Genetic Diversity of Clover by SRAP
Runfang LI Hanfeng DING Cune WANG Lingyun LU Xiaodong ZHANG



Abstract In the present study, genetic diversity and genetic relationship of 11materials including red clover (Trifolium pretense L.), white clover (T. repen L.) and alsike clover (T. hybridum L.) and leaf-type variants of white clover were investigated by SRAP (sequence related amplified polymorphism). Forty SRAP primers were screened, and 792 reliable bands were amplified, among which 426 (53.8%) were polymorphic. The number of polymorphic bands per pair of primers ranged from 3 to 38 with an average of 10.6. SRAP cluster analysis showed that the similarity coefficients between white clover materials were from 0.465 to 0.997 with an average of 0.812, indicating that there was certain genetic diversity among white clover. Specific bands appeared in white clover variants, indicating that there were certain differences in DNA sequence between normal while clover plants and their leaf-type variants. The similarity coefficients between red clover materials were from 0.457 to 0.827 with an average of 0.597, indicating that the different red clover germplasms had genetic diversity at the DNA molecular level. The results also showed that the genetic relationship of the alsike clover with the red clover was closer than that with the white clover. This study shows that SRAP technique can be effectively used for the analysis on interspecific and intraspecific relationship, germplasm resource identification and genetic diversity of clover.
Key words White clover (Trifolium repen L.); Red clover (Trifolium pratense L.); Alsike clover (Trifolium hybridum L.); SRAP
Red clover (Trifolium pretense L.), white clover (T. repen L.) and alsike clover (T. hybridum L.), belonging to Trifolium of Faboideae in Leguminosae, are excellent forages and green manures. Among them, the medicinal plant red clover has received much attention due to the inclusion of isoflavonoids in recent years[1]. There are few reports on the genetic diversity of clover germplasm resources by molecular markers. At present, only Han et al.[2]optimized the RAPD reaction system of clover. SRAP (sequence related amplified polymorphism), is a new molecular marker technique originally developed in 2001 by Dr. Li and Quiros from the Department of Vegetables, University of California, USA, which is characterized by amplifying ORFs (Open Reading Frames) through unique primer design. This marker technique is simple, stable, efficient, and reproducible. It has been successfully applied in other crops such as persimmon[3], cotton[4], rapeseed[5], tomato[6], safflower[7], corn[8]and ramie[9]. SRAP technique has been successfully applied to the study on the genetic diversity of Ganoderma lucidum[10], okra[11]and alfalfa[12]. There are significant differences in the content of medicinal ingredients, fiber yield and protein quality among different clover germplasms[1]. The degree of difference in the genetic basis of varieties cannot be identified only from agronomic traits such as appearance, leaf type, plant height and stem color of clover. Therefore, the application of modern molecular biological techniques, especially molecular marker techniques, to study the genetic diversity and genetic relationship of clover is particularly important. In this study, the SRAP marker technique was applied for the first time to the investigation of the interspecific and intraspecific genetic diversity and genetic relationship of clover, aiming to provide a molecular basis for the classification and genetic diversity of clover germplasm resources.
Materials and Methods
Materials
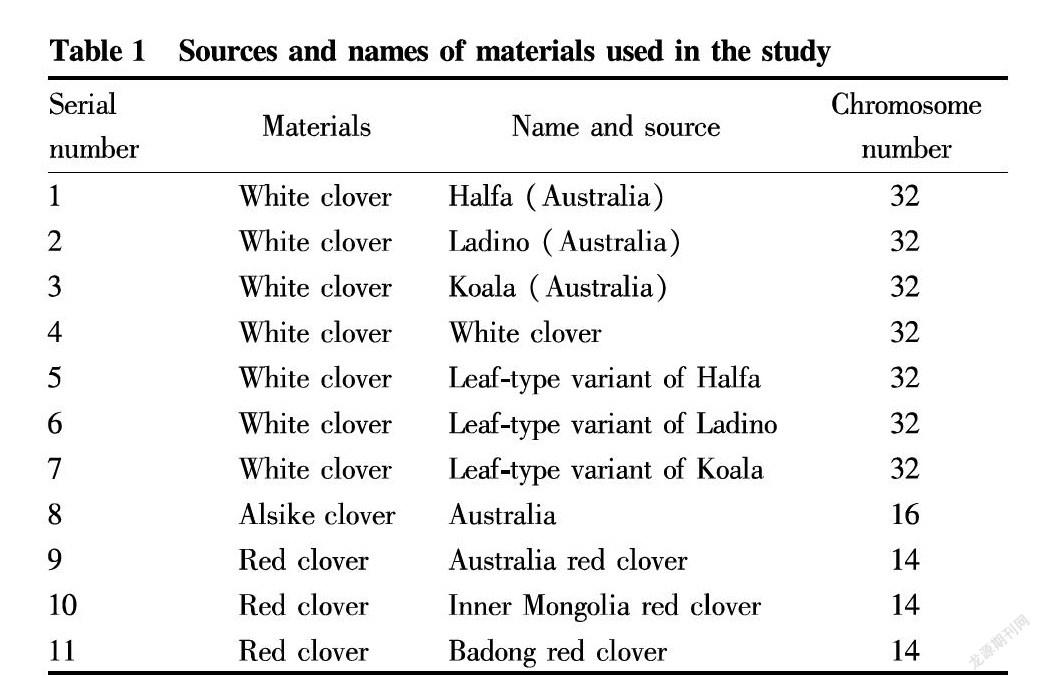
In this experiment, 11 clover materials were used. The chromosome number, name and source information are shown in Table 1. There were 3 red clover materials, 1 alsike clover material and 7 white clover materials, including leaf-type variants of white clover (i.e., the number of leaflets on the compound leaves is larger than the number of normal three leaflets, and the same leaf-type variant may have three-leaflet, four-leaflet, five-leaflet and six-leaflet leaves).
Methods
DNA extraction
About 1 g of leaf with undeveloped leaflets was collected from the field. The total DNA of different materials was extracted according to the CTAB method of Paterson et al.[13], respectively, and RNA was removed by RNase.
PCR amplification system and program
The 13 primers and PCR amplification used in this experiment were based on the method of Lin et al.[4]. Five forward primers and eight reverse primers were combined to form 40 pairs (Table 2). The primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd. PCR amplification was performed on a GeneAmp 9700 thermal cycler. The PCR reaction system contained DNA template 60 ng, primers 30 ng each, dNTPs 200 μmol/L each, 1×Reaction Buffer 1.5 mmol/L, MgCl2 1.5 mmol/L, and Taq enzyme 1 U, and the total volume was 20 μl. The DNA amplification program was conducted based on the method of Li and Quiros[14].
The amplified product was separated by 6% PAGE gel with 0.5×TBE as the running buffer. During electrophoresis, pre- electrophoresis was carried out with a voltage of 2 000 V until the current was 30 mA. After sample loading, it was electrophoresed with 2 500 V for 1.5-2.0 h, and silver staining was carried out after electrophoresis.
Data analysis
According to the results of PCR amplification, for amplified bands of each sample, the 1, 0 type data were constructed by assigning 1 for the occurrence of a band and 0 for no band, excluding bands without polymorphism. The similarity coefficients were calculated using the NTSYS software, and a similarity coefficient matrix was obtained. Cluster analysis was performed by the UPGMA (Unweighted pair group method arithmetic average), constructing a dendrogram.
Results and Analysis
SRAP molecular polymorphism of clover
Using the 40 pairs of primers, a total of 792 bands were amplified, of which 426 bands were polymorphic. The percentage of polymorphic sites was 53.8%, and the number of polymorphic bands per pair of primers was 3-28, with an average of 10.6. The amplification results of partial primer are shown in Fig. 1.
Interspecific genetic relationship of clover
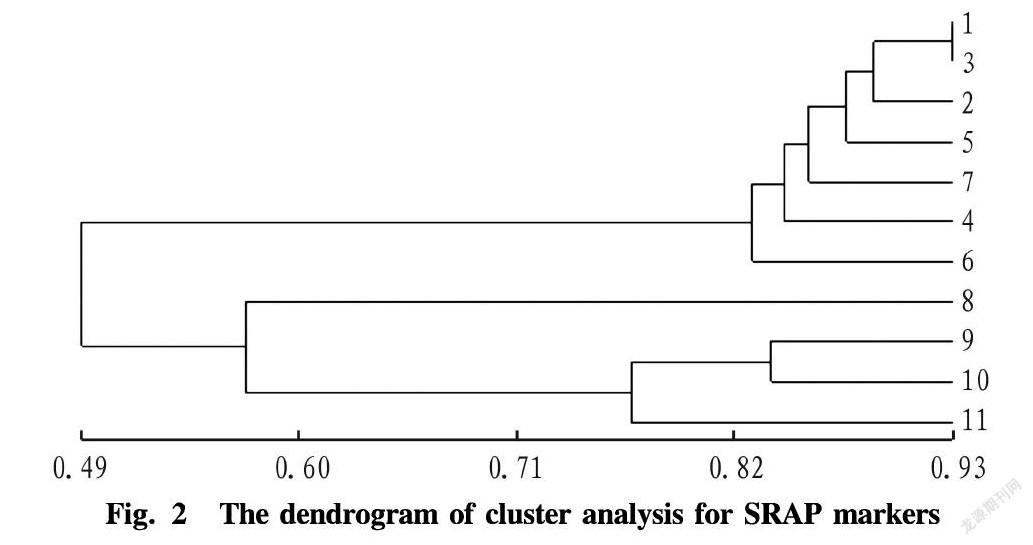
Based on the genetic similarity coefficients of 11 materials (Table 3), according to the UPGMA method, cluster analysis was performed, constructing a dendrogram (Fig. 2). The similarity coefficients were between 0.425 and 0.997. At the threshold of 0.568, the test materials can be divided into three major groups: white clover, red clover and alsike clover. The similarity coefficients of white clover with 32 chromosomes ranged from 0.465 to 0.997, and the average similarity coefficient was 0.812. The similarity coefficients of red clover with 14 chromosomes were in the range of 0.457-0.827, and the average similarity coefficient was 0.597, indicating that red clover was abundant in genetic diversity. The alsike clover had a relatively closer genetic relationship with the red clover, and the average similarity coefficient was 0.559; and the genetic relationship with the white clover was relatively farther, and the similarity coefficient was 0.504. The genetic similarity between the white clover Ladino and the red clover was the lowest, and the relationship was the farthest.
Intraspecific genetic relationship of clover
The genetic differences between the red clover of different germplasms were larger. The genetic similarity coefficients of Badang red clover with Australia red clover and Inner Mongolia red clover were 0.457 and 0.508, respectively (Table 3).
Among the 11 materials of white clover, when the threshold was 0.896, they can be divided into 6 groups. The genetic distances between the normal white clover materials Halfa, Koala and Ladino were the closest, and they were clustered into one group; and the genetic similarity between the white clover Halfa and Koala was the highest, and the genetic distance was the closest. White clover No. 4 formed a separate group. The normal leaves and mutational leaves of Halfa s leaf-type variant A were clustered into one group. The normal leaves and mutational leaves of Koala s leaf-type variant D were clustered into one group. The normal leaves and mutational leaves of Ladinos leaf-type variant B were clustered into one group. The normal leaves and mutational leaves of Ladinos leaf-type variant C were also clustered into one group. The latter two groups formed a large group. The normal and mutational leaves of the leaf-type variants of Halfa, Koala and Ladino all showed specific bands that were not found in normal plants, and they were not preferentially clustered with their respective normal plants (Fig. 2).
In this study, the 14 white clover test materials were all tested to have the chromosome number 2n=32. The results of the SRAP molecular marker analysis showed that white clover No.1, 2, and 3 that are three varieties cultivated in China, were very similar in biological characteristics, and it was concluded that their genetic similarity coefficients were high and their genetic relationships were close, indicating that the three varieties had little difference at the DNA molecule level. However, white clover No. 4 was relatively farther from these three materials, which showed that there was certain genetic diversity among different white clover germplasms with the same number of chromosomes.
The cluster results of white clover showed that the normal leaf type of Halfa was clustered with the normal leaf type of Koala prior to the normal leaf type cluster of Ladino, and among the leaf type variants, Halfas leaf-type variant was clustered with Koalas leaf-type variant prior to Ladinos leaf-type variant, indicating that the relationship of Halfa with Koala were relatively closer, and that with Ladino was farther. The clustering of the normal plants of Halfa, Ladino and Koala was prior to the clustering with their respective variants, and their variants all showed specific bands, indicating that there were some differences in DNA sequence between the leaf-type variants and the normal leaf-type plants, which provides strong evidence for exploring the cause of the leaf type variation of white clover.
It is reported that the number of red clover chromosomes is 2n=16, 28, 32[15], but in this study, the red clover test materials have their chromosome number 2n=14. The SRAP molecular marker results showed that although their chromosome numbers are the same, the genetic similarity coefficients of material No. 18 Batong red clover with materials No.16 and No.17 were 0.457 and 0.508, respectively, indicating that there was genetic polymorphism at the molecular level in different red clover germplasms.
In this study, SRAP technique was applied to investigate the genetic diversity of clover germplasm resources. With the 40 pairs of primers, the polymorphic bands were up to 10.6 for each pair of primers, and the percentage of polymorphic bands was 53.8%. The good amplification effect indicated that there were abundant interspecific and intraspecific genetic diversity of clover, which provides a theoretical basis for the study of germplasm resource classification, genetic diversity and genetic relationship of clover.
References
[1]HUANG SY, TU PF. Isolation and identification of isoflavones from Trifolium pratense[J]. Acta Scientiarum Naturalium Universitatis Pekinensis, 2004, 40(4): 544-549. (in Chinese)
[2]HAN WB, TANG FL, ZHANG YX, et al. Optimization of RAPD reaction system for clover[J]. Heilongjiang Agricultural Sciences, 2006(1): 10-12. (in Chinese)
[3]GUO DL. Establishment of several molecular makers and analysis of genetic relationships in Diospyros Linn.[D]. Wuhan: Huazhong Agricultural University, 1998. (in Chinese)
[4]LIN ZX. Linkage maps constitution in cotton and QTL mapping for yield and fiber-related traits[D]. Wuhan: Huazhong Agricultural University, 2005. (in Chinese)
[5]WEN YC, WANG HZ. Analysis of genetic diversity and genetic basis of Chinese rapeseed cultivars (Brassica napus L.) by sequence-related amplified polymorphism markers[J]. Scientia Agricultura Sinica, 2006, 39(2): 246-256. (in Chinese)
[6]WANG Y, GONG YQ. Optimization of SPAP-PCR system and cultivar molecular identification in tomato (Lycopersicon esculentum L.)[J]. Journal of Nanjing Agricultural University, 2007, 30(1): 23-29. (in Chinese)
[7]PENG X, GUO ML, CHEN YH. Establishment and optimization of sequence-related amplified polymorphism amplification system for Carthamus tinctorius L.[J]. Academic Journal of Second Military Medical University, 2006, 27(5): 544-577. (in Chinese)
[8]ZHAO XL, MA Q. Optimization of SRAP system in maize[J]. Journal of Anhui Agricultural Sciences, 2006, 34(15): 3619-3620. (in Chinese)
[9]LIU LJ, MENG ZQ. Optimization for SRAP reaction system in ramie (Boehmeria nivea L. Guad)[J]. Molecular Plant Breeding, 2006, 4(5): 726-730. (in Chinese)
[10]SUN SJ, GAO W, LIN SQ. Analysis of genetic diversity in Ganoderma population with a novel molecular marker SRAP[J]. Appl Microbiol Biotechnol, 2006(72): 537-543.
[11]OSMAN GULSEN, SULEYMAN KARAGUL, KAZIM ABAK. Diversity and relationships among Turkish okra germplasm by SRAP and phenotypic marker polymorphism[J]. Biologgia, Bratislava, 2007, 62(1): 41-45.
[12]VANDEMARK GJ, ARISS JJ, BAUCHAN GA, et al. Estimating genetic relationships among historical sources of alfalfa germplasm and selected cultivars with sequence related amplified polymorphisms[J]. Euphytica, 2006(152): 9-16.
[13]PATERSON AH, CURT LB, WENDEL JF. A rapid method for extraction of cotton (Gossypium spp) genomic DNA suitable for RFLP and PCR analysis[J]. Plant Mol Bil Rep, 1993(11):112-127.
[14]LI G, QUIROS CF. Sequence related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: Its application to mapping and gene tagging in Brassica[J]. Theor Appl Genet, 2001(103): 455-461.
[15]ZHANG ZP, WU LH, KANG YF. Karyotype analysis of red clover (Trifolium pratense) and white clover (Trifolium repers L)[J]. Grassland of China, 1993(3): 65-66. (in Chinese)
Editor: Yingzhi GUANG Proofreader: Xinxiu ZHU
- 农业生物技术(英文版)的其它文章
- Analysis of the Southern China Tilapia Production and Economic Benefits of Different Breeding Patterns in 2018
- Dynamic Monitoring and Control Measures of Spodoptera frugiperda (J.E.Smmith) in Low Latitude Plateau Sugarcane Areas
- Control Effects of a New Sex Pheromone Trap and Biological Agents on Sesamia inferens Walker and Argyroploce schistaceana (Snellen)
- Comparative Study on Grain Cadmium Content and Yield in Different Rice Varieties
- Simulation Experiment of Air Temperature Variation in Multi-film Covering at Night
- Identification of Growth-promoting Bacteria from Rhizosphere of Pastures and Their Effects on Growth of Lotus corniculatus L.

