Screening and Verification of Genes Specifically Responding to Continuous Cropping Obstacle in Rehmannia glutinosa L.
Xiaoran WANG Zheng LI1, Fangming LIU Weixi LI Yuhong WANG Xinjian CHEN




Abstract Rehmannia glutinosa L. is one of the important medicinal crops in China. Continuous cropping obstacle severely restricts the yield and quality of R. glutinosa, but its molecular mechanism is still unclear. In this study, with widely-planted "Wen 85-5" as an experiment material, based on the digital gene expression profiling (DGE) data of previous five stress treatments (continuous cropping, phenolic acid, salt, drought and waterlogging) and the first cropping and continuous cropping treatments of R. glutinosa in five different periods (seedling period, elongation period, early expanding period, middle expanding period and later expanding period), 80 candidate genes (|log2 ratio|≥1, FDR<0.001) specifically responding to continuous cropping obstacle in R. glutinosa were screened. Functional analysis revealed that the differentially expressed genes were involved in the secretion and endocytosis of root cells, which may suggest that the recognition and absorption of allelopathic autotoxins by the roots of R. glutinosa is an important factor that restricts the development of roots in continuous cropping of R. glutinosa. In order to accurately lock genes specifically responding to continuous cropping obstacle in R. glutinosa, continuous cropping soil extract and ferulic acid and p-hydroxybenzonic acid were used to treat aseptic plantlets of R. glutinosa, respectively, and it was confirmed through qRT-PCR that the expression levels of some genes under phenolic acid treatment changed more severely than that under the continuous cropping soil extract treatment, and four key genes involved in the response of R. glutinosa to continuous cropping were finally locked. This study lays a foundation for further exploration of the molecular mechanism of continuous cropping obstacle.
Key words Rehmannia glutinosa L.; Continuous cropping obstacle; Response gene; Soil extract; Phenolic acid treatment
Rehmannia glutinosa L. is a perennial plant of Scrophulariaceae. It is one of the most important medicinal herbs in China. Its cultivation has long been affected by continuous cropping obstacle. Continuous cropping results in poor resistance in R. glutinosa, severe pests and diseases and decreased yield and quality[1]. In addition, a variety of medicinal plants also have problems with continuous cropping[2-4], which seriously restricts the sustainable development of agriculture and Chinese medicine industry.
A large number of studies have confirmed that the toxic effects of allelochemicals are key factors causing plant continuous cropping obstacle[5-7]. Exogenous phenolic acids were added to the soil to treat R. glutinosa, the results showed that the five exogenous phenolic acids and their mixtures had a certain degree of toxic effect on R. glutinosa, and the inhibition of the phenolic acid mixtures on R. glutinosa was similar to that of R. glutinosa suffered from continuous cropping obstacle[8]. Zhang et al.[9]applied HPLC to track the dynamic changes of five phenolic acids closely related to allelopathy in the rhizosphere soil first planted with R. glutinosa and those planted with R. glutinosa for different time. It was found that ferulic acid had the strongest inhibitory effect on R. glutinosa suffered from continuous cropping obstacle. The accumulation of allelochemicals not only reduces root activity, inhibits root elongation and hinders normal expansion of R. glutinosa, but also stimulates or induces changes in soil microbial populations, alters micro-ecological environment, and indirectly affects the growth of R. glutinosa[10-12].
Omics studies have found that continuous cropping obstacle leads to disturbances in multiple cell signaling systems, including the "calcium signaling" system known as the second messenger and the ethylene signaling system, and interferes normal expression of R. glutinosa genes and miRNAs[13-15]. Meanwhile, continuous cropping obstacle shows typical "autotoxicity", that is, continuous cropping toxicity only directs at the same plant, but not at other kinds of plants[16]. Therefore, it is speculated that there are genes specifically responding to continuous cropping in R. glutinosa, which participates in the transmission of continuous cropping obstacle signaling system and affects the normal growth and development of R. glutinosa. In this study, with the omics analysis data of previous five stress treatments (continuous cropping, phenolic acid, salt, drought and waterlogging) and the first cropping and continuous cropping treatments of R. glutinosa in five periods[4,17], 80 candidate genes (|log2 ratio|≥1, FDR<0.001) for specific response to continuous cropping obstacle in R. glutinosa were screened, and functional analysis revealed that continuous cropping disrupted the endocytosis of R. glutinosa root cells and affected the exchange process between root cells and foreign substances. With aseptic plantlets treated with soil extracts and phenolic acid as verification materials, qRT-PCR technology was applied to further accurately lock the genes that specifically respond to the continuous cropping obstacle in R. glutinosa, which lays a foundation for further analysis of the molecular mechanism of R. glutinosa.
Materials and Methods
Preparation of aseptic R. glutinosa plantlets
The R. glutinosa cultivar "Wen 85-5" was used as an experimental material. The roots at the middle expanding period (about 1-2 cm in diameter) were thoroughly cleaned, soaked in Tween-20 for more than 30 min, and rinsed with running water (about 1 h). The roots were cut into small pieces with bud eyes, which were disinfected with 75% ethanol for 10 min, rinsed with sterile water for 2 times, treated with 0.1% HgCl2 for 10 min, and finally rinsed with sterile water for 5-6 times. In a sterile environment, damaged tissues on the surface were cut off during the surface sterilization process, and the parts with bud eyes were cut into 1.5-2.0 cm pieces, which were put on the filter paper to absorb excess water, and then put into 1/2 MS basal medium. The regenerated buds were transferred to a new MS basal medium 15 d later and subcultured once 30 d.
Soil extract stress treatment
The field soil that was not planted with R. glutinosa for ten years was used as control (first cropping soil), and the field soil that had been planted with R. glutinosa for one year served as a treatment (continuous cropping soil). The soils were dried naturally in a cool place, and then smashed through a 40 mesh sieve (with a pore diameter of 0.45 mm). Each soil was mixed with distilled water at room temperature at 5∶4 (500 g of soil plus 400 ml of distilled water), obtaining the mixture, which was shaken at 180 rpm in a shaker for 24 h, and stood for 24 h. Each supernatant was filtered with filter paper, giving a soil extract, which was evaporated and concentrated into 50 ml. The concentrates were vacuum filtered with 0.22 μm filter membrane on a clean bench for 3 times. The 50 ml of filtrates were preheated in a 65 ℃ water bath and mixed with 250 ml of MS medium which had been sterilized at 121 ℃ for 20 min, respectively. The mixtures were added into sterile culture flasks. Aseptic plantlets were selected from the rooting medium 1/2MS+NAA (0.5 mg/L) and transplanted into the media added with the extracts. The transplanted plants were cultured in an incubator at 25 ℃ with 2 000 lx illumination (14 h/d) (GXZ280, Ningbo Jiangnan Instrument Factory) and observed. Sampling was performed after one month of culture, during which three plants were taken from each treatment, quickly frozen in liquid nitrogen and stored at -80 ℃.
Phenolic acid stress treatment
The MS basal medium was used as a control, and MS+16.0 mg/L ferulic acid and MS+24.0 mg/L p-hydroxybenzoic acid were used to treat R. glutinosa. Aseptic plantlets with uniform growth vigor were transplanted to the control and treatment media. The transplanted plants were cultured in an incubator at 25 ℃ with 2 000 lx illumination (14 h/d) and observed. Sampling was performed after one month of culture, during which three plants were taken from each treatment, quickly frozen in liquid nitrogen and stored at -80 ℃.
Total RNA extraction and reverse transcription
Total RNA was extracted from R. glutinosa by the Trizol (Invitrogen) method and was detected for its integrity by routine electrophoresis on 1% agarose gel. The concentration was measured with an ultra-micro UV spectrophotometer (Nanodrop2000, Thermo), and used for synthesizing cDNA according to the reverse transcription kit (TransGen).
Real-time fluorescence quantitative PCR (qRT-PCR)
The primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd. (Attached list 1). According to the instructions of real-time PCR kit (TransGen), the system included 2×TransStart Top Green qPCR Mix 10 μl, cDNA 1 μl, Forward Primer (10 mM) 0.4 μl, Reverse Primer (10 mM) 0.4 μl, and water added to 20 μl, and each reaction was repeated 3 times in parallel. The system was mixed well and centrifuged instantly, for qRT-PCR detection (IQ5, Bio-Rad). The PCR was started with 95 ℃ for 2 min, followed by 40 cycles of 95℃ for 10 s, 59 ℃ for 30 s and 72 ℃ for 30 s. The dissolution curve was plotted with 95 ℃ for 1 min, 60 ℃ for 1 min and 95 ℃ for 10 s. For the plotting, the temperature increased at a rate of 0.5 ℃ per cycle, and each temperature was held for 30 s. Each sample was set with three biological replicates, and the log2 treatment/control method was used to calculate the relative expression levels of genes between different samples.
Results and Analysis
Screening and functional analysis of specific response genes of R. glutinosa
In the previous study, the comparative analysis on the DGE data of R. glutinosa cultivar "Wen 85-5" between the first cropping and continuous cropping treatments in five periods (seedling period, elongation period, early expanding period, middle expanding period and later expanding period) confirmed that the number of genes showing differential expression in continuously cropped R. glutinosa in all the five periods was 4 398 (Fig. 1A)[4]. Furthermore, we carried out five stress treatments on R. glutinosa (continuous cropping, phenolic acid, salt, drought and waterlogging), and performed DGE analysis on samples of the various treatments in the early expanding period. Through comparisons between multiple groups, continuous cropping triggered changes in the expression of 2 502 genes, and excluding 1 824 genes that were differentially expressed in other four stress treatments (phenolic acid, salt, drought and waterlogging), 678 candidate genes for specific response to continuous cropping were screened (Fig. 1B)[17]. In order to further narrow the candidate gene screening range, taking the intersection of above two sets of analysis results, with |log2 ratio|≥1, FDR<0.001 as the screening conditions, 80 candidate genes for specific response to continuous cropping obstacle in R. glutinosa were finally screened (Fig. 1).
The clustering analysis of the 80 differential genes of the first cropping and continuous cropping treatments in the five periods (Fig. 1C) showed that the expression patterns of the differentially expressed genes in the first three periods of the first cropping treatment (T1-T3) were consistent, indicating that the expression of these genes was relatively stable in the early growth and development period of R. glutinosa. However, continuously cropped R. glutinosa remodeled the expression patterns of above differential genes. Meanwhile, the expression patterns of differentially expressed genes in T1 and L1 (the first period of the continuous cropping treatment) were significantly different. It can be seen that although cytological analysis confirms that the early expanding period is a critical period for continuous cropping obstacle[18], the gene response may have been produced as early as the seedling period (L1).
Agricultural Biotechnology2019
Gene Ontology (GO) function annotation was performed on the 80 differentially expressed genes, and the physiological metabolic processes involving the encoded products of the differential genes were analyzed from such three aspects as biological processes, cellular component and molecular function (Fig. 2). It was found that in the biological process analysis, the products were significantly enriched in metabolic and stress response processes; in the cellular component analysis, the products were significantly enriched in organelles and cell membrane; and in the molecular function analysis, they were significantly enriched in the aspects including metabolic process, enzyme catalytic activity and binding action. It was thus confirmed that the enzymes and proteins related to metabolism and stress response in the cell membrane and organelles significantly changed under continuous cropping conditions. Among the response genes, Unigene7699_All gene was involved in protein secretion and participated in the transmembrane transport of kinesin; CL9284.contig2_All gene was associated with root tip plasma membrane transport and endocytosis; Unigene 1702_All gene was associated with nitrate transmembrane transport activity; and CL8319.Contig2_All gene was homologous with the encoding gene of WRKY transcription factor and was involved in stress response. The results showed that continuous cropping obstacle resulted in the variation of genes related to metabolism and transport of a large amount of substances in R. glutinosa root cells, which destroyed the normal physiological metabolic process of root cells, which led to the inhibition of R. glutinosa growth and development.
Response detection of candidate genes to water-soluble soil extracts
MS basal medium was added with the first cropping and continuous cropping soil extracts (VMS/Vextract: 5/1) to stress aseptic R. glutinosa plantlets. After 5 d of culture, the growth of the aboveground part of the aseptic plantlets treated with the first cropping and continuous cropping soil extracts was basically the same, but the aseptic plantlets cultured with the first cropping soil extract rooted obviously (Fig. 3A); and after 30 d of cultivation, with the continuous cropping soil extract, the leaves of the aboveground part yellowed, and the root development was significantly inhibited (Fig. 3A). The results confirmed that the water-soluble extracts of continuous cropping soils contain allelochemicals that are capable of inhibiting the growth of R. glutinosa, and can be used to simulate continuous cropping environments in in-vitro culture for the screening of candidate genes.
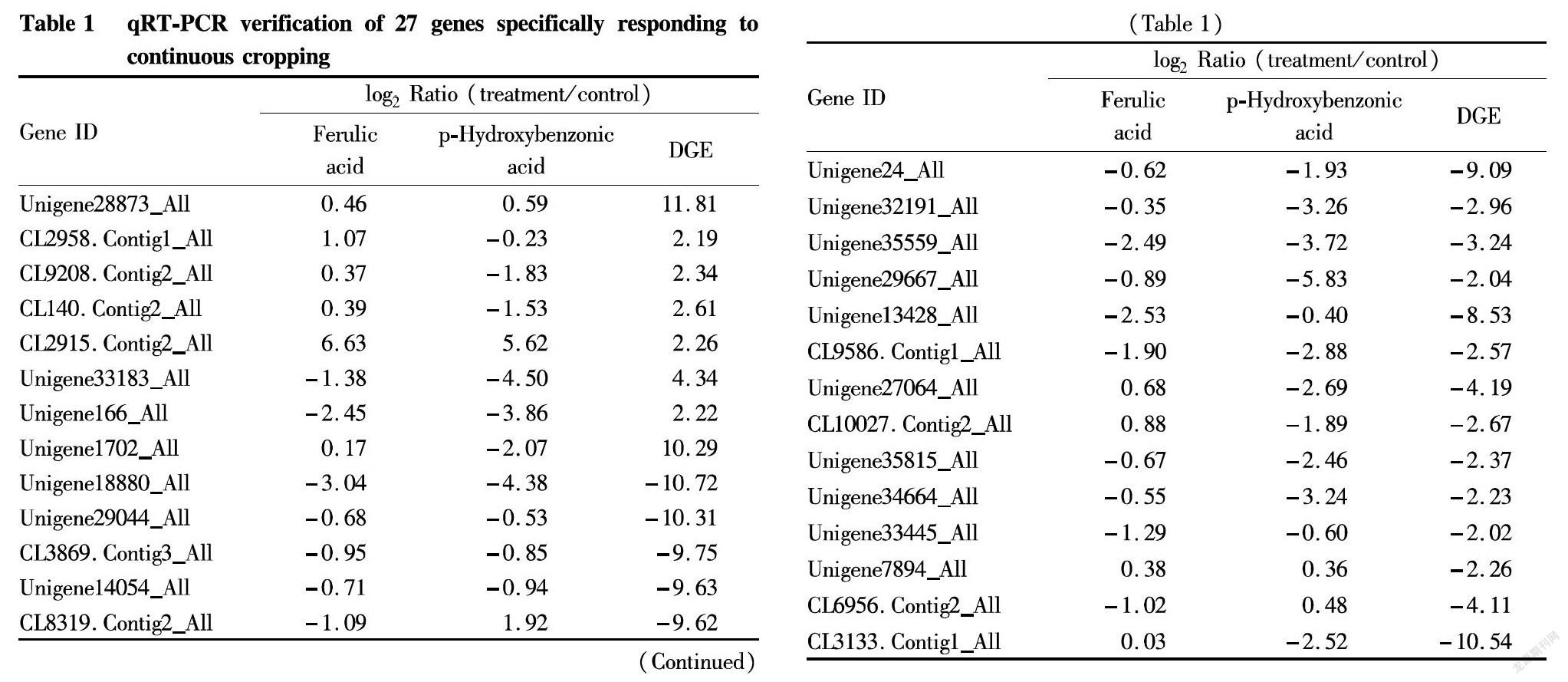
To accurately verify the candidate genes for the specific response in R. glutinosa, gene expression detection was performed on 49 of the 80 candidate genes (|log2 ratio|≥2) by qRT-PCR (Fig. 3B). It was verified that the expression trends of the 40 genes in aseptic plantlets cultured with continuous cropping soil extract was consistent with the DGE results. Since the variation multiples detected by qRT-PCR were generally lower than DGE detection results, in order to improve the accuracy of detection results, with |log2 ratio|≥0.4 determined as the screening threshold for qRT-PCR detection, 27 candidate genes that can respond to continuous cropping soil extract were finally screened (Table 1).
Response detection of candidate genes to phenolic acids
Studies have confirmed that ferulic acid and p-hydroxybenzoic acid are the main allelochemicals against R. glutinosa[16,19-21]. It was found from the aseptic plantlets cultured with MS basal medium added with ferulic acid (16.0 mg/L) or p-hydroxybenzoic acid (24.0 mg/L) that ferulic acid and p-hydroxybenzoic acid inhibited the root growth of R. glutinosa after 10 d of culture. After 30 d of culture, the treatment with ferulic acid and p-hydroxybenzoic acid resulted in changes in both aboveground and underground parts of R. glutinosa, and the effect of ferulic acid was more obvious than that of p-hydroxybenzoic acid (Fig. 4A).
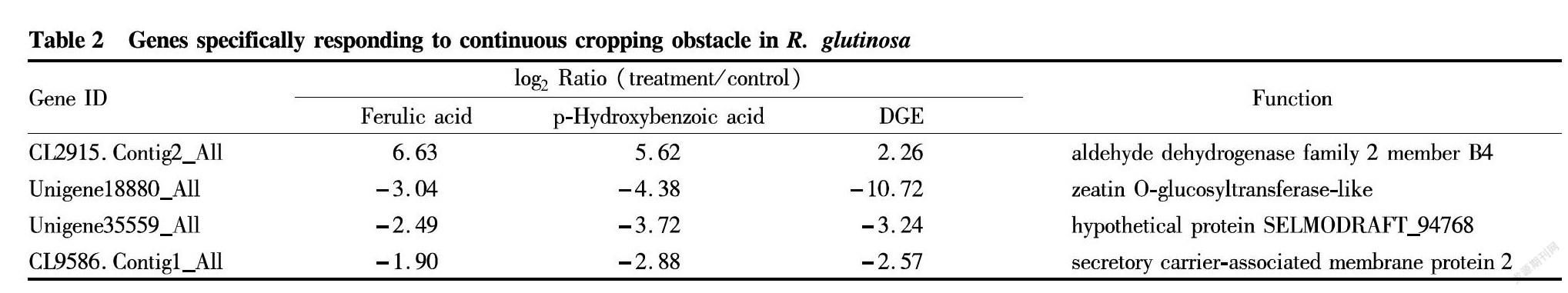
In order to accurately target the specific response genes of R. glutinosa, qRT-PCR was applied to detect the 27 genes that responded to the continuous cropping soil extract in R. glutinosa treated with ferulic acid and p-hydroxybenzoic acid (Table 1). The results showed that 21 of the 27 differentially expressed genes were consistent with the DGE results (Fig. 4B), but the changes in the expression levels of some genes after phenolic acid treatment were severer than those of the continuous cropping soil extract treatment. By comparing the verification results of the samples treated with ferulic acid and p-hydroxybenzoic acid, four genes specifically responding to continuous cropping obstacle were finally locked (Table 2).
Conclusions and Discussion
The lack of genomic information has become one of the important factors restricting the rapid development of the research on the mechanism of continuous cropping obstacle in R. glutinosa. The development of high-throughput sequencing technology provides a wealth of information for the study on mechanisms of continuous cropping obstacle. However, the in-depth mining and rigorous verification of omics analysis data is a key link to accurately reveal the mechanism of continuous cropping obstacle in R. glutinosa[22-23]. In the preliminary work of this research group, the digital gene expression (DGE) analysis of five different stress treatments (continuous cropping, phenolic acid, salt, drought and waterlogging) and first cropping and continuous cropping in five different periods (seedling period, elongation period, pre-expanding period, mid-expanding period and late-expanding period) were completed[4,17], and combining the results of two sets of analysis, 80 candidate genes (|log2 ratio|≥1, FDR<0.001) for specific response to continuous cropping obstacle in R. glutinosa were screened. In order to accurately lock the response genes for continuous cropping obstacle in R. glutinosa, continuous cropping soil extract and phenolic acids were used to treat aseptic plantlets to verify the above-mentioned candidate genes for specific response to continuous cropping, and four target genes were locked (Table 1), laying a foundation for further exploration of the continuous cropping obstacle mechanism in R. glutinosa.
Cell membrane receptor proteins can recognize extracellular signals, interact with other proteins during endosomal transport and trigger signal transduction processes, thereby adapting cells to environmental changes and promoting information exchange between cells, and ultimately maintaining normal growth and development of living organisms[24-27]. It was found from the research on the early response to salt stress in soybean leaves that stress-related membrane proteins were activated to regulate the expression of downstream proteins in the early stage of stress treatment[28-29]. Under salt stress, a variety of membrane proteins involved in salt stress response were isolated from rice roots, which participated in rice salt stress response in different varieties through different molecular mechanisms[30]. The calcium signaling system known as the "second messenger" also senses external environmental stimulation through the membrane receptor protein[31-33]. Our previous study showed that continuous cropping caused a significant change in the calcium signaling system, suggesting that allelopathic autotoxins may trigger a specific response signal to continuous cropping obstacle in R. glutinosa roots[4]. Multiple genes specifically responding to continuous cropping obstacle obtained by screening were involved in root tip plasma membrane transport and endocytosis, cell membrane transport, and the transport of nutrients such as nitrate and phosphate. The finally locked CL9586.Contig1_All gene belongs to the secretory carrier membrane proteins, and its expression variation once again confirmed that the cell membrane transport system of R. glutinosa root cells was abnormal and affected the normal metabolism of the root cells.
Screening and functional analysis of the genes specifically responding to continuous cropping obstacle in R. glutinosa showed that continuous cropping obstacle affected the growth and development of R. glutinosa by destroying the signal transduction, material transport and stress response system of R. glutinosa root cells. With the genes specifically responding to continuous cropping obstacle in R. glutinosa obtained by screening and identification, further revealing their functions and regulation network by means of reverse genetics will provide theoretical guidance for solving the problem of continuous cropping obstacle in R. glutinosa.
References
[1]ZHAO YJ. The importance and application prospect of allelopathy in the cultivation of medicinal plant[J]. Chinese Traditional and Herbal Drugs, 2000(8): 81. (in Chinese)
[2]LIU J, ZHU JK. A calcium sensor homolog required for plant salt tolerance[J]. Science, 1998, 280(5371): 1943-1945.
[3]HE CN, GAO WW, YANG JX, et al. Identification of autotoxic compounds from fibrous roots of Panax quinquefolium L[J]. Plant and Soil, 2009, 318(1):63-72.
[4]YANG YH, LI MJ, LI XY, et al. Transcriptome-wide identification of the genes responding to replanting disease in Rehmannia glutinosa L. roots[J]. Molecular Biology Reports, 2015, 42(5):881-892.
[5]KITAZAWA H, ASAO T, BAN T, et al. Autotoxicity of root exudates from strawberry in hydroponic culture[J]. The Journal of Horticultural Science and Biotechnology, 2005, 80(6): 677-680.
[6]BOUHAOUEL I, GFELLER A, FAUCONNIER ML, et al. Allelopathic and autotoxicity effects of barley (Hordeum vulgare L. ssp. vulgare) root exudates[J]. BioControl, 2015, 60(3): 425-436.
[7]BU RF, XIE JM, YU JH, et al. Autotoxicity in cucumber (Cucumis sativus L.) seedlings is alleviated by silicon through an increase in the activity of antioxidant enzymes and by mitigating lipid peroxidation[J]. Journal of Plant Biology, 2016, 59(3): 247-259.
[8]ZHANG B, LI XZ, WANG FQ, et al. Assaying the potential autotoxins and microbial community associated with Rehmannia glutinosa replant problems based on its ‘autotoxic circle’[J]. Plant and Soil, 2016, 407(1):307-322.
[9]DU JF, YIN WJ, ZHANG ZY. Autotoxicity and phenolic acids content in soils with different planting interval years of Rehmannia glutinosa[J]. Chinese Journal of Ecology, 2009(3): 71. (in Chinese)
[10]WU LK, LI FZ, LI J, et al. Assessment of shifts in microbial community structure and catabolic diversity in response to Rehmannia glutinosa monoculture[J]. Applied Soil Ecology, 2013(67): 1-9.
[11]ZHOU XG, GAO DM, LIU J, et al. Changes in rhizosphere soil microbial communities in a continuously monocropped cucumber (Cucumis sativus L.) system[J]. European Journal of Soil Biology, 2014(60):1-8.
[12]WU LK, WANG JY, HUANG WM, et al. Plant-microbe rhizosphere interactions mediated by Rehmannia glutinosa root exudates under consecutive monoculture[J]. Scientific Reports, 2015(5):15871.
[13]LI MJ, YANG YH, CHEN XJ, et al. Transcriptome/Degradome-wide identification of Rehmannia glutinosa miRNAs and their targets: The role of miRNA activity in the replanting disease[J]. PLOS ONE, 2013, 8(7): e68531.
[14]YANG YH, LI MJ, CHEN XJ, et al. De novo characterization of the Rehmannia glutinosa leaf transcriptome and analysis of gene expression associated with replanting disease[J]. Molecular Breeding, 2014, 34(3):905-915.
[15]YANG M, ZHANG XD, XU YG, et al. Autotoxic ginsenosides in the rhizosphere contribute to the replant failure of Panax notoginseng[J]. PLOS ONE, 2015, 10(2):e0118555.
[16]SINGH HP, BATISH DR, KOHLI RK. Autotoxicity: Concept, organisms, and ecological significance[J]. Critical Reviews in Plant Sciences, 1999, 18(6):757-772.
[17]TIAN YH, FENG FJ, ZHANG B, et al. Transcriptome analysis reveals metabolic alteration due to consecutive monoculture and abiotic stress stimuli in Rehamannia glutinosa Libosch[J]. Plant Cell Reports, 2017, 36(6):859-875.
[18]LI MJ, YANG YH, LI XY, et al. Analysis of integrated multiple ‘omics’ datasets reveals the initiation and determination mechanisms of tuberous root formation in Rehamannia glutinosa [J]. Journal of Experimental Botany, 2015, 66(19):?5837-5851.
[19]ZHOU X, YU G, WU F. Soil phenolics in a continuously mono-cropped cucumber (Cucumis sativus L.) system and their effects on cucumber seedling growth and soil microbial communities[J]. European Journal of Soil Science. 2012, 63(3):332-340.
[20]KATO-NOGUCHI H, NAKAMURA K, OHNO O, et al. Asparagus decline: Autotoxicity and autotoxic compounds in asparagus rhizomes[J]. Journal of Plant Physiology, 2017(213):23-29.
[21]REN X, YAN ZQ, HE XF, et al. Allelochemicals from rhizosphere soils of Glycyrrhiza uralensis Fisch: Discovery of the autotoxic compounds of a traditional herbal medicine[J]. Industrial Crops and Products, 2017(97):302-307.
[22]LAY JO, LIYANAGE R, BORGMANN S, et al. Problems with the "omics"[J]. TrAC Trends in Analytical Chemistry, 2006, 25(11):1046-1056.
[23]IOANNIDIS JPA, KHOURY MJ. Improving Validation Practices in "Omics" Research[J]. Science, 2011, 334(6060):1230-1232.
[24]STONE JM, WALKER JC. Plant protein kinase families and signal transduction[J]. Plant Physiology, 1995, 108(2):451-457.
[25]ZHU JK. Salt and drought stress signal transduction in plants[J]. Annual Review of Plant Biology, 2002, 53(1):247-273.
[26]LIPIEC J, DOUSSAN C, NOSALEWICZ A, et al. Effect of drought and heat stresses on plant growth and yield: A review[J]. International Agrophysics, 2013, 27(4):463-477.
[27]HARUTA M, SABAT G, STECKER K, et al. A peptide hormone and its receptor protein kinase regulate plant cell expansion[J]. Science, 2014, 343(6169): 408-411.
[28]PHANG TH, SHAO G, LAM HM. Salt tolerance in soybean[J]. Journal of Integrative Plant Biology. 2008, 50(10):1196-1212.
[29]XU XY, FAN R, ZHENG R, et al. Proteomic analysis of seed germination under salt stress in soybeans[J]. Journal of Zhejiang University SCIENCE B, 2011, 12(7):507-517.
[30]CHENG YW, QI YC, ZHU Q, et al. New changes in the plasma-membrane-associated proteome of rice roots under salt stress[J]. Proteomics, 2009, 9(11):3100-3114.
[31]DOBNEY S, CHIASSON D, LAM P, et al. The calmodulin-related calcium sensor CML42 plays a role in trichome branching[J]. The Journal of biological chemistry, 2009, 284(46):31647-31657.
[32]LUAN S. The CBL-CIPK network in plant calcium signaling[J]. Trends in plant science, 2009, 14(1):37-42.
[33]SOBOLOFF J, ROTHBERG BS, MADESH M, et al. STIM proteins: dynamic calcium signal transducers[J]. Nature reviews Molecular cell biology, 2012, 13(9):549-565.
Editor: Yingzhi GUANG Proofreader: Xinxiu ZHU
- 农业生物技术(英文版)的其它文章
- Analysis of the Southern China Tilapia Production and Economic Benefits of Different Breeding Patterns in 2018
- Dynamic Monitoring and Control Measures of Spodoptera frugiperda (J.E.Smmith) in Low Latitude Plateau Sugarcane Areas
- Control Effects of a New Sex Pheromone Trap and Biological Agents on Sesamia inferens Walker and Argyroploce schistaceana (Snellen)
- Comparative Study on Grain Cadmium Content and Yield in Different Rice Varieties
- Simulation Experiment of Air Temperature Variation in Multi-film Covering at Night
- Identification of Growth-promoting Bacteria from Rhizosphere of Pastures and Their Effects on Growth of Lotus corniculatus L.

