE提高下游细胞伸长基因H3K27me3水平抑制其表达,从而抑制下胚轴伸长[91]。另外, PICKLE分别通过与PIF3、BZR1和DELLAs相互作用参与暗形态建成、BR和GA信号传导过程[93], 从而将后三者整合在一起。进一步研究表明, PICKLE通过抑制DELLAs的活性参与植物生长发育阶段的转化[89]。除GA信号外,PICKLE还通过直接促进/的H3K27me3水平抑制其表达从而促进植物营养阶段转变(vegetative phase change)(从童年到成熟)[90]。除调控H3K27me3水平外, PICKLE还通过参与RNA指导的DNA甲基化(RdDM, RNA-directed DNA methylation)抑制下游基因表达[88]。
2.4 ISWI 复合体
ISWI蛋白(imitation SWI)最早从果蝇胚胎细胞提取的核小体重塑活性过程中分离鉴定的,其为一类DEAD/H-related ATP酶,且还含有SANT和SLIDE结构域。在体外加入模板,果蝇ISWI复合体促进转录,而在体内ISWI复合体亚基突变稳定抑制同源基因(homeotic gene)表达。目前对植物ISWI类复合体的组成还知之甚少,对催化亚基的研究表明其参与多种生理过程。其中CHR11和CHR17通过共同调控基因区(gene body)核小体之间的“距离”(nucleosome spacing)调节下游基因表达[99]。同时CHR11和CHR17还分别通过与含DDT结构域蛋白RINGLET1 (RLT1)和RLT2相互作用调控开花时间和花器官发育[100]。对该家族另一个成员DECREASE IN DNA METHYLATION 1 (DDM1)的研究表明,其主要参与DNA的甲基化过程。DDM1通过与Methyl- CpG binding domains (MBDs)相互作用改变后者在染色质上的定位影响DNA的甲基化[94],而一般认为MBDs通过影响组蛋白的甲基化参与DNA的甲基化。与RdDM不同,DDM1参与的DNA甲基化主要由DNA甲基转移酶CMT2介导[95],二者通过提高异染色质基因(如)不同区段的DNA甲基化水平维持基因沉默。事实上, DDM1是通过影响异染色质H1构象使CMT2更加容易结合于异染色质DNA而实现DNA甲基化修饰的[95]。进一步研究表明,DDM1还可促进甲基化的DNA在核小体上缠绕形成更为紧密的染色质结构[96]。有意思的是,突变体在连续种植5代内,其端粒大小与野生型相似,而在第6和以后的世代中端粒大小显著减小[97],这似乎说明在介导的端粒维持有一定的时间剂量效应。同时突变体亦出现叶片延迟衰老的表型,提示还参与叶片发育,然而对于具体机制仍不清楚[98]。
2.5 其他Snf2-like蛋白
目前对其他Snf2-like蛋白的功能还不是十分清楚,根据同源性分析这些蛋白(41个中的大部分)并不能归于以上复合体中的任意一种。但通过突变体表型分析表明某些成员也有重要的功能。Rad54- like家族成员DRD1/CHR35 (DEFECTIVE IN RNA- DIRECTED DNA METHYLATION1)与RDM1 (RNA-DIRECTED DNA METHYLATION 1)和DMS3 (DEFECTIVE IN MERISTEM SILENCIN G3)形成复合体(DDR complex),通过polymerase V介导参与RdDM[101]。而该家族另一成员Rad54/CHR25则通过同源重组途径参与DNA的修复[102]。
与拟南芥相比,其他物种仅有少数Snf2-like蛋白被研究。如水稻()同源基因和也参与DNA的甲基化[115],而CHD3家族成员通过GA信号途径调控水稻幼苗发育[116]。小番茄(‘Micro-Tom’)过表达基因()可抑制生长发育[117]。
3 结语
染色质重塑作为表观遗传调控的重要内容在真核生物DNA复制、转录、重组和DNA修复等过程中起到重要的作用。对植物染色质重塑复合体亚基(染色质重塑因子)的研究表明,它们参与细胞分化、器官发育和激素信号转导等多种生理过程。然而相对于酵母和动物,植物染色质重塑的研究还相对滞后,其作用机制并不十分清楚,最主要的问题是植物染色质重塑是如何识别其作用位点的。在酵母和动物中的研究结果表明该过程与组蛋白的修饰有关,这就为我们后续研究植物染色质重塑和组蛋白修饰之间的Cross-Talk指明了方向。同时综上所述,染色质重塑因子在不同的发育(或基因调节)过程与不同的因子(包括转录因子和核蛋白等)相互作用以及染色质重塑因子的翻译后修饰将极大拓展其调控基因表达的内涵。
[1] LI B, CAREY M, WORKMAN J L. The role of chromatin during transcription [J]. Cell, 2007, 128(4): 707–719. doi: 10.1016/j.cell.2007. 01.015.
[2] CLAPIER R, CAIRNS B R. The biology of chromatin remodeling complexes [J]. Annu Rev Biochem, 2009, 78(1): 273–304. doi: 10. 1146/annurev.biochem.77.062706.153223.
[3] HARGREAVES D C, CRABTREE G R. ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms [J]. Cell Res, 2011, 21(3): 396–420. doi: 10.1038/cr.2011.32.
[4] SMITH C L, HOROWITZ-SCHERER R, FLANAGAN J F, et al. Structural analysis of the yeast SWI/SNF chromatin remodeling complex [J]. Nat Struct Biol, 2003, 10(2): 141–145. doi: 10.1038/nsb888.
[5] EISEN J A, SWEDER K S, HANAWALT P C. Evolution of the SNF2 family of proteins: Subfamilies with distinct sequences and functions [J]. Nucl Acids Res, 1995, 23(14): 2715–2723. doi: 10.1093/nar/23. 14.2715.
[6] KIDDER B L, PALMER S, KNOTT J G. SWI/SNF-brg1 regulates self- renewal and occupies core pluripotency-related genes in embryonic stem cells [J]. Stem Cells, 2009, 27(2): 317–328. doi: 10.1634/stem cells.2008-0710.
[7] KASTEN M, SZERLONG H, ERDJUMENT-BROMAGE H, et al. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14 [J]. EMBO J, 2004, 23(6): 1348–1359. doi: 10.1038/sj.emboj.7600143.
[8] ZHOU Y G, GRUMMT I. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing [J]. Curr Biol, 2005, 15(15): 1434–1438. doi: 10.1016/j.cub. 2005.06.057.
[9] BOYER L A, LATEK R R, PETERSON C L. The SANT domain: A unique histone-tail-binding module? [J]. Nat Rev Mol Cell Biol, 2004, 5(2): 158–163. doi: 10.1038/nrm1314.
[10] THOMPSON P M, GOTOH T, KOK M, et al., a new member of the chromodomain gene family, is preferentially expressed in the nervous system [J]. Oncogene, 2003, 22(7): 1002–1011. doi: 10.1038/sj. onc.1206211.
[11] PRAY-GRANT M G, DANIEL J A, SCHIELTZ D, et al. Chd1 chromo- domain links histone H3 methylation with SAGA- and SLIK- dependent acetylation [J]. Nature, 2005, 433(7024): 434–438. doi: 10. 1038/nature03242.
[12] FLANAGAN J F, MI L Z, CHRUSZCZ M, et al. Double chromo- domains cooperate to recognize the methylated histone H3 tail [J]. Nature, 2005, 438(7071): 1181–1185. doi: 10.1038/nature04290.
[13] BAO Y H, SHEN X T. INO80 subfamily of chromatin remodeling complexes [J]. Mutat Res, 2007, 618(1/2): 18–29. doi: 10.1016/j.mrf mmm.2006.10.006.
[14] CORONA D F V, TAMKUN J W. Multiple roles for ISWI in trans- cription, chromosome organization and DNA replication [J]. Biochim Biophys Acta, 2004, 1677(1/2/3): 113–119. doi: 10.1016/j.bbaexp. 2003.09.018.
[15] WHITEHOUSE I, FLAUS A, CAIRNS B R, et al. Nucleosome mobilization catalysed by the yeast SWI/SNF complex [J]. Nature, 1999, 400(6746): 784–787. doi: 10.1038/23506.
[16] PHELAN M L, SCHNITZLER G R, KINGSTON R E. Octamer transfer and creation of stably remodeled nucleosomes by human SWI- SNF and its isolated ATPases [J]. Mol Cell Biochem, 2000, 20(17): 6380–6389. doi: 10.1128/mcb.20.17.6380-6389.2000.
[17] LORCH Y, ZHANG M C, KORNBERG R D. Histone octamer transfer by a chromatin-remodeling complex [J]. Cell, 1999, 96(3): 389–392. doi: 10.1016/s0092-8674(00)80551-6.
[18] MIZUGUCHI G, SHEN X T, LANDRY J, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex [J]. Science, 2004, 303(5656): 343–348. doi: 10.1126/science. 1090701.
[19] PAPAMICHOS-CHRONAKIS M, WATANABE S, RANDO O J, et al. Global regulation of H2A.Z localization by the INO80 chromatin- remodeling enzyme is essential for genome integrity [J]. Cell, 2011, 144(2): 200–213. doi: 10.1016/j.cell.2010.12.021.
[20] LUK E, RANJAN A, FITZGERALD P C, et al. Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome [J]. Cell, 2010, 143(5): 725–736. doi: 10.1016/j. cell.2010.10.019.
[21] JIN C Y, FELSENFELD G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z [J]. Genes Dev, 2007, 21(12): 1519–1529. doi: 10.1101/gad.1547707.
[22] JIN C Y, ZANG C Z, WEI G, et al. H3.3/H2A.Z double variant- containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions in the human genome [J]. Nat Genet, 2009, 41(8): 941–945. doi: 10.1038/ng.409.
[23] KUMAR S V, WIGGE P A. H2A.Z-containing nucleosomes mediate the thermosensory response in[J]. Cell, 2010, 140(1): 136–147. doi: 10.1016/j.cell.2009.11.006.
[24] FITZGERALD D J, DELUCA C, BERGER I, et al. Reaction cycle of the yeast Isw2 chromatin remodeling complex [J]. EMBO J, 2004, 23 (19): 3836–3843. doi: 10.1038/sj.emboj.7600364.
[25] SAHA A, WITTMEYER J, AIRNS B R. Chromatin remodelling: The industrial revolution of DNA around histones [J]. Nat Rev Mol Cell Biol, 2006, 7(6): 437–447. doi: 10.1038/nrm1945.
[26] ZOFALL M, PERSINGER J, KASSABOV S R, et al. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome [J]. Nat Struct Mol Biol, 2006, 13(4): 339–346. doi: 10. 1038/nsmb1071.
[27] STROHNER R, WACHSMUTH M, DACHAUER K, et al. A ‘loop recapture’ mechanism for ACF-dependent nucleosome remodeling [J]. Nat Struct Mol Biol, 2005, 12(8): 683–690. doi: 10.1038/nsmb966.
[28] ZHANG Y L, SMITH C L, SAHA A, et al. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC [J]. Mol Cell, 2006, 24(4): 559–568. doi: 10.1016/j.molcel.2006.10.025.
[29] CLAPIER C R, LÄNGST G, CORONA D F V, et al. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI [J]. Mol Cell Biol, 2001, 21(3): 875–883. doi: 10.1128/mcb.21.3.875-883.2001.
[30] SHIBA T, KAKUDA S, OKA S, et al. Molecular mechanisms in acceptor substrate recognition of a human glucuronyltransferase, GlcAT-P, an enzyme critical in the biosynthesis of the carbohydrate epitope HNK-1 [J]. Seikagaku, 2005, 77(2): 153–158.
[31] FERREIRA H, FLAUS A, OWEN-HUGHES T. Histone modifications influence the action of Snf2 family remodelling enzymes by different mechanisms [J]. J Mol Biol, 2007, 374(3): 563–579. doi: 10.1016/j. jmb.2007.09.059.
[32] KNIZEWSKI L, GINALSKI K, JERZMANOWSKI A. Snf2 proteins in plants: Gene silencing and beyond [J]. Trends Plant Sci, 2008, 13 (10): 557–565. doi: 10.1016/j.tplants.2008.08.004.
[33] JERZMANOWSKI A. SWI/SNF chromatin remodeling and linker histones in plants [J]. BBA Gene Struct Expr,2007, 1769(5/6): 330– 345. doi: 10.1016/j.bbaexp.2006.12.003.
[34] FARRONA S, HURTADO L, BOWMAN J L, et al. Thethaliana SNF2 homolog AtBRM controls shoot development and flowering [J]. Development, 2004, 131(20): 4965–4975. doi: 10.1242/ dev.01363.
[35] HURTADO L, FARRONA S, REYES J C. The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in[J]. Plant Mol Biol, 2006, 62(1/2): 291–304. doi: 10.1007/s11103-006-9021-2.
[36] ZHAO M L, YANG S G, CHEN C Y, et al.BREVI- PEDICELLUS interacts with the SWI2/SNF2 chromatin remodeling ATPase BRAHMA to regulateandexpression in control of inflorescence architecture [J]. PLoS Genet, 2015, 11(3): e1005125. doi: 10.1371/journal.pgen.1005125.
[37] WU M F, SANG Y, BEZHANI S, et al. SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors [J]. Proc Natl Acad Sci USA, 2012, 109(9): 3576–3581. doi: 10.1073/ pnas.1113409109.
[38] EFRONI I, HAN S K, KIM H J, et al. Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses [J]. Dev Cell, 2013, 24(4): 438–445. doi: 10.1016/j.devcel.2013.01.019.
[39] VERCRUYSSEN L, VERKEST A, GONZALEZ N, et al. ANGUSTI- FOLIA3 binds to swi/snf chromatin remodeling complexes to regulate transcription duringleaf development [J]. Plant Cell, 2014, 26(1): 210–229. doi: 10.1105/tpc.113.115907.
[40] ZHANG D, LI Y H, ZHANG X Y, et al. The SWI2/SNF2 chromatin- remodeling ATPase BRAHMA regulates chlorophyll biosynthesis in[J]. Mol Plant, 2017, 10(1): 155–167. doi: 10.1016/j.molp. 2016.11.003.
[41] LI C L, GU L F, GAO L, et al. Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in[J]. Nat Genet, 2016, 48(6): 687–693. doi: 10.1038/ng.3555.
[42] BRZEZINKA K, ALTMANN S, CZESNICK H, et al.FORGETTER1 mediates stress-induced chromatin memory through nucleosome remodeling [J]. Elife, 2016, 5: e17061. doi: 10.7554/elife. 17061.
[43] YANG S G, Li C L, ZHAO L M, et al. TheSWI2/SNF2 chromatin remodeling ATPase BRAHMA targets directly toand is required for root stem cell niche maintenance [J]. Plant Cell, 2015, 27(6): 1670–1680. doi: 10.1105/tpc.15.00091.
[44] ZHANG J J, LAI J B, WANG F G, et al. A SUMO ligase AtMMS21 regulates the stability of the chromatin remodeler BRAHMA in root development [J]. Plant Physiol, 2017, 173(3): 1574–1582. doi: 10. 1104/pp.17.00014.
[45] HAN S K, SANG Y, RODRIGUES A, et al. The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in[J]. Plant Cell, 2012, 24 (12): 4892–4906. doi: 10.1105/tpc.112.105114.
[46] PEIRATS-LLOBET M, HAN S K, GONZALEZ-GUZMAN M, et al. A direct link between abscisic acid sensing and the chromatin-remodeling ATPase BRAHMA via core ABA signaling pathway components [J]. Mol Plant, 2016, 9(1): 136–147. doi: 10.1016/j.molp.2015.10.003.
[47] WANG Z Y, MA Z Y, CASTILLO-GONZÁLEZ C, et al. SWI2/SNF2 ATPase CHR2 remodels pri-miRNAsserrate to impede miRNA production [J]. Nature, 2018, 557(7706): 516–521. doi: 10.1038/s415 86-018-0135-x.
[48] HAN P, LI Q, ZHU Y X. Mutation ofcauses meristem defects by failing to confineexpression to the organizing center [J]. Plant Cell, 2008, 20(6): 1482–1493. doi: 10. 1105/tpc.108.058867.
[49] KWON C S, CHEN C B, WAGNER D.is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in[J]. Genes Dev, 2005, 19(8): 992–1003. doi: 10. 1101/gad.1276305.
[50] LEEGGANGERS H A C F, FOLTA A, MURAS A, et al. Reduced seed germination inover-expressing SWI/SNF2 ATPase genes [J]. Physiol Plant, 2015, 153(2): 318–326. doi: 10.1111/ppl.12231.
[51] FOLTA A, SEVERING E I, KRAUSKOPF J, et al. Over-expression ofchromatin remodeling ATPase results in increased variability of growth and gene expression [J]. BMC Plant Biol, 2014, 14: 76. doi: 10.1186/1471-2229-14-76.
[52] MLYNÁROVÁ L, NAP J P, BISSELING T. The SWI/SNF chromatin- remodeling genemediates temporary growth arrest inupon perceiving environmental stress [J]. Plant J, 2007, 51(5): 874–885. doi: 10.1111/j.1365-313x.2007.03185.x.
[53] SARNOWSKI T J, RIOS G, JASIK J, et al. SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles duringdevelopment [J]. Plant Cell, 2005, 17(9): 2454–2472. doi: 10.1105/tpc.105.031203.
[54] BEZHANI S, WINTER C, HERSHMAN S, et al. Unique, shared, and redundant roles for theSWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED [J]. Plant Cell, 2007, 19(2): 403– 416. doi: 10.1105/tpc.106.048272.
[55] SAEZ A, RODRIGUES A, SANTIAGO J, et al. HAB1-SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in[J]. Plant Cell, 2008, 20(11): 2972–2988. doi: 10.1105/tpc.107.056705.
[56] ZHU Y Y, ROWLEY M J, BÖHMDORFER G, et al. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing [J]. Mol Cell, 2013, 49(2): 298–309. doi: 10. 1016/j.molcel.2012.11.011.
[57] LIU Z W, ZHOU J X, HUANG H W, et al. Two components of the RNA-directed DNA methylation pathway associate with MORC6 and silence loci targeted by MORC6 in[J]. PLoS Genet, 2016, 12(5): e1006026. doi: 10.1371/journal.pgen.1006026.
[58] HAN W X, HAN D L, HE Z P, et al. The SWI/SNF subunit SWI3B regulates IAMT1 expressionchromatin remodeling inleaf development [J]. Plant Sci, 2018, 271: 127–132. doi: 10.1016/j. plantsci.2018.03.021.
[59] SARNOWSKA E A, ROLICKA A T, BUCIOR E, et al. DELLA-inter- acting SWI3C core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk in[J]. Plant Physiol, 2013, 163(1): 305–317. doi: 10.1104/pp.113.223933
[60] TANG X R, HOU A F, BABU M, et al. TheBRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves [J]. Plant Physiol, 2008, 147(3): 1143–1157. doi: 10.1104/pp.108.121996.
[61] BRZESKI J, PODSTOLSKI W, OLCZAK K, et al. Identification and analysis of thegene, a member of thegene family [J]. Nucl Acids Res, 1999, 27(11): 2393–2399. doi: 10. 1093/nar/27.11.2393.
[62] SACHAROWSKI S P, GRATKOWSKA D M, SARNOWSKA E A, et al. SWP73 subunits ofSWI/SNF chromatin remodeling complexes play distinct roles in leaf and flower development [J]. Plant Cell, 2015, 27(7): 1889–1906. doi: 10.1105/tpc.15.00233.
[63] JÉGU T, DOMENICHINI S, BLEIN T, et al. A swi/snf chromatin remodelling protein controls cytokinin production through the regu- lation of chromatin architecture [J]. PLoS One, 2015, 10(10): e0138276. doi: 10.1371/journal.pone.0138276.
[64] JÉGU T, LATRASSE D, DELARUE M, et al. The BAF60 subunit of the swi/snf chromatin-remodeling complex directly controls the formation of a gene loop atin[J]. Plant Cell, 2014, 26(2): 538–551. doi: 10.1105/tpc.113.114454.
[65] JÉGU T, VELUCHAMY A, RAMIREZ-PRADO J S, et al. TheSWI/SNF protein BAF60 mediates seedling growth control by modulating DNA accessibility [J]. Genome Biol, 2017, 18: 114. doi: 10.1186/s13059-017-1246-7.
[66] CAMPI M, D’ANDREA L, EMILIANI J, et al. Participation of chromatin- remodeling proteins in the repair of ultraviolet-b-damaged DNA [J]. Plant Physiol, 2012, 158(2): 981–995. doi: 10.1104/pp.111.191452.
[67] KANDASAMY M K, DEAL R B, MCKINNEY E C, et al. Silencing the nuclear actin-related protein AtARP4 inhas multiple effects on plant development, including early flowering and delayed floral senescence [J]. Plant J, 2005, 41(6): 845–858. doi: 10.1111/j. 1365-313x.2005.02345.x.
[68] KANDASAMY M K, MCKINNEY E C, DEAL R B, et al.ARP7 is an essential actin-related protein required for normal embryo- genesis, plant architecture, and floral organ abscission [J]. Plant Physiol, 2005, 138(4): 2019–2032. doi: 10.1104/pp.105.065326.
[69] FRITSCH O, BENVENUTO G, BOWLER C, et al. The INO80 protein controls homologous recombination in[J]. Mol Cell, 2004, 16(3): 479–485. doi: 10.1016/j.molcel.2004.09.034.
[70] ZHANG C, CAO L, RONG L, et al. The chromatin-remodeling factor AtINO80 plays crucial roles in genome stability maintenance and in plant development [J]. Plant J, 2015, 82(4): 655–668. doi: 10.1111/ tpj.12840.
[71] HAN Y F, DOU K, MA Z Y, et al. SUVR2 is involved in trans- criptional gene silencing by associating with SNF2-related chromatin- remodeling proteins in[J]. Cell Res, 2014, 24(12): 1445– 1465. doi: 10.1038/cr.2014.156.
[72] DOKLÁDAL L, BENKOVÁ E, HONYS D, et al. An armadillo- domain protein participates in a telomerase interaction network [J]. Plant Mol Biol, 2018, 97(4/5): 407–420. doi: 10.1007/s11103-018- 0747-4.
[73] CHOI K, PARK C, LEE J, et al.homologs of components of the SWR1 complex regulate flowering and plant development [J]. Development, 2007, 134(10): 1931–1941. doi: 10.1242/dev.001891.
[74] MARCH-DÍAZ R, GARCIA-DOMÍNGUEZ M, FLORENCIO F J, et al. SEF, a new protein required for flowering repression in, interacts with PIE1 and ARP6 [J]. Plant Physiol, 2007, 143(2): 893– 901. doi: 10.1104/pp.106.092270.
[75] GÓMEZ-ZAMBRANO Á, CREVILLÉN P, FRANCO-ZORRILLA J M, et al.SWC4 binds DNA and recruits the SWR1 complex to modulate histone H2A.Z deposition at key regulatory genes [J]. Mol Plant, 2018, 11(6): 815–832. doi: 10.1016/j.molp.2018.03.014.
[76] BERRIRI S, GANGAPPA S N, KUMAR S V. SWR1 chromatin- remodeling complex subunits and H2A.Z have non-overlapping functions in immunity and gene regulation in[J]. Mol Plant, 2016, 9(7): 1051–1065. doi: 10.1016/j.molp.2016.04.003.
[77] CHOI K, KIM J, MULLER S Y, et al. Regulation of microRNA- mediated developmental changes by the SWR1 chromatin remodeling complex [J]. Plant Physiol, 2016, 171(2): 1128–1143. doi: 10.1104/pp. 16.00332.
[78] CUI Z B, TONG A Z, HUO Y Q, et al. SKIP controls flowering timethe alternative splicing ofpre-mRNA in[J]. BMC Biol, 2017, 15: 80. doi: 10.1186/s12915-017-0422-2.
[79] QIN Y, ZHAO L H, SKAGGS M I, et al. Actin-related protein6 regulates female meiosis by modulating meiotic gene expression in[J]. Plant Cell, 2014, 26(4): 1612–1628. doi: 10.1105/tpc. 113.120576.
[80] ZHAO L H, CAI H Y, SU Z X, et al.suppresses megasporocyte cell fate through SWR1-mediated activation ofexpression in[J]. Proc Natl Acad Sci USA, 2018, 115(3): E526-E535. doi: 10.1073/pnas.1716054115..
[81] ZACHARAKI V, BENHAMED M, POULIOS S, et al. Theortholog of the YEATS domain containing protein YAF9a regu- lates flowering by controlling H4 acetylation levels at thelocus [J]. Plant Sci, 2012, 196: 44–52. doi: 10.1016/j.plantsci.2012.07.010.
[82] CREVILLÉN P, GÓMEZ-ZAMBRANO Á, LÓPEZ J A, et al.YAF9 histone readers modulate flowering time through nuA4- complex-dependent H4 and H2A.Z histone acetylation atchromatin [J]. New Phytol, 2019, 222(4): 1893–1908. doi: 10.1111/nph.15737.
[83] SHEN Y, DEVIC M, LEPINIEC L, et al. Chromodomain, helicase and DNA-binding CHD1 protein, CHR5, are involved in establishing active chromatin state of seed maturation genes [J]. Plant Biotechnol J, 2015, 13(6): 811–820. doi: 10.1111/pbi.12315.
[84] ZOU B H, SUN Q, ZHANG W L, et al. Thechromatin- remodeling factor CHR5 regulates plant immune responses and nucleosome occupancy [J]. Plant Cell Physiol, 2017, 58(12): 2202– 2216. doi: 10.1093/pcp/pcx155.
[85] AICHINGER E, VILLAR C B R, DI MAMBRO R, et al. The CHD3 chromatin remodeler PICKLE and polycomb group proteins antago- nistically regulate meristem activity in theroot [J]. Plant Cell, 2011, 23(3): 1047–1060. doi: 10.1105/tpc.111.083352.
[86] FUKAKI H, TANIGUCHI N, TASAKA M. PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity duringlateral root initiation [J]. Plant J, 2006, 48(3): 380–389. doi: 10.1111/j.1365-313x.2006.02882.x.
[87] ZHANG H, BISHOP B, RINGENBERG W, et al. The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27 [J]. Plant Physiol, 2012, 159(1): 418–432. doi: 10.1104/pp.112.194878.
[88] YANG R, ZHENG Z M, CHEN Q, et al. The developmental regulator PKL is required to maintain correct DNA methylation patterns at RNA- directed DNA methylation loci [J]. Genome Biol, 2017, 18(1): 103. doi: 10.1186/s13059-017-1226-y.
[89] PARK J, OH D H, DASSANAYAKE M, et al. Gibberellin signaling requires chromatin remodeler PICKLE to promote vegetative growth and phase transitions [J]. Plant Physiol, 2017, 173(2): 1463–1474. doi: 10.1104/pp.16.01471.
[90] XU M L, HU T Q, SMITH M R, et al. Epigenetic regulation of vegetative phase change in[J]. Plant Cell, 2016, 28(1): 28–41. doi: 10.1105/tpc.15.00854.
[91] JING Y J, ZHANG D, WANG X, et al.chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation [J]. Plant Cell, 2013, 25(1): 242–256. doi: 10.1105/tpc.112.105742.
[92] SMACZNIAK C, IMMINK R G H, MUIÑO J M, et al. Charac- terization of MADS-domain transcription factor complexes inflower development [J]. Proc Natl Acad Sci USA, 2012, 109(5): 1560–1565. doi: 10.1073/pnas.1112871109.
[93] ZHANG D, JING Y J, JIANG Z M, et al. The chromatin-remodeling factor PICKLE integrates brassinosteroid and gibberellin signaling during skotomorphogenic growth in[J]. Plant Cell, 2014, 26(6): 2472–2485. doi: 10.1105/tpc.113.121848.
[94] ZEMACH A, LI Y, WAYBURN B, et al. DDM1 bindsmethyl-CpG binding domain proteins and affects their subnuclear localization [J]. Plant Cell, 2005, 17(5): 1549–1558. doi: 10.1105/tpc. 105.031567
[95] ZEMACH A, KIM M Y, HSIEH P H, et al. Thenucleo- some remodeler DDM1 allows DNA methyltransferases to access h1- containing heterochromatin [J]. Cell, 2013, 153(1): 193–205. doi: 10. 1016/j.cell.2013.02.033.
[96] LYONS D B, ZILBERMAN D. DDM1 and lsh remodelers allow methylation of DNA wrapped in nucleosomes [J]. Elife, 2017, 6: e30 674. doi: 10.7554/eLife.30674.
[97] XIE X Y, SHIPPEN D E. DDM1 guards against telomere truncation in[J]. Plant Cell Rep, 2018, 37(3): 501–513. doi: 10.1007/ s00299-017-2245-6.
[98] CHO E J, CHOI S H, KIM J H, et al. A mutation in plant-specific swi2/snf2-like chromatin-remodeling proteins, DRD1 and DDM1, delays leaf senescence in[J]. PLoS One, 2016, 11(1): e0146826. doi: 10.1371/journal.pone.0146826.
[99] LI G, LIU S J, WANG J W, et al. ISWI proteins participate in the genome-wide nucleosome distribution in[J]. Plant J, 2014, 78(4): 706–714. doi: 10.1111/tpj.12499.
[100] LI G, ZHANG J W, LI J Q, et al. Imitation switch chromatin remo- deling factors and their interacting RINGLET proteins act together in controlling the plant vegetative phase in[J]. Plant J, 2012, 72(2): 261–270. doi: 10.1111/j.1365-313X.2012.05074.x.
[101] LAW J A, AUSIN I, JOHNSON L M, et al. A protein complex required for polymerase V transcripts and RNA-directed DNA methy- lation in[J]. Curr Biol, 2010, 20(10): 951–956. doi: 10. 1016/j.cub.2010.03.062.
[102] HIRAKAWA T, HASEGAWA J, WHITE C I, et al. RAD54 forms DNA repair foci in response to DNA damage in living plant cells [J]. Plant J, 2017, 90(2): 372–382. doi: 10.1111/tpj.13499.
[103] ARCHACKI R, BUSZEWICZ D, SARNOWSKI T J, et al. BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin-mediated responses in[J]. PLoS One, 2013, 8(3): e58588. doi: 10.1371/journal.pone.0058588.
[104] FARRONA S, HURTADO L, MARCH-DÍAZ R, et al. Brahma is required for proper expression of the floral repressorin[J]. PLoS One, 2011, 6(3): e17997. doi: 10.1371/journal.pone. 0017997.
[105] LI C L, CHEN C, GAO L, et al. Theswi2/snf2 chromatin remodeler BRAHMA regulates polycomb function during vegetative development and directly activates the flowering repressor gene[J]. PLoS Genet, 2015, 11(1): e1004944. doi: 10.1371/journal.pgen. 1004944.
[106] WALLEY J W, ROWE H C, XIAO Y M, et al. The chromatin remo- deler SPLAYED regulates specific stress signaling pathways [J]. PLoS Pathog, 2008, 4(12): e1000237. doi: 10.1371/journal.ppat.1000237.
[107] JOHNSON K C M, XIA S T, FENG X Q, et al. The chromatin remo- deler SPLAYED negatively regulates SNC1-mediated immunity [J]. Plant Cell Physiol, 2015, 56(8): 1616–1623. doi: 10.1093/pcp/pcv087.
[108] GENG F, CAO Y, LAURENT B C. Essential roles of snf5p in snf-swi chromatin remodeling[J]. Mol Cell Biol, 2001, 21(13): 4311– 4320. doi: 10.1128/MCB.21.13.4311-4320.2001.
[109] MULLER J, OMA Y, VALLAR L, et al. Sequence and comparative genomic analysis of actin-related proteins [J]. Mol Biol Cell, 2005, 16(12): 5736–5748. doi: 10.1091/mbc.e05-06-0508.
[110] KANDASAMY M K, DEAL R B, MCKINNEY E C, et al. Plant actin-related proteins [J]. Trends Plant Sci, 2004, 9(4): 196–202. doi: 10.1016/j.tplants.2004.02.004.
[111] KANDASAMY M K, MCKINNEY E C, MEAGHER R B. Cell cycle-dependent association ofactin-related proteins AtARP4 and AtARP7 with the nucleus [J]. Plant J, 2003, 33(5): 939– 948. doi: 10.1046/j.1365-313x.2003.01691.x.
[112] LI H C, CHUANG K, HENDERSON J T, et al. PICKLE acts during germination to repress expression of embryonic traits [J]. Plant J, 2005, 44(6): 1010–1022. doi: 10.1111/j.1365-313x.2005.02602.x.
[113] AICHINGER E, VILLAR C B R, FARRONR S, et al. CHD3 proteins and polycomb group proteins antagonistically determine cell identity in[J]. PLoS Genet, 2009, 5(8): e1000605. doi: 10.1371/ journal.pgen.1000605.
[114] CARTER B, HENDERSON J T, SVEDIN E, et al. Cross-Talk between sporophyte and gametophyte generations is promoted by CHD3 chromatin remodelers in[J]. Genetics, 2016, 203(2): 817–829. doi: 10.1534/genetics.115.180141
[115] HIGO H, TAHIR M, TAKASHIMA K, et al.(decrease in DNA methylation) genes in rice () [J]. Mol Genet Genom, 2012, 287(10): 785–792. doi: 10.1007/s00438-012-0717-5.
[116] MA X D, MA J, ZHAI H H, et al. CHR729 is a CHD3 protein that controls seedling development in rice [J]. PLoS One, 2015, 10(9): e0138934. doi: 10.1371/journal.pone.0138934.
[117] FOLTA A, BARGSTEN J W, BISSELING T, et al. Compact tomato seedlings and plants upon overexpression of a tomato chromatin remodelling ATPase gene [J]. Plant Biotechnol J, 2016, 14(2): 581– 591. doi: 10.1111/pbi.12400.
Functions of ATP-dependent Chromatin Remodeling Factors in Plant Development
PENG Xiu-ling1,2, WANG Jian-hao1,2, YANG Song-guang1*
(1. Key Laboratory of South China Agricultural Plant Molecular Analysis and Genetic Improvement, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou 510650, China; 2.University of Chinese Academy of Sciences, Chinese Academy of Sciences,Beijing 100049, China)
In eukaryotic cells, the ATP-dependent chromatin remodeling complexes utilize the energy of ATP to disrupt nucleosome DNA contacts, move nucleosomes along DNA, and remove or exchange nucleosomes. They thus make DNA/chromatin available to proteins that need to access DNA or histones directly during cellular processes. The first chromatin remodeling complex was found in yeast, containing at least 11 subunits by biochemical analysis. However, the chromatin remodeling complexes in plants are less known. The studies on plant chromatin remodeling factors were reviewed, which would provide insights into the involvement of plant chromatin remodeling in development.
Chromatin remodeling factor; Epigenetic regulation; Function; Plant
10.11926/jtsb.4070
2019–03–25
2019–05–17
广东省杰出青年基金项目(2016A030306047); 广州市珠江科技新星项目(201610010138); 国家自然科学基金项目(31672161)资助
This work was supported by the Guangdong Natural Science Funds for Distinguished Young Scholars (Grant No. 2016A030306047); the Pearl River Science and Technology Nova Program of Guangzhou (Grant No. 201610010138), and the National Natural Science Foundation of China (Grant No. 31672161).
E-mail: yangsongguang@scbg.ac.cn

