Nrf2-inducing and HMG-CoA reductase inhibitory activities of a polyphenol-rich fraction of Guazuma ulmifolia leaves
Sulistiyani✉, Syamsul Falah, Wulan Tri Wahyuni, Dimas Andrianto, Arthur Ario Lelono, Waras Nurcholis,Valeri Mossine, Mark HanninkTropical Biopharmaca Research Center of Excellence, IPB University (Bogor Agricultural University), Bogor, Indonesia Department of Biochemistry, Faculty of Mathematics and Natural Sciences, IPB University (Bogor Agricultural University), Bogor, Indonesia Department of Chemistry, Faculty of Mathematics and Natural Sciences, IPB University (Bogor Agricultural University), Bogor, Indonesia Chemistry Research Center, Indonesian Institute of Sciences, Serpong, Indonesia Biochemistry Department, Christopher S. Bond Life Sciences Center, University of Missouri-Columbia, Columbia-MO, USA
Keywords:Antioxidant signaling HMG-CoA reductase inhibitor Polyphenols Antioxidant-related transcription factor Nrf2 Reporter gene cell-based assay
ABSTRACT Objective: To fractionate and identify polyphenols from Guazuma ulmifolia Lam. leaves, and to explore their antioxidant, 5-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase inhibitory, and Nrf2 modulatory activities.Methods: The 1,1-diphenyl-2-picrylhydrazyl assay was used to evaluate the antioxidant activity of a polyphenolic fraction of the extract of Guazuma ulmifolia Lam. leaves. THP-1 gene reporter cell lines constructed with a transcriptional response element specific for Nrf2 and a minimal promoter for the firefly luciferase-green fluorescent protein transgene were used to determine the effect of the polyphenolic fraction on the Nrf2 signaling pathway.Furthermore, an assay of HMG-CoA reductase inhibitory activity was performed by using a commercial enzyme kit. Polyphenolic compounds were identified by liquid chromatographytandem mass spectrometry.Results: The polyphenolic fraction showed fairly strong antioxidant activity [IC50 = (14.90± 4.70) μg/mL] and inhibited HMG-CoA reductase activity by 69.10%, which was slightly lower than that by pravastatin (84.37%) and quercetin (84.25%). Additionally, the polyphenolic fraction activated the Nrf2 antioxidant signaling pathway at 500 μg/mL. Eleven subfractions resulting from the column chromatography separation of the polyphenolic fraction also showed relatively strong antioxidant activities (IC50: 17.46-217.14 μg/mL). The subfraction(F6) stimulated the Nrf2 signaling pathway and had HMG-CoA reductase inhibitory activity (65.43%). Moreover, the subfraction contained two main flavonoids: quercetin and quercimeritrin.Conclusions: The polyphenolic fraction of Guazuma ulmifolia could induce antioxidant genes via the Nrf2/antioxidant regulatory elements pathway, and is a promising candidate for an inhibitor of HMG-CoA reductase.
1. Introduction
Extensive research has demonstrated that oxidative stress has a critical role in the pathogenesis of atherosclerosis, the underlying cause of coronary heart disease and stroke. Previously, antioxidants have been shown to decrease the progression of experimental atherosclerosis[1]. Recent advances in understanding the molecular aspects of oxidative stress signaling in human cells have led to the identification of plant-derived compounds that target the intracellular antioxidant signaling pathway controlled by the nuclear factor erythroid 2-related factor 2 (Nrf2) transcription factor[2].The Nrf2 transcription factor itself binds to DNA elements known as antioxidant regulatory elements (AREs), which control gene expression associated with the cellular response toward antioxidants.The pharmacological activation of Nrf2 by various compounds, such as flavonoids and polyphenols among others, has been proposed for use in the prevention of disease related to oxidative stress[3]. Plantderived polyphenolic compounds are being investigated in clinical trials[4], which has led to heightened awareness in our society of the beneficial use of plant-derived antioxidants for treatment of disease.Guazuma ulmifolia (G. ulmifolia) Lam., called “Jati Belanda” in Indonesia, has been used in various Indonesian herbal products[5].Traditionally, G. ulmifolia has been used to treat diabetes[6],inflammation[7], and gastrointestinal diseases[8]that have been correlated with oxidative stress[9]. Pharmacologically, G. ulmifolia has been reported to possess antioxidant[10], antidiabetic[11], antiinflammatory[12], anticholinesterase and antifungal[13], hypotensive and vasorelaxant[14], and anticancer[15]activities. Recently,phytochemicals extracted from G. ulmifolia have been reported in the literature and identified as polyphenolic compounds, such as phenols, flavonoids[16], and flavanocoumarin[12]. Therefore, the objective of this study was to fractionate and identify polyphenolic compounds from G. ulmifolia leaves, and to explore their antioxidant,5-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase inhibitory, and Nrf2-modulatory activities.
2. Materials and methods
2.1. Chemicals and reagents
The 1,1-diphenyl-2-picrylhydrazyl (DPPH), pravastatin, and quercetin were purchased from Sigma-Aldrich (USA). Extraction solvents and phytochemical reagents were of analytical grade and purchased from Merck (Germany). Dimethyl sulfoxide (DMSO) and the HMG-CoA reductase kit were purchased from Sigma-Aldrich(Catalog No.CS1090).
2.2. Plant materials, extraction, and fractionation
Leaves of G. ulmifolia were obtained from the collections of the Tropical Biopharmaca Research Center, Bogor Agricultural University (IPB University), Bogor, Indonesia. G. ulmifolia leaves were identified by Mr. Taopik Ridwan, and voucher specimen has been deposited at IPB University (BMK2017020001). The leaves were collected on February 2017 during a wet season. After washing,the leaves were dried at 50 ℃ and then pulverized into powder form(100 mesh).
The dried leaf powder of G. ulmifolia (225 g) was macerated (3 × 24 h) in 96% ethanol (1:5). Subsequently, the filtrates were concentrated by using a vacuum rotary evaporator to determine recovery. The concentrated ethanolic crude extracts were partitioned sequentially at room temperature with n-hexane to remove the nonpolar fraction and with chloroform to remove the semi-polar ones. The remaining ethanolic fractions, which contained the crude polyphenolic compounds (8.55 g), were stored in a dark bottle until use. The ethanolic extract and crude polyphenolic fraction were analyzed by qualitative phytochemical screening using a standard procedure[17].The total flavonoids content was then determined in the crude polyphenolic fraction by using aluminum trichloride reagents[18],and the value was expressed in mg quercetin equivalent per g crude polyphenol fraction (mg QE/g). The radical scavenging activities of the crude polyphenolic fraction were measured according to a previous method with slight modification[19]by using DPPH as a radical model, and the absorbance at 517 nm was measured by using a microplate reader (EPOCH). Quercetin was used as the standard.Radical scavenging activity was expressed as the half maximal inhibitory concentration (IC50) of DPPH.
Before the fractionation by silica gel column chromatography,the crude polyphenolic fraction was subjected to thin layer chromatography using several solvent systems to determine the most suitable chromatographic method by optimization of the mobile phase. Subsequently, a portion of the polyphenol-containing ethanolic fractions of G. ulmifolia was chromatographed on a silica gel column (1.5 × 50 cm) by using a stepwise gradient with an ethyl acetate/acetone (1:1)-methanol solvent system. A total of 11 fractions were collected, and their antioxidant activities were measured by DPPH assay, as described previously. The subfraction with the best results in both antioxidant activities and yield was selected for use in various analyses of HMG-CoA reductase inhibition, Nrf2 signaling pathway activation, and bioactivity by liquid chromatographytandem mass spectrometry (LC-MS/MS).
2.3. HMG-CoA reductase inhibitory activity of G. ulmifolia fractions
The in vitro HMG-CoA reductase inhibitory activities of the polyphenolic fractions and selected subfraction (F6) (50 μg/mL)were determined by spectrophotometric assay using quercetin and pravastatin as the reference compounds. The activity of the enzyme was analyzed by using an HMG-CoA reductase assay kit from Sigma-Aldrich (St. Louis, MO, USA) according to the manufacturer’s recommendations. The concentration of the purified enzyme (Sigma) was 0.50-0.70 mg protein/mL. For evaluation of HMG-CoA reductase inhibitory activity, 3 μL of a sample [50 μg/mL in DMSO] or control (absence) was added into 546 μL of a buffer solution mixture, pH 7.5. Subsequently, 12 μL of nicotinamide adenine dinucleotide phosphate, 36 μL of HMG-CoA substrate,and 3 μL of HMG-CoA reductase were added together and diluted to a total volume of 600 μL. The rates of nicotinamide adenine dinucleotide phosphate consumed were expressed as the HMGCoA reductase activity and were monitored every 15 s for up to 6 min at 37 ℃ by spectrophotometry (340 nm). The percent inhibitory enzymatic activity was calculated by using the following formula:
% Inhibition=[(Δ Absorbance control- Δ Absorbance sample)/Δ Absorbance control]×100%
2.4. Cell-based assay for Nrf2-signaling pathway of G.ulmifolia fractions
The polyphenolic fractions and selected subfraction (F6) were assayed by a reporter gene cell-based assay using a cultured THP-1 human monocytic cell line[20]. THP-1 cells represent a non-adherent monocytic cell line widely used as a model of hematopoietic progenitor cells. In this study, we used the original THP-1 and THP-1 reporter cell line THP.R05F containing the Nrf2 activation reporter. The original THP-1 cell line was used as a blank control. As a positive control, a Michael addition acceptor triterpenoid CDDOMe, as a specific inducer of Nrf2-mediated ARE activity, was used.Briefly, a suspension of the original THP-1 or the other reporter cell line was cultured in complete RPMI 1640 medium at densities between 200 000 and 1 000 000 cells/mL. All cultured cells were incubated in a cell culture incubator set at 37 ℃, 5% CO2, and 100%humidity. To transform THP monocytes into adherent macrophages,the cells were resuspended in differentiation Gibco Serum-Free Macrophage Medium (SFM) and seeded at 500 000 cells/mL in 96-well culture plates. The cells were then treated with phorbol-12-myristate-13-acetate (50 ng/mL) in the SFM for 2 d, and all cells were transformed into macrophages and became adherent cells. The modified macrophages were further incubated for 1 d in adaptation medium (Corning Phenol Red-free, SFM) before experimentation.On the day of experimentation, solutions of the experimental compounds were prepared in Corning Phenol Red-free SFM. In a typical experiment, differentiated THP reporters seeded at 0.5 ×105- 1 × 105cells/well in 96-well plates were treated with samples in DMSO at increasing concentrations (100-2 500 μg/mL for polyphenolic fraction, 5-5 000 μg/mL for selected subfraction F6)or quercetin (20 μM) and bardoxolone (CDDO) (20 μM) in 100 μL of Corning Phenol Red-free SMF for 14-24 h in the standard cell culture condition (37 ℃, 5% CO2, 100% humidity). Samples without the fraction or subfraction and those with DMSO were used as controls. After treatment, the reporter cells were washed with phosphate buffer saline and lysed in 100 μL of the luciferase reporter lysing buffer (Promega) by keeping the cells overnight in the refrigerator. The lysates’ fluorescence was measured the following day by using a fluorescence/bioluminescence plate reader. The lysates’ fluorescence was measured at 482 (9)/512(17)nm wavelength (width) set up by using a black U-bottomed 96-well plate. After the fluorescence was read, aliquots of the lysates were transferred into a white U-bottomed 96-well plate followed by addition of the luciferase substrate (Promega) and kinetic luminescence measurements in the wells (the plate was read four times at 1-minute intervals). All of the analyses were performed by using a fluorescence and luminescence plate reader with Enspire Manager software (Perkin-Elmer).
Fluorescence values of green fluorescent protein were used for both evaluation of relative cell proliferation and normalization of the reporter luciferase activities in wells. The cellular proliferation rate was calculated from the fluorescence data of green fluorescent protein: cellular proliferation rate = [sample relative fluorescence units (RFU)-blanks RFU]/(control RFU-blank RFU). The transcription factor activity was derived from the luminescence data normalized by the fluorescence of the same wells: transcription factor activity = [(sample relative luminescence units (RLU) - blanks RLU) / (sample RFU - blanks RFU)] / [(control RLU - blank RLU) /(control RFU - blank RFU)].
2.5. LC-MS/MS characterization of G. ulmifolia subfractions
The subfraction F6 that showed the best result in both radical scavenging activity and yield was subjected to LC-MS/MS at the instrumentation research facility, Indonesian Institute of Sciences,Indonesia. The LC-MS/MS analyses were performed on a Waters Xevo G2-XS QT of, Acquity UPLC I CLASS equipped with a highperformance tandem of mass analyzer (MS2), autosampler, column oven, in-line degasser, and binary pump. The LC-MS/MS spectra were compared with standards and literature to tentatively identify peaks and bioactive compounds.
2.6. Statistical analysis
Values were expressed as mean ± SD and the results were subjected to ANOVA and the Tukey test. Statistical significance was accepted for P < 0.05.
3. Results
3.1. Phytochemical screening of the ethanolic extract and crude polyphenolic fraction of G. ulmifolia
Phytochemical screening results showed that flavonoids, tannins,saponins, and steroids were present except for alkaloids and triterpenoids in the ethanolic extract of G. ulmifolia leaves. Further fractionation which yielded a crude polyphenolic fraction still contained phytochemical constituents including flavonoids and tannins, whereas the alkaloids, saponin, steroids, and triterpenoids were not detected. The total flavonoids content of the crude polyphenolic fraction was (15.91 ± 0.14) mg QE/g extract.

Table 1. Radical scavenging activity and yield of polyphenolic fractions of Guazuma ulmifolia leaves.
3.2. Radical scavenging activity of G. ulmifolia fractions
The fractionation process using column chromatography yielded 160 subfractions of G. ulmifolia, which then were pooled according to the similarity of the components, as verified by silica gel thin layer chromatography. The pooled fractions resulted in 11 pooled subfractions. The antioxidant activities of the crude polyphenolic fraction and 11 pooled subfraction are presented in Table 1. The polyphenolic fraction and subfractions demonstrated lower radical scavenging activity than that of the quercetin standard (IC50of 3.08 μg/mL). The results showed that the polyphenolic fraction had the highest antioxidant activity and fraction yield, with values of 14.90 μg/mL and 518 mg, respectively. In the pooled subfractions, F6 was selected for subsequent experiments because of its relatively strong antioxidant activity (IC50of 23.19 μg/mL) and yield (58.3 mg).
3.3. HMG-CoA reductase inhibitory activity of G. ulmifolia fractions
The HMG-CoA reductase inhibitory activities of the polyphenolic fraction and F6 subfraction were 69.10% and 65.43%, respectively,whereas the HMG-CoA reductase inhibitory activities of the pravastatin(84.37%) and quercetin (84.25%) standards were significantly higher(P < 0.05) than the polyphenolic fraction and F6 subfraction.
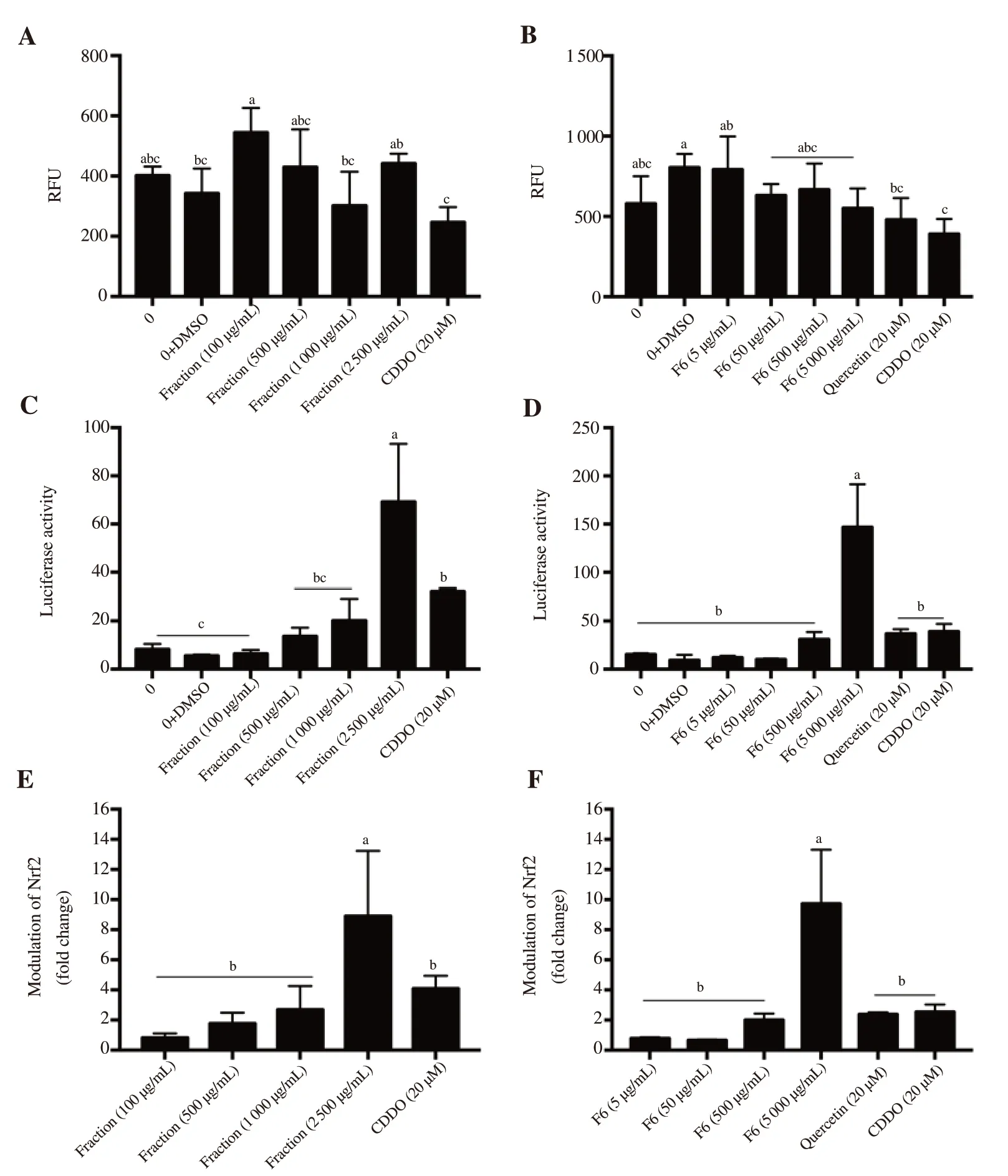
Figure 1. Effect of (A, C, E) polyphenolic fraction and (B, D, F) selected subfraction (F6) of Guazuma ulmifolia leaves on relative fluorescence unit (RFU),Nfr2 transcription factor activity and activation of the Nrf2 signaling pathway. Values are presented as mean ± SD (n = 4) and are followed by letters indicating significant differences determined by the Tukey test at P < 0.05. DMSO: dimethyl sulfoxide. CDDO: bardoxolone.
3.4. Cell-based assay for activation of the Nrf2-signaling pathway of the G. ulmifolia fractions
The results of the polyphenolic fraction and selected subfraction(F6) activating the Nrf2 signaling pathway are shown in Figure 1. There was no significant (P < 0.05) difference in the RFU of polyphenolic fraction and subfraction F6 when compared with the control (concentration of 0 μg/mL). This indicated that the polyphenolic fraction and subfraction F6 of G. ulmifolia leaves did not affect the cell viability. The polyphenolic fraction and selected subfraction (F6) showed a concentration-dependent increase in Nrf2 transcription activity. Significantly higher (P < 0.05)Nrf2 transcription activity was recorded for 2 500 μg/mL of the polyphenolic fraction (8.90-fold) and 5 000 μg/mL of the selected subfraction (F6, 9.72-fold) than that of the quercetin (2.38-fold)and CDDO (4.09-fold and 2.54-fold for Figure 1E and Figure 1F,respectively) standards.
3.5. LC-MS/MS characterization of active subfractions selected
Several peaks in the chromatogram and the structures of the compounds of subfraction F6 are shown in Figure 2. Quercetin and quercimeritrin, flavonoid glucosides, were identified in F6 of G.ulmifolia leaves (Table 2).

Table 2. Identification and mass spectrometric properties of the flavonoid compounds from subfraction F6 of Guazuma ulmifolia leaves.
4. Discussion
G. ulmifolia has been declared to possess biological activities and to be a rich source of polyphenol compounds. In this study, we investigated the radical scavenging activity, HMG-CoA reductase inhibitory activity, and Nrf2 signaling pathway activation of a polyphenolic fraction from an ethanolic extract of G. ulmifolia leaves.Flavonoids were identified in the ethanolic extract and polyphenolic fractions. Quercetin and quercimeritrin, which are flavonoid compounds, were identified in subfraction F6. Polyphenolic fractions and subfraction F6 showed radical scavenging, HMG-CoA reductase inhibitory, and modulation of Nrf2 activities.
Secondary metabolites, such as polyphenolic compounds, are abundant in the plant, which potentially have pharmacological activities that may be useful in human health[21]. Preliminary phytochemical analyses of a methanolic extract of G. ulmifolia leaves were reported by Kaneria et al[22]which showed the presence of tannins, triterpenoids, steroids, saponins, and cardiac glycosides. In the present study, the ethanolic extract obtained from G. ulmifolia leaves showed the presence of tannins, flavonoids, saponins, and steroid compounds, and the polyphenolic fraction showed the presence of flavonoids and tannins. These results were consistent with those of a previous study that reported the presence of flavonoids, saponins, steroids, and tannins in an ethanolic extract of G. ulmifolia[5]. In addition, the total flavonoids value reported in this study [(15.91 ± 0.14) mg QE/g extract] was much higher than that in a previous study (0.80 mg QE/g extract)[13]using a sample from Brazil. The differences in the flavonoid content observed between this study and studies in the literature might be because of genetic and geo-agricultural factors.
The antioxidant activities of plant extracts generally have been determined by scavenging free radicals, such as DPPH[23]. In this study,the antioxidant activities in polyphenolic fractions and subfractions ofG. ulmifolia were evaluated by determining their capacity to scavenge DPPH radicals. The polyphenolic fraction showed higher radical scavenging activity (IC50= 14.90 μg/mL) than those of subfractions(IC50values ranged from 17.46 to 217.14 μg/mL). Recently, a study has also presented that G. ulmifolia possessed antioxidant activities, which were in agreement with these results[24]. Another study reported that the essential oils of G. ulmifolia leaves showed higher antioxidant activity (IC50= 7.61 μg/mL) than that found in the present study[25].The yields of the subfractions ranged from 1.1 to 306.0 mg. Based on the antioxidant activities and subfraction yields, we concluded that subfraction F6 had the best result of radical scavenging activity and subfraction yield among all tested subfractions. Therefore, F6 was used as a sample for HMG-CoA reductase inhibitory activity,Nrf2 signaling pathway activation, and LC-MS/MS analysis.

Figure 2. MS base peak chromatograms of selected subfraction (F6) from (A) Guazuma ulmifolia leaves, (B) quercetin standard, (C) and quercimeritrin standard as well as the chemical structures of the compounds.
HMG-CoA reductase, a key enzyme in the regulation of cholesterol biosynthesis, has often been targeted by drugs, such as statins, in clinical studies of hypercholesterolemia and atherosclerosis[26].In vitro HMG-CoA reductase inhibitory activity measured in the present study demonstrated potent HMG-CoA reductase inhibitory potential of the polyphenolic fraction of G. ulmifolia leaves and its subfraction F6, with values of 69.10% and 65.43%, respectively.However, the HMG-CoA reductase inhibitory activities of the polyphenolic fraction of G. ulmifolia leaves and its subfraction F6 were significantly lower than those of pravastatin (84.37%) and quercetin (84.25%). Rahmania et al.[27] reported higher HMG-CoA reductase inhibitory activity (79.85%-94.42%) in the methanolic fraction of G. ulmifolia leaves. These findings show that the polyphenolic fraction and subfraction F6 of G. ulmifolia leaves also had reasonably potent HMG-CoA reductase inhibitory activity. The present in vitro data for quercetin is consistent with the in vivo data reported previously[28], which showed that the effect of quercetin as an inhibitor of the HMG-CoA enzyme improved the lipid profiles of hypercholesterolemic rats (decreased concentrations). The most relevant observation to our experiments, however, was that the antioxidant activities protecting against lipid peroxidation increased in the treated animals[29]. This latter effect can be attributed to the effect of quercetin via the Nrf2 antioxidant signaling mechanism,which will induce activation of some antioxidant enzymes within the cells.
Nrf2 is a transcription factor that has a vital role in maintaining cellular homeostasis, particularly when cells are exposed to chemical or oxidative stress. Nrf2 is able to regulate expression of various antioxidant proteins, detoxification enzymes, and xenobiotic transporters as well as contributes to several cellular functions, such as lipid synthesis, inflammation, proliferation, and differentiation[30].In this study, the Nrf2 transcription factor was evaluated by using THP-1-derived reporter gene cells. The THP-1 cells are the most commonly used human monocytic leukemia line and are very versatile since they can be further differentiated into a number of phenotypes, including macrophages[31]. Moreover, the use of THP-1 cells in this research is particularly relevant and appropriate since THP-1 macrophages also have previously been used in a foam cell formation study in the pathogenesis of atherosclerosis[32]. Induction of the Nrf2 signaling mechanism in these cells will contribute to a better understanding of the mechanism of antioxidation in the prevention of macrophage foam cell formation in the early stage of atherosclerotic lesion development. The pharmacological activation of Nrf2 by various compounds, such as flavonoids and polyphenols among others, has been proposed for use in the prevention of diseases related to oxidative stress[33]. Compounds derived from medicinal plants, such as curcumin and resveratrol, have been reported to induce the activity of Nrf2[34].
Thus, experiments were conducted to obtain a better understanding of how a polyphenolic fraction and a subfraction F6 of G. ulmifolia leaves could modulate a molecular signaling pathway, which may yield a novel therapeutic strategy for stress-related disease, such as cardiovascular disease. The polyphenolic fraction and a subfraction F6 of G. ulmifolia leaves exhibited modulation of Nrf2 transcription factor at concentrations ≥ 500 μg/mL. At 500 μg/mL, the result of subfraction was more similar to those of positive controls than polyphenolic fraction. This effect was concentration dependent, as demonstrated by the increasing response when the polyphenolic fractions and subfraction F6 were increased. The flavonoids contained in polyphenolic fractions and subfraction F6 of G. ulmifolia leaves might serve as activators of the Nrf2 signaling pathway.Several studies have shown that flavonoids activate Nrf2 nuclear factor through some targets of the signaling pathway, including phosphatidylinositol-3 kinase, protein kinase B, and protein kinase A[35,36]. Several studies have reported the mechanism of translocation of Nrf2 into the nucleus, such as through stress signals[37]and the ARE-linked cytoprotective transcriptional response[38]. The mechanism by which Nrf2 mediates the transcriptional response of cells to oxidative stress is through its translocation into the nucleus following or accompanying its activation by electrophiles or reactive oxygen species[39]. Curcumin has been reported to upregulate Nrf2 nuclear translocation by increasing the nuclear expression level of Nrf2[40]. Therefore, it will be interesting for future research to investigate if this same mechanism also applies to the flavonoids of G. ulmifolia.
The modulation of Nrf2 factor has been shown to be correlated with cellular pathologies, including cardiovascular, cancer, and neurodegeneration disease[41]. Therefore, Nrf2 overexpression can be caused by cancer inducers. A previous experiment demonstrated that an ethanolic extract of G. ulmifolia was able to induce apoptosis in yeast cells by increasing the gene expression responsible for the yeast apoptosis mechanism, thus suggesting that our samples were not cancer inducers[42].
Further, subfraction F6 of G. ulmifolia leaves was subjected to LCMS/MS, and two flavonoid compounds were identified by their mass spectra: quercetin (m/z 303.050) and quercimeritrin (m/z 465.102).These compounds were confirmed against LC-MS/MS standards.As reported by recent studies, quercetin and quercimeritrin possess antioxidant[43], vasorelaxant[44], and HMG-CoA reductase inhibitory[45]activities. Moreover, a work has also reported that quercetin activated the Nrf2 antioxidant response element signaling pathway[46]. Therefore, the antioxidant, HMG-CoA reductase, and Nrf2 modulatory properties observed in our study might have been due to the presence of quercetin and quercimeritrin in subfraction F6 of G. ulmifolia leaves.
In conclusion, the polyphenolic fraction of an ethanolic extract of G. ulmifolia leaves and subfraction F6 showed strong radical scavenging activity, inhibited HMG-CoA reductase activity and modulated Nrf2 activity. In addition, metabolite identification of the F6 subfraction by LC-MS/MS revealed the presence of quercetin and quercimeritrin.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Funding
This work was funded by an International Collaboration Research Grant provided by the Ministry of Research, Technology, and Higher Education of the Republic of Indonesia (No. 011/SP2H/LT/DRPM/VIII/2017), and University of Missouri-Columbia, MO (USA).
 Asian Pacific Journal of Tropical Biomedicine2019年9期
Asian Pacific Journal of Tropical Biomedicine2019年9期
- Asian Pacific Journal of Tropical Biomedicine的其它文章
- Determining endemic values of cutaneous leishmaniasis in Iranian Fars province by retrospectively detected clusters and receiver operating characteristic curve analysis
- Smallanthus sonchifolius roots ameliorate non-alcoholic fatty liver disease by reducing redox imbalance and hepatocyte damage in rats fed with a high fructose diet
- Pyrrolidine dithiocarbamate and saxagliptin ameliorate ulcerative colitis in rats
- Antidiabetic effect of Opuntia dillenii seed oil on streptozotocin-induced diabetic rats
- Phytochemical analysis and antibacterial activities of Eleutherine bulbosa (Mill.) Urb.extract against Vibrio parahaemolyticus
- Information for Authors Asian Pacific Journal of Tropical Biomedcine
