Cloning and Bioinformatics Analysis of TLR4 Gene from Wuzhishan Pig in Hainan
Yan ZHANG Lili HUANG Hailong LIU Zhemin LIN Shuyi TAN






Abstract In order to obtain the TLR4 gene sequence of Hainan Wuzhishan pig and analyze its molecular structure characteristics, its genomic sequence was cloned and sequenced by PCR and subjected to related bioinformatics analysis with the genomic sequence of the TLR4 gene of pig in GenBank as a reference sequence. The full-length DNA sequence of Wuzhishan pig TLR4 gene was 10 435 bp in length, of which CDS was 2 526 bp in length and encoded 841 amino acids. Compared with the common pig TLR4 gene sequence published by GenBank, there were 38 mutations, of which two mutations were in the CDS region. The results of homology analysis showed that the amino acid sequence of the TLR4 gene of Wuzhishan pig shared 99.9%, 80.7%, 79.9%, 74.8%, 74.7%, 73.1%, 73.0%, 72.7%, 62.5% and 43.4% homology with those of common pig, cattle, sheep, dog, horse, macaque, human, chimpanzee, mouse and chicken, respectively. The protein encoded by the TLR4 of Wuzhishan pig belonged to secreted protein and transmembrane protein. It was hydrophilic, and had 8 glycosylation modification sites, 17 serines (ser), 7 threonines (Thr) and 8 tyrosine (TYR) phosphorylation sites. The amino acid phylogenetic tree analysis showed that the TLR4 genes of different species are highly conserved during evolution. This result lays a foundation for further study of the function of TLR4 gene.
Key words Wuzhishan pig; TLR4 gene; Clone; Bioinformatics analysis
Toll-like receptor 4 (TLR4) is a kind of pattern recognition receptor that specifically recognizes lipopolysaccharide (LPS) or lipid A (the active component of LPS) expressed by Gram-negative bacteria, rapidly initiate immune response, induce release of various inflammatory mediators, and activate relevant immune cells to resist microbial invasion, thereby playing an important role in the immune response and inflammation of animal organisms[1]. Currently, the TLR4 gene is a research hotspot in the field of immunity and infection. Numerous studies have shown that deletion or mutation of the TLR4 gene may induce a decrease or loss of responsiveness of cells to LPS, resulting in increased susceptibility to disease in the body. For example, the loss of TLR4 gene may aggravate lung injury in patients with chronic obstructive pneumonia[2]; the occurrence of proximal tumor and metastatic disease is associated with mutation of TLR4 gene[3]; and TLR4 gene mutation increases the risk of chronic pyelonephritis in children[4].
Hainan Wuzhishan pig is one of the important experimental animal resources, as well as the first choice for the Experimental Miniature Swine Germplasm Resource Center, with high social and economic value. At present, there have been no reports on the cloning of TLR4 gene and prediction of related functional structures in Hainan Wuzhishan pig. Therefore, in this study, in June 2016, the mutation sites in the TLR4 gene of Wuzhishan pig as the research object were searched by clone sequencing method, and its sequence was analyzed and functionally predicted by bioinformatics software. This study will provide reference for further study on the function of the TLR4 gene in Wuzhishan pig and the relationship between its genetic variation and immune diseases.
Materials and Methods
Materials
Test animals and sample collection
Ten Wuzhishan pigs from Yongfa Pig Farm of Hainan Academy of Agricultural Sciences were selected as the research object. Blood samples were collected and stored in the Hainan Key Lab of Tropical Animal Reproduction & Breeding and Epidemic Disease Research at -20 ℃.
Main reagents
Blood genomic DNA extraction kit, PCR product purification and recovery kit and recombinant plasmid extraction kit, all purchased from Tiangen Biotech (Beijing) Co., Ltd.; Ex Taq DNA polymerase, T4 DNA ligase, EcoR I, Hind III and pMD18-T vector kit, all purchased from TaKaRa Biotechnology (Dalian) Co. Ltd.; cloning host strain JM109, provided by the laboratory of Institute of Animal Science and Veterinary Medicine, Hainan Academy of Agricultural Sciences.
Primer Design and Synthesis
According to the TLR4 gene sequence in GenBank (accession number: AY753179.1), 11 pairs of primers (Table 1) were designed and synthesized by Sangon Biotech (Shanghai) Co., Ltd.
Methods
DNA extraction
The genomic DNA was extracted according to the instructions of the blood genomic DNA extraction kit, and a portion of the DNA sample was diluted to 100 ng/μl and stored at -20 ℃ until use.
PCR amplification
The PCR amplification system included DNA template 2.0 μl, 10 × PCR Buffer (without Mg2+) 2.5 μl, dNTP Mixture (2.5 mmol/L) 2.0 μl, MgCl2 (25 mmol/L) 2.0 μl, Ex Taq polymerase (5 U/μl) 0.5 μl, P1 (10 pmol/L) 0.5 μl, P2 (10 pmol/L) 0.5 μl and sterile deionized H2O 15 μl. The PCR reaction procedure was started with pre-denaturation at 95 ℃ for 4 min, followed by 30 cycles of denaturation at 94 ℃ for 50 s, annealing for 50 s (the annealing temperature was shown in Table 1) and extension at 72 ℃ for 2 min, and completed by extension at 72 ℃ for 10 min. The PCR product was detected by 1.2% agarose gel electrophoresis.
Gene cloning and sequence determination
The purified PCR product was recovered by a gel recovery kit and ligated into the pMD18-T easy vector, which was then transformed into JM109 competent cells. The obtained JM109 competent cells were coated onto the plates containing IPTG, X-gal and ampicillin, and cultured at 37 ℃. White spots were inoculated in LB medium, and the culture liquid was subjected to PCR and digestion. The correct clones were transported to and sequenced by Sangon Biotech (Shanghai) Co., Ltd.
Bioinformatics analysis
The DNAMAN software was used to align and splice the DNA sequences. NCBIs ORF Finder program was used to find the open reading frame of the sequence. A phylogenetic tree was constructed with the MEGA (6.0) software. The ProtParam (http://web.expasy.org/protparam/) Program, Protscale software, NetPhos 2.0 TMHMM program and NetNGlyc (http: //www.cbs.dtu.dk /services/NetPhos/) program were used to analyze protein characteristics. The prediction of signal peptides was performed by the Signal P 4.1 Server program. The transmembrane domains were predicted using TMHMM (http://www.cbs.dtu.dk/services/TMHMM) software. The SOPMA program (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl Page = npsa_sopma.html) and SWISS-MODEL (http://swissmodel.expasy.org/) were used to analyze protein structure. Subcellular localization was performed using the PSORT I1 program.
Results and Analysis
Cloning of Wuzhishan pig TLR4 gene
A total of 11 pairs of primers were used to amplify the TLR4 gene of Wuzhishan pig, and DNA sequences of 370, 1 406, 1 477, 1 960, 1 428, 1 661, 1 618, 1 284, 1 248, 1 396, and 1 433 bp were obtained, respectively (Fig. 1).
Genomic sequence analysis of Wuzhishan pig TLR4 gene
After sequencing and splicing, the full-length 10 435 bp DNA sequence of Wuzhishan pig TLR4 gene was obtained, of which CDS was 2 526 bp in length and encoded 841 amino acids (Fig. 2). Compared with the common pig TLR4 gene sequence published by GenBank, there were 38 mutations (Table 2). There were two mutations in the CDS region sequence, of which one was a nonsense mutation at the 7 779 site where G was mutated to A, causing no amino acid changes, and the other was a sense mutation, at the 7 781 site where G was mutated to A, causing the amino acid change from arginine to histidine.
Homology alignment of TLR4 amino acid sequence in Wuzhishan pig and phylogenetic tree construction
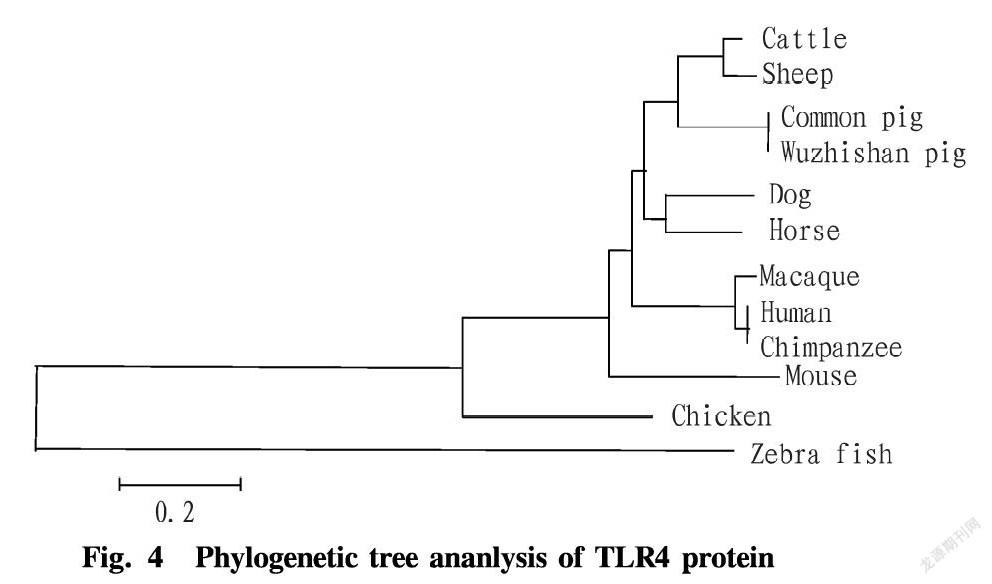
As can be seen from Fig. 3, the amino acid sequence of the TLR4 gene of Wuzhishan pig shared higher homology with the amino acid sequences of common pigs (accession number: AAW82895.1), cattle (accession number NP_776623.5), and sheep (accession number: NP_001129402.1) published in GenBank, at 99.9%, 80.7% and 79.9%, respectively. It shared slightly lower homology with dog (accession number: NP_001002950.2), horse (accession number: NP_001093239.2), macaque (registration number: NP_001032169.1), human (accession number: NP_003257.1), chimpanzee (accession number: BAG55036.1) and mouse (accession number: NP_067272.1), being 74.8%, 74.7%, 73.1%, 73%, 72.7% and 62.5%, respectively. And the homology with chickens ((accession number: ACR26315.1) was the lowest at 43.4%. The phylogenetic tree was constructed using the NJ method of MEGA 6.0 software. The results (Fig. 3) showed that the TLR4 gene of Wuzhishan pig was closely related to common pigs, sequentially followed by sheep, cattle, dog, horse, macaque, human, chimpanzee, mouse and chicken.
Analysis of physical and chemical properties of TLR4 protein in Wuzhishan pig
According to the online prediction of ProtParam software, TLR4 protein of Wuzhishan pig had the molecular weight of 96 312.93 u, with the isoelectric point of 6.12, and the molecular formula was C4 378H6 808N1 146O1 241S30. Its amino acid composition and proportions were alanine (Ala, 4.4%), arginine (Arg, 3.4%), asparagine (Asn, 6.7%), aspartic acid (Asp, 4.6%), cysteine (Cys, 2.3%), glutamine (Gln, 5.2%), glutamic acid (Glu, 5.7%),
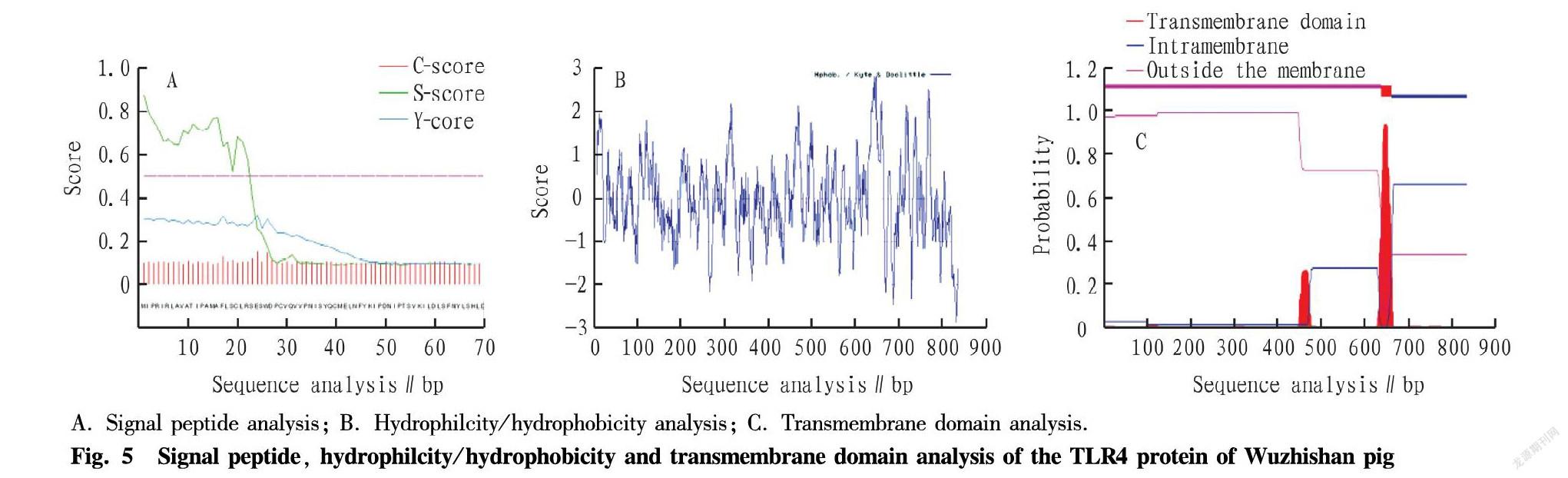
glycine (Gly, 3.7%), histidine (Hi, 3.9%), isoleucine (Ile, 5.6%), leucine (Leu, 16.1%), lysine (Lys, 5.1%), methionine (Met, 1.3%), phenylalanine (Phe, 6.7%), proline (Pro, 3.7%), serine (Ser, 8.6%), threonine (Thr, 3.8%), tryptophan (Trp, 1.1%), tyrosine (Tyr, 2.5%), valine (Val, 5.7%), pyrrolysine (Pyl, 0.0) and selenocysteine (Sec, 0.0). SignalP-4.1 software predicted that the TLR4 protein of Wuzhishan pig had a signal peptide sequence, indicating that it was a secreted protein (Fig. 5A). According to the analysis of Prot Scale software, the maximum value of hydrophobic amino acids among the 841 amino acids of the TLR4 protein of Wuzhishan pig was 2.811, and the minimum value of the hydrophilic amino acid was -2.867. Most of the amino acids were hydrophilic, and the protein belonged to the hydrophilic protein (Fig. 5B). Analyzing with the Tmhmm software, it was found that Wuzhishan pig TLR4 had a transmembrane domain and belonged to transmembrane protein (Fig. 5C).
Prediction of TLR4 protein structure in Wuzhishan pig
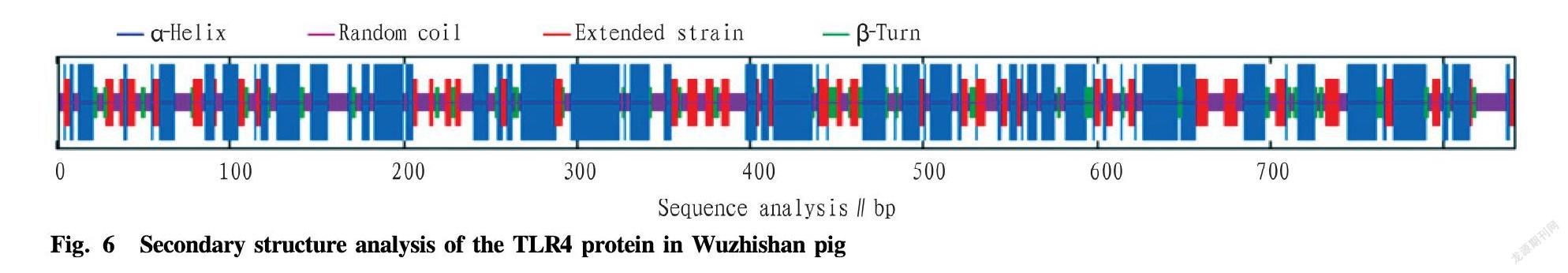
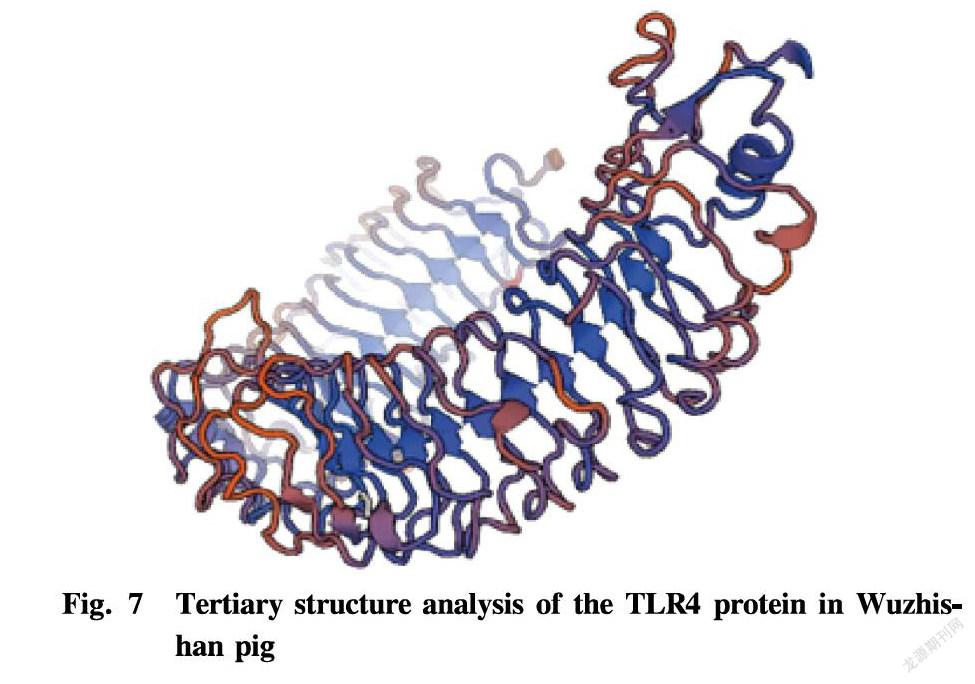
The SOPMA program was used to predict the secondary structure of the TLR4 protein in Wuzhishan pig. The TLR4 protein of Wuzhishan pig was mainly composed of α-helix (47.44%), extended strain (17.36%), β-turn (8.44%) and random coil (26.75%) (Fig. 6). The tertiary structure of the TLR4 protein in Wuzhishan pig was predicted with the SWISS-MODEL online software, and it was found that the TLR4 protein was mainly composed of α-helix and random coil (Fig. 7), which is consistent with the prediction of the secondary structure.
Subcellular localization, phosphorylation and glycosylation sites of the TLR4 protein in Wuzhishan pig
The functional prediction analysis and localization analysis showed that the TLR4 protein of Wuzhishan pig was distributed in endoplasmic reticulum, mitochondria, vesicular secretion system, cytoplasm, extracellular matrix and plasma membrane, and the proportions were 33.30%, 22.20%, 11.10%, 11.10%, 11.10% and 11.11%, respectively, with the most distribution in the endoplasmic reticulum. The phosphorylation analysis showed that there were 17 serine (ser), 7 threonine (Thr) and 8 tyrosine (TYR) phosphorylation sites in the TLR4 protein of Wuzhishan pig. The glycosylation site analysis indicated that there were 8 glycosylation sites in the TLR4 protein of Wuzhishan pig, which were located at the 35, 205, 238, 282, 309, 526, 575 and 625 positions of the amino acid sequence, respectively.
Discussion and Conclusions
TLR4 plays a key role in innate immune system regulation and inflammation and tissue damage repair. It not only can identify bacterial lipopolysaccharides, but also can induce endogenous mediator activation[5]. Although a large number of studies have selected the TLR4 gene as a candidate gene for disease resistance, the exact relationship between the mutation of TLR4 gene and diseases is still unclear.
In this study, the TLR4 gene of Wuzhishan pig was cloned by PCR. The sequence analysis showed that the full-length DNA sequence of Wuzhishan pig TLR4 gene was 10 435 bp in length, of which CDS was 2 526 bp in length and encoded 841 amino acids. Compared with the common pig TLR4 gene sequence published by GenBank, there were 38 mutations. There were two mutations in the CDS region sequence, of which one was a nonsense mutation, and the other was a sense mutation, causing the amino acid change from arginine to histidine, suggesting that there might be some differences in the biological functions of the TLR4 gene between Wuzhishan pig and normal pig. The results of homology analysis showed that the amino acid sequence of TLR4 gene of Wuzhishan pig shared 99.9%, 80.7%, 79.9%, 74.8%, 74.7%, 73.1%, 73.0%, 72.7%, 62.5%, 43.4% homology with those of common pig, cattle, sheep, dog, horse, macaque, human, chimpanzee, mouse and chicken, respectively, indicating that the TLR4 gene is relatively conserved among different species, but there is still some species specificity.
The function of proteins is closely related to their spatial structures[6]. Studies have shown that the TLR4 protein of Wuzhishan pig is mainly composed of α-helix and random coil. Among them, α-helix is the most common secondary element in proteins, and random coil is usually constructed into a special functional site or an enzyme active site[7]. The TLR4 protein of Wuzhishan pig had a signal peptide sequence, indicating that it was a secreted protein; there was a transmembrane region, indicating that it was a transmembrane protein, which might be involved in signal transduction; and there are many hydrophilic amino acids in the protein, so the protein was hydrophilic.
Phosphorylation and glycosylation are common modifications after the translation of biological proteins, and are widely involved in the regulation of many life activities[8-9], such as interfering with the bodys response to antigens and affecting the proliferation of viruses in vitro[10]. After phosphorylation and glycosylation site analysis of the TLR4 protein in Wuzhishan pig, it was found that there were 17 serine, 7 threonine and 8 tyrosine phosphorylation sites in the TLR4 protein of Wuzhishan pig, and there were 8 glycosylation modification sites, but the role of these phosphorylation and glycosylation sites in this gene needs further investigation.
References
[1]ZHOU Y, LI YB, ZHOU B, et al. Inflammation and apoptosis: dual mediator role for toll-like receptor 4 in the development of necrotizing enterocolitis[J]. Inflammatory Bowel Diseases, 2017, 23(1): 44-56.
[2]KNOBLOCH J, CHIKOSI SJ, YANIK S, et al. A systemic defect in Toll-like receptor 4 signaling increases lipopolysaccharide-induced suppression of IL-2-dependent T-cell proliferation in COPD[J]. American Journal of Physiology. Lung Cellular and Molecular Physiology, 2016, 310(1): L24- L39.
[3]ELLIOTT DR, PERNER J, LI XA, et al. Impact of mutations in Toll-like receptor pathway genes on esophageal carcinogenesis[J]. PLoS Genetics, 2017, 13(5): e1006808.
[4]HARSHMAN VP, KRYUCHKO TO, KOLENKO IO, et al. Role of genetic mutations in development of immunological and clinical disorders in children with chronic pyelonephritis[J]. Wiadomosci Lekarskie, 2017, 70(1): 47-51.
[5]MCKEOWN-LONGO PJ, HIGGINS PJ. Integration of canonical and noncanonical pathways in TLR4 signaling:complex regulation of the wound repair program[J]. Advances in Wound Care, 2017, 6(10): 320-329.
[6]WANG JY, ZHU SG, XU CF. Biochemistry[M]. Beijing: Higher Education Press, 2002. (In Chinese)
[7]DING D, LIU ZZ, LI YY, et al. Cloning and bioinformatics analysis of Ross 308 chicken IL-2 gene[J]. Heilongjiang Animal Science and Veterinary Medicine, 2015, 12(23): 90-94, 314. (In Chinese)
[8]ZHOU L, GU JX. N-glycosylation site identification method and non-canonical N-glycosylation sequence[J]. Chinese Bulletin of Life Sciences, 2011, 23(6): 605-611. (In Chinese)
[9]RUAN BJ, DAI P, WANG W, et al. Progress in post-translational modification of proteins[J]. Chinese Journal of Cell Biology, 2014, 36(7): 1027-1037. (In Chinese)
[10]WEI W, XIANG JZ, LUO JH. Study and evaluation of glycosylation of recombinant protein expression in eukaryotic cells[J]. Chinese Journal of New Drugs, 2014, 23(15): 1743-1748. (In Chinese)
- 农业生物技术(英文版)的其它文章
- Analysis of the Southern China Tilapia Production and Economic Benefits of Different Breeding Patterns in 2018
- Dynamic Monitoring and Control Measures of Spodoptera frugiperda (J.E.Smmith) in Low Latitude Plateau Sugarcane Areas
- Control Effects of a New Sex Pheromone Trap and Biological Agents on Sesamia inferens Walker and Argyroploce schistaceana (Snellen)
- Comparative Study on Grain Cadmium Content and Yield in Different Rice Varieties
- Simulation Experiment of Air Temperature Variation in Multi-film Covering at Night
- Identification of Growth-promoting Bacteria from Rhizosphere of Pastures and Their Effects on Growth of Lotus corniculatus L.

