Liquid biopsy in lymphomas: a potential tool for refining diagnosis and disease monitoring
Riccardo Moia, Chiara Favini, Silvia Rasi, Clara Deambrogi, Valentina Ferri, Mattia Schipani, Sruthi Sagiraju, Abdurraouf Mokhtar Mahmoud, Ahad Ahmed Kodipad, Ramesh Adhinaveni, Andrea Patriarca,Luca Nassi, Gianluca Gaidano
Division of Hematology, Department of Translational Medicine, University of Eastern Piedmont and Ospedale Maggiore della Carità, Novara 28100, Italy.
Abstract Liquid biopsy consists in a simple blood sampling that allows to analyze cell free DNA (cfDNA), containing specific genomic clues released by the tumor into the bloodstream. In this review, we shall focus on the analysis of cfDNA in lymphoma and, in particular, on its application in the genotyping and monitoring of two common types of B-cell lymphoma, i.e., diffuse large B-cell lymphoma (DLBCL) and classical Hodgkin lymphoma (cHL). From a diagnostic standpoint and based upon the current international guidelines, lymphoma diagnosis has so far relied on the analysis of the tissue biopsy. From a molecular viewpoint, though, the tissue biopsy does not reflect the entire molecular heterogeneity of lymphomas. In fact, in an individual patient, lymph nodes at different anatomical sites,as well as different areas of the same lymph node, may show different genetic profiles. Consequently, molecular analysis of genomic DNA extracted from a single lymph node biopsy may not recapitulate the whole mutational landscape of the disease. Liquid biopsy may overcome this hurdle, since cfDNA is released by all tumoral cells and can reveal the entire molecular complexity of lymphomas. From a translational perspective, liquid biopsy may also be used to evaluate clonal evolution, response to therapy and minimal residual disease. Consistently, in DLBCL as well in cHL, the drop of the mutational burden during the treatment course provides complementary information to conventional imaging techniques. The integration of liquid biopsy with imaging techniques may prove useful for a better prediction of patients’ outcome and for a better treatment tailoring.
Keywords: Liquid biopsy, lymphoma, precision medicine
GENERAL CONCEPT ON CIRCULATING CELL FREE DNA
Liquid biopsy consists in a simple and easy sampling of peripheral blood, that can be subjected to the molecular analysis of specific genomic clues released by the tumor[1,2]. Because circulating tumor cells are very rare or absent in many types of lymphomas, liquid biopsy approaches have been focused on circulating tumoral DNA (ctDNA) released by lymphoma cells into the bloodstream[3-5]. However, the ctDNA represents only a fraction of the total amount of cell free DNA (cfDNA), that is derived also from healthy cells. The cfDNA circulates in plasma at a low concentration as double-stranded DNA fragments with a dimension < 200 base pairs[6]. In individuals without malignancies, plasma cfDNA derives mainly from the apoptosis of normal hematopoietic cells. cfDNA levels may also rise in para-physiological conditions, such as trauma, burns or high exercise. In healthy subjects, the cfDNA concentration ranges between 1 and 10 ng/mL and can reach 30ng/mL in lymphoma patients[7].
CFDNA ISOLATION
It is possible to selectively isolate cfDNA from plasma in order to proceed with subsequent molecular analysis[8]. To isolate cfDNA, a few very important technical precautions need to be observed in order to avoid white blood cell lysis that may contaminate the cfDNA with genomic DNA (gDNA)[9]. If using standard collecting tubes containing ethylenediaminetetraacetic-acid (EDTA) as anticoagulant, plasma extraction is recommended within 3 h from the collection of peripheral blood[9]. EDTA tubes, however, are not able to preserve the integrity of cfDNA and are not able to prevent the lysis of nucleated cells for more than 3 h. Therefore, in order to allow the shipment of biological material, specific tubes, namely cfDNA BCT Streck tubes (Streck), can stabilize cfDNA and can prevent the contamination by gDNA from white blood cells for up to 14 days at a temperature between 6 and 37 °C[10]. After collection, plasma is separated from the corpuscular blood by centrifugation of blood samples at 800 rpm at 4 °C for 10 min. A second centrifugation at 13,000 rpm at 4 °C for 10 min allows to pellet and remove any remaining cells. Plasma samples are then stored in 1 mL aliquots at -80 °C until cfDNA extraction[8].
cfDNA can be selectively extracted using two different methods. The first method, named QIAamp Circulating Nucleic Acid (Qiagen), relies on the use of ion-exchange resins that bind the plasma cfDNA[11].The second approach relies on an automatic method that utilizes the Maxwell RSC Instrument coupled with Maxwell RSC ccfDNA Plasma Kit (Promega). This method consists in an automated nucleic acid purification platform that processes up to 16 samples simultaneously and allows to obtain high quality cfDNA starting from 1 mL of plasma[12]. After extraction, cfDNA is usually quantified by a fluorometric assay. Approximately 30-40 ng of cfDNA are needed for subsequent molecular analysis[8].
CAPP-SEQ ANALYSIS AS FOR DEFINING THE LYMPHOMA GENOTYPE ON CFDNA
Since lymphoid malignancies harbor a unique molecular marker, namely the immunoglobulin (Ig)gene rearrangement, the identification of presence of this biomarker in the cfDNA may be used to track minimal residual disease (MRD) during the course of treatment. Once the Ig gene rearrangement has been identified on the lymph node biopsy, it is possible to evaluate the amount of this rearrangement in the cfDNA using Next-Generation-Sequencing (NGS) or polymerase chain reaction based methods[13,14]. Pivotal studies have shown that the proportion of cfDNA carrying the lymphoma-specific Ig rearrangement decreases rapidly in patients who respond to therapy and tends to remain negative in those who maintain the response[13,14]. Conversely, the Ig rearrangement remains high in patients who do not respond to treatment[13,14]. As expected, these patients experience both a worse progression-free survival and a worse overall survival compared to patients with undetectable Ig gene rearrangement[13,14]. This method is very effective in monitoring MRD but has some pitfalls. First, this method does not work in a biopsy free manner since it requires, at the time of diagnosis, the identification of the individual patient’s specific Ig rearrangement on the lymph node tissue biopsy. Once the patient’s unique Ig rearrangement has been identified on the tissue biopsy, it is then possible to track it on the liquid biopsy to evaluate MRD. Second,this method does not allow a comprehensive disease genotyping on the liquid biopsy[15].
To overcome these limits, the use of Cancer Personalized Profiling by Deep Sequencing (CAPP-Seq) has been established as a reliable tool to genotype cancer patients[16]. This molecular strategy utilizes disease specific probes that selectively capture a set of exonic and intronic regions known to be recurrently affected in a particular cancer type. The selected targets are then amplified and sequenced by NGS allowing the detection of the specific tumor mutational profile of a patient. This method, linked to a stringent bioinformatic analysis, allows to detect mutations in cfDNA with a high sensitivity, identifying mutations with allelic frequency as low as 0.02%. In addition, CAPP-Seq allows the identification of single nucleotide variants, as well as insertion/deletions, copy number alterations and rearrangements, thus covering a large variety of genomic alterations of human cancers[16].
APPLICATION OF LIQUID BIOPSY IN LYMPHOMA
Lymph node biopsy is the gold standard for the diagnosis of lymphoma, but genetic lesions identified in the tissue biopsy may not reflect the entire molecular complexity of every single patient with lymphoma[17,18].Consistently, in an individual patient, lymph nodes at different anatomical sites, as well as different areas of the same lymph node, may show different genetic profiles[19]. However, outside of clinical studies aimed at investigating the clonal heterogeneity of lymphomas, multiple biopsies are not routinely performed in lymphoma patients for both practical and ethical concerns. On these grounds, once a diagnosis of lymphoma is performed on a tissue biopsy, liquid biopsy may be used to explore the entire mutational landscape of lymphoma, since this approach has the potential to collect the tumor cfDNA deriving from most, or potentially all, of the different sites of tumor involvement in the body [Figure 1][15]. In particular,a liquid biopsy CAPP-Seq approach may be used to: (1) genotype lymphoma patients; and to (2) evaluate treatment response [Figure 2].
LIQUID BIOPSY AS A TOOL FOR GENOTYPING LYMPHOMA PATIENTS
In DLBCL, liquid biopsy allows to identify at least one somatic non-synonymous mutation per patient in over 70% of cases[20,21]. The mutational profile identified by liquid biopsy reflects that revealed by DLBCL genomic studies performed on the lymph node biopsies[22]. More precisely, the most common genetic lesions found by liquid biopsy reflect those detected on the tissue biopsy, and comprise alterations ofKMT2D, followed byTP53,CREBBP, andPIM1. In addition,EZH2andBCL2mutations, as expected, are more frequently found in germinal center (GC) DLBCL, whereasTNFAIP3andPIM1mutations are more frequent in non-GC DLBCL[20]. In addition, by comparing the mutations identified in cfDNA with those identified in gDNA extracted from the tissue biopsy, cfDNA appears to be representative of most of the mutations that occur in > 20% of the alleles of the tumor biopsy, with a sensitivity > 90% and a specificity of ~100%. Furthermore, a fraction of mutations has been found exclusively in cfDNA, conceivably because,due to spatial tumor heterogeneity, they are restricted to clones that are anatomically distant from the biopsy site[20].
Regarding cHL, the rarity of neoplastic Hodgkin and Reed-Sternberg (HRS) cells in the biopsy has limited the assessments of the genetic landscape of the disease[23]. In this context, liquid biopsy may be a reliable tool to overcome this limit. Analogous to DLBCL, it has been demonstrated that cfDNA analysis in cHL mirrors the genetics of HRS cells micro-dissected from the tissue biopsy. Consistently, most of cHL mutations are identified in the tissue biopsy as well as in the cfDNA and some more mutations are identified only in cfDNA, conceivably because they are derived from different anatomical sites[23].Genotyping of cHL on the liquid biopsy has allowed to refine the current knowledge of cHL genetics.STAT6, not previously reported in exome sequencing studies of this lymphoma, has been identified as the most frequently mutated gene in cHL, underlying the importance of cytokine signaling in the biology of this tumor[23]. Also, liquid biopsy has documented that different histologic subtypes of cHL are biologically distinct.STAT6andTNFAIP3mutations are enriched in nodular sclerosis cHL compared with mixed cellularity cases. Also, NF-kB, PI3K-AKT, cytokine and NOTCH signaling pathways, are frequently deregulated by gene mutations, and might be relevant for the biology of the disease and for the identification of potential therapeutic targets[23].
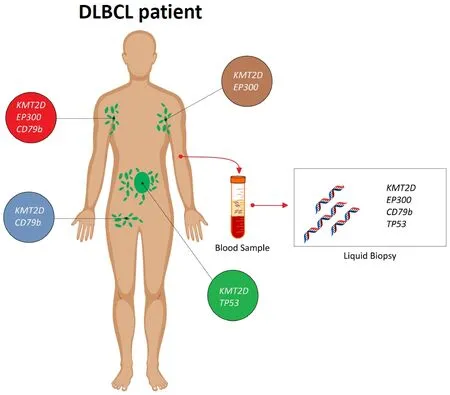
Figure 1. Liquid biopsy may recapitulate the entire mutational complexity of diffuse large B-cell lymphoma (DLBCL) patients. In every single DLBCL patient, the mutational profile of the lymphoma may vary in different anatomical sites. For instance, a DLBCL patient may harbor a EP300 and KMT2D mutation in the left axillary lymph node (in brown), a EP300, KMT2D and CD79b mutation in the right axillary lymph node (in red), a KMT2D and CD79b mutation in the right inguinal lymph node (in blue), and a KMT2D and TP53 mutation in a lymph node deep in the abdomen (in green). Consistently, if the biopsy targets a superficial lymph node (e.g., the left axillary lymph node; in brown), in order to avoid unnecessary surgical risks, a certain number of mutations (i.e., CD79b and TP53 mutation) present in the lymphoma genome would go undetected. Importantly, those mutations, if detected, might serve as predictive biomarkers as well as molecular markers allowing the monitoring of the disease during treatment. Conversely, cfDNA analysis on the liquid biopsy may overcome these limitations, since it is representative of all the different anatomical sites of the disease
LIQUID BIOPSY AS A TOOL TO EVALUATE TREATMENT RESPONSE
In cHL, imaging techniques, such as interim positron emission tomography/computer tomography(PET/CT) scan after 2 cycles of chemotherapy, provide a powerful tool to predict cHL outcome before completion of chemotherapy[24-26]. However, interim PET/CT results are inconsistent with the final outcome in 20% to 30% of patients[24-26]. This lack of specificity and/or sensibility may be corrected, at least in part,by monitoring ctDNA concentration during the course of treatment. A recent study prospectively analyzed ctDNA during treatment in a cohort of 24 cHL patients homogenously treated with adriamycin, bleomycin,vinblastine, dacarbazine (ABVD)[23]. The study showed that ctDNA analysis may complement interim PET/CT in predicting patients’ outcome[23]. More precisely, patients achieving a complete response had a larger drop in the ctDNA load after 2 ABVD courses compared to refractory patients. Also, a 2-log drop in ctDNA after 2 chemotherapy courses sorted out as the best cutoff to predict progression. Indeed, cured patients who were inconsistently judged as interim PET/CT-positive achieved more than a 2-log drop in ctDNA, whereas relapsing patients who were inconsistently judged as interim PET/CT negative achieved less than a 2-log drop in ctDNA[23].
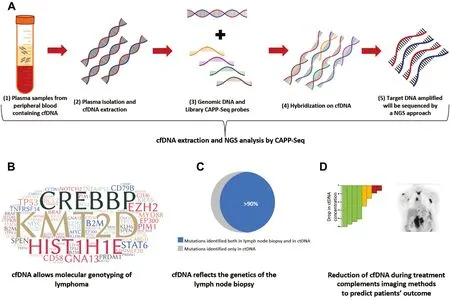
Figure 2. Applications of liquid biopsy in lymphomas. A: cell free DNA (cfDNA) can be selectively isolated and extracted from plasma samples. cfDNA may be analysed with the Cancer Personalized Profiling by Deep Sequencing (CAPP-Seq) method, that exploits magnetic probes that selectively capture and isolate specific genomic regions of interests, namely the target region. The target region is then subsequently sequenced by a Next-Generation-Sequencing (NGS) approach; B: consistently, cfDNA analysis on the liquid biopsy allows the genotyping of lymphomas and allows a better understanding of lymphoma pathogenesis; C: liquid biopsy mirrors the mutational landscape of the lymph node biopsy in most of cases and, in addition, allows the identification of mutations not identified in the tissue biopsy; D: the integration of imaging methods with the monitoring of the kinetics of the mutations identified by liquid biopsy on cfDNA may allow a more sensitive and specific prediction of patients’ outcome. The colored bars indicate the various levels of cfDNA drop during therapy. A higher drop in the cfDNA concentration (in green) may predict a favorable response to therapy, whereas a smaller drop may predict resistance to therapy
Similarly, ctDNA analysis by CAPP-Seq methodology also allows to evaluate treatment response in DLBCL patients. ctDNA has been analyzed at baseline and during the course of treatment in a multicenter cohort of 217 patients with DLBCL treated with rituximab, cyclophosphamide, doxorubicin, vincristine,prednisone (R-CHOP) or R-CHOP-like chemo-immunotherapy[21]. Two different thresholds have been identified to optimally predict patients’ outcome, namely early molecular response (EMR) and major molecular response (MMR). These thresholds, including a 2-log drop in ctDNA after one cycle (EMR) and a 2.5-log drop after two cycles (MMR), predict event free survival after front-line therapy in the training cohort and in two validation sets[21]. Interestingly, the prognostic value of molecular response maintained an independent association with an increased risk of progression and death also in multivariate analyses[21].Recently, a new prediction tool for DLBCL patients, namely Continuous Individualized Risk Index (CIRI),has been documented to dynamically determine outcome probabilities for individual patients utilizing risk predictors acquired over time, including clinical, radiological and molecular markers identified with a liquid biopsy approach[27].
CONCLUSIONS AND FUTURE PERSPECTIVES
In conclusion, liquid biopsy is a non-invasive method that may be used in lymphomas to achieve several objectives: (1) better characterize the biology and pathogenesis of the lymphoma through the analysis of biological markers that reflect the entire molecular heterogeneity of the disease in its different anatomical sites; (2) identify new prognostic and predictive markers; and (3) prospectively evaluate the dynamics of ctDNA during treatment in order to predict the probability of response to treatment in a more sensitive and specific manner and, if necessary, the need for an early switch to a more efficacious treatment. In order to validate these initial results, several ongoing clinical trials aim at assessing whether cfDNA genotyping can improve outcome prediction in lymphoid malignancies and whether it might identify mutations predisposing to treatment resistance (NCT03280394[28], NCT02883517[29], NCT03702309[30]).
In addition to the previously mentioned application of liquid biopsy in lymphoma, preliminary evidence also suggests that cfDNA analysis might reflect the methylation profile identified in the tissue biopsy[31,32].More precisely, global hypomethylation of the genome, as well as aberrant methylation of specific regions of interest detected on the liquid biopsy, namely DAPK1 promoter hypermethylation, might represent potential prognostic biomarkers in lymphomas[31,32].
The liquid biopsy of lymphoma, as described in this review, aims at analysing mutations of somatic origin,and therefore does not pose major ethical issues beyond the conventional ethical requirements. If the patient plasma is stored in a biobank for subsequent analysis, the conventional guidelines for biobanking should be followed[33,34].
DECLARATIONS
Authors’ contributions
Writing, reviewing and editing the manuscript: Moia R, Favini C, Rasi S, Deambrogi C, Ferri V, Schipani M,Sagiraju S, Mokhtar Mahmoud A, Kodipad AA, Adhinaveni R, Patriarca A, Nassi L, Gaidano G
Availability of data and materials
Not applicable.
Financial support and sponsorship
Work by the authors described in this review has been supported by Molecular bases of disease dissemination in lymphoid malignancies to optimize curative therapeutic strategies, (5 ´ 1000 No. 21198),Associazione Italiana per la Ricerca sul Cancro Foundation Milan, Italy; Progetto Ricerca Finalizzata RF-2011-02349712, Ministero della Salute, Rome, Italy; PRIN 2015ZMRFEA_004, MIUR, Rome, Italy; and AGING Project - Department of Excellence - DIMET, Università del Piemonte Orientale, Novara, Italy.
Conflicts of interest
All authors declared that there are no conflicts of interest.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Copyright
© The Author(s) 2019.
 Journal of Cancer Metastasis and Treatment2019年9期
Journal of Cancer Metastasis and Treatment2019年9期
- Journal of Cancer Metastasis and Treatment的其它文章
- AUTHOR INSTRUCTIONS
- Micromanaging autophagy with microRNAs to drive cancer metastasis
- Role of autophagy in therapeutic resistance of glioblastoma
- Membrane lipid binding molecules for the isolation of bona fide extraceIIuIar vesicIe types and associated biomarkers in Iiquid biopsy
- The immunological regulation of cancer cachexia and its therapeutic implications
