Variation in expression of small ubiquitin-like modifiers in injured sciatic nerve of mice
Dian-Ying Zhang , Kai Yu , Zhong Yang , Xiao-Zhi Liu, Xiao-Fang Ma, Yan-Xia Li
1 Department of Orthopedics and Trauma, People's Hospital, Peking University, Beijing, China
2 Department of Orthopedics, Tianjin Fifth Central Hospital, Tianjin, China
3 Central Laboratory, Tianjin Fifth Central Hospital, Tianjin, China
Abstract Small ubiquitin-like modifiers (SUMOs) have been shown to regulate axonal regeneration, signal transduction, neuronal migration, and myelination, by covalently and reversibly attaching to the protein substrates during neuronal cell growth, development, and differentiation. It has not been reported whether SUMOs play a role in peripheral nerve injury and regeneration. To investigate any association between SUMOylation and potential neuroprotective effects during peripheral nerve injury and regeneration, C57/BL mice were randomly divided into sham and experimental groups. The sciatic nerve was exposed only in the sham group. The experimental group underwent neurotomy and epineurial neurorrhaphy. Real-time quantitative polymerase chain reaction and western blot assay results revealed different mRNA and protein expression levels of SUMO1, SUMO2, SUMO3 and UBC9 in sciatic nerve tissue (containing both 5 mm of proximal and distal stumps at the injury site) at various time points after injury. Compared with the sham group, protein levels of SUMO1 and SUMO2/3 increased in both their covalent and free states after sciatic nerve injury in the experimental group, especially in the covalent state. UBC9 protein levels showed similar changes to those of SUMO1 and SUMO2/3 in the covalent states. Immunohistochemical staining demonstrated that SUMO1 and SUMO2/3 immunopositivities were higher in the experimental group than in the sham group. Our results verified that during the repair of sciatic nerve injury, the mRNA and protein expression of SUMO1, SUMO2, SUMO3 and UBC9 in injured nerve tissues changed in varying patterns and there were clear changes in the expression of SUMO-related proteins.These findings reveal that SUMOs possibly play an important role in the repair of peripheral nerve injury. All animal protocols were approved by the Institutional Animal Care and Use Committee of Tianjin Fifth Central Hospital, China (approval No. TJWZXLL2018041)on November 8, 2018.
Key Words: nerve regeneration; peripheral nerve injury; sciatic nerve; SUMO1; SUMO2/3; UBC9; SUMOylation; epineurial neurorrhaphy;neural regeneration
Graphical Abstract
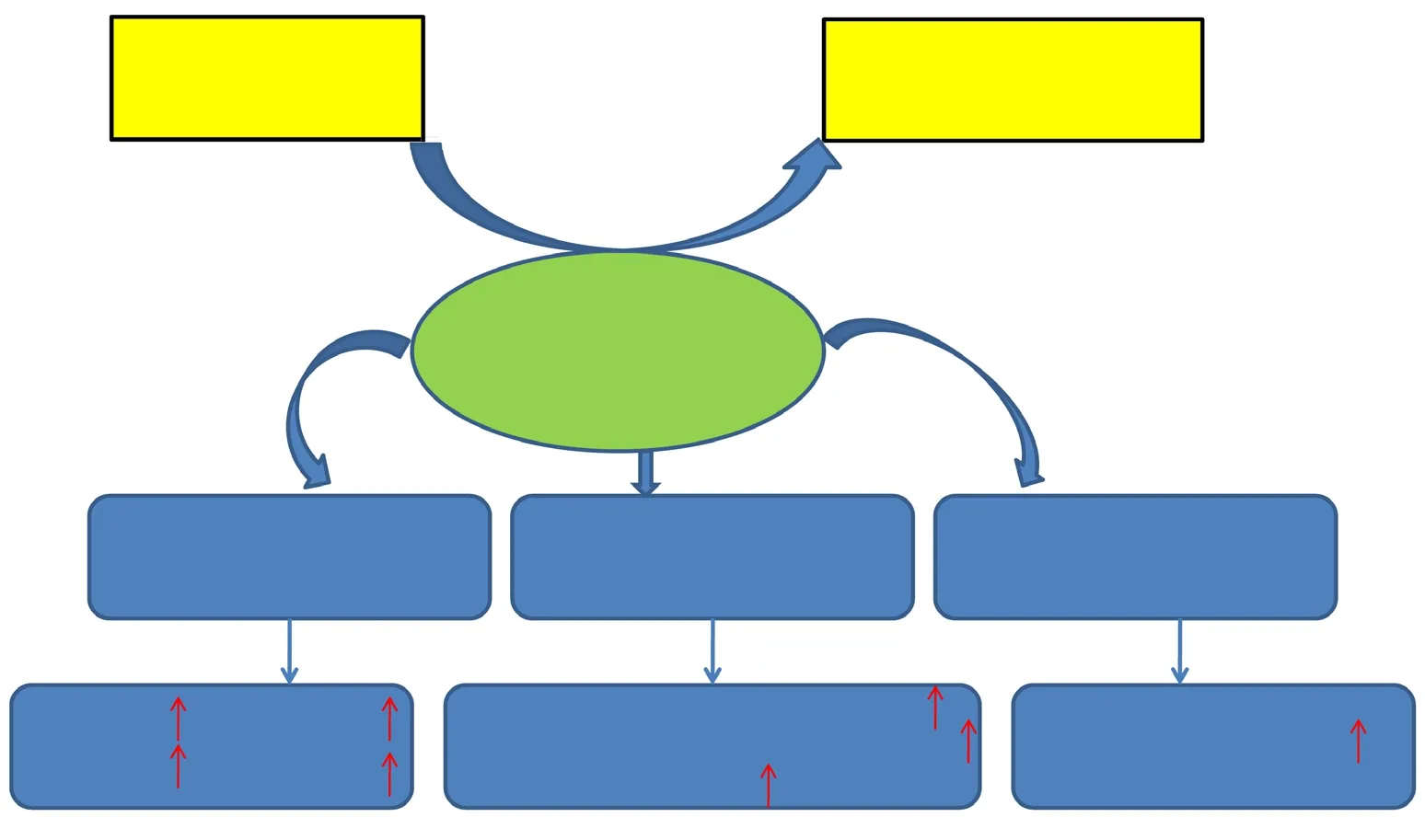
Expression of small ubiquitin-like modifier (SUMO) protein during repair of peripheral nerve injury
Introduction
Peripheral nerve injury is commonly presented in clinical practice and current treatments have limited success. Consequently patients' quality of life is affected, causing economic or social disability (Robinson, 2000; Taylor et al., 2008; Grinsell and Keating, 2014). Numerous previous studies have reported that the peripheral nervous system has the capability to regenerate spontaneously in response to traumatic injury.However, recovery is always unsatisfactory with a poor functional outcomes following repair of nerve injuries (Webber and Zochodne, 2010; Paprottka et al., 2013; Deng et al., 2017;Egawa et al., 2017; Navarro et al., 2018). Most investigations into the regeneration following peripheral nerve injury have focused on the morphological changes (Schmidhammer et al., 2007; Webber and Zochodne, 2010; Liu et al., 2015; Wang et al., 2017). The changes observed, such as demyelination,degeneration, remyelination, and regeneration, are associated with physiological and pathological changes to the nerve and surrounding tissues. Thus, it is important to thoroughly understand the molecular bases of the morphological change and local microenvironment regulated by gene expression during peripheral nerve injury and regeneration (Bosse et al., 2001,2006; Navarro et al., 2007; Qin et al., 2016; Liu et al., 2018).
SUMOylation (analogous to the ubiquitination process)is an evolutionarily conserved enzymatic pathway, which covalently and reversibly conjugates a small polypeptide of 97 amino acids, ubiquitin-like modifier (SUMO), to lysine residues of target proteins (Matunis et al., 1996; Mahajan et al., 1997; Henley et al., 2018; Zhao, 2018). SUMOylation plays essential roles in various biological functions, including cell growth, migration, and cellular responses (Cuijpers et al.,2017; Yang et al., 2017). Recently, the posttranslational modification, SUMOylation, has emerged as a major regulator of synapse formation, synaptic function and plasticity (Martin et al., 2007; Anderson et al., 2009, 2017). Many researchers have suggested that SUMO may be neuroprotective (Craig and Henley, 2012; Henley et al., 2014; Liu et al., 2017a). This was supported by the long-term effects on neuronal function of SUMOylation by regulation of transcription and nuclear traffic (Wilkinson et al., 2010). However, the research into SUMOylation and biological processes during peripheral nerve injury and regeneration has been sparse. It was especially tempting to associate the SUMOylation and potential neuroprotective effects to the site of the nerve injury. In the present study, the expressive variety of SUMOs was investigated following sciatic nerve transection injury in mice to discover the potential mechanism that may mediate peripheral nerve regeneration after injury. This study investigated SUMO1,SUMO2/3 and UBC9 expression, which may further improve therapeutic and preventative approaches to peripheral nerve injury.
Materials and Methods
Animals
All animal protocols were approved by the Institutional Animal Care and Use Committee of Tianjin Fifth Central Hospital, China (approval No. TJWZXLL2018041) on November 8,2018. Eighty-four C57/BL male mice aged 8 weeks and weighing 20-25 g were purchased from SPF Biotechnology Co., Ltd.(Beijing, China) (license No. SCXK (Jing) 2016-0002). The mice were randomly divided into a sham group and an experimental group. The sham group underwent sham operation(n = 12). The experimental group underwent neurotomy and epineurial neurorrhaphy (n = 72). Sciatic nerves (1 cm sciatic nerves containing both 5 mm proximal and distal stumps at the injury site) were harvested at the required time after surgery (1, 5, 7, 14, 21 and 28 days).
Group assignment and modeling
Mice were intraperitoneally anesthetized with 75 mg/kg pentobarbital (Sigma-Aldrich Inc., Shanghai, China). Using aseptic technique, the right sciatic nerve was exposed through a gluteal muscle-splitting incision, and transected at the sciatic notch with fine scissors. The sciatic nerve was sutured with 11-0 nylon suture at 10× microscopes by epineurial neurorrhaphy (Figure 1). Finally, the incision was closed with sutures. After resuscitation, the mice were returned to their cages. Mice whose sciatic nerves were successfully damaged presented with severe right hind paw drop with deep flexion and inversion of the toes. The mice were sacrificed by CO2inhalation at the required time after surgery (1, 5, 7, 14, 21, and 28 days). The procedure of sham operation was that mice only had surgical exposure of the sciatic nerve without any transection of the nerve. The sciatic nerves (1 cm sciatic nerves containing both 5 mm of proximal and distal stumps at the injury site) of experimental group and (1 cm sciatic nerves around the sciatic notch) of sham group were dissected from the suture point, and harvested for immunohistochemistry,real-time quantitative polymerase chain reaction (RT-qPCR)and western blot assay at the designated time after surgery.
Immunohistochemistry
At each designated time, four mice from each group were deeply anesthetized with pentobarbital and transcardially perfused with phosphate-buffered saline (0.1 M phosphate buffered saline) followed by 4% paraformaldehyde. The samples (1 cm sciatic nerves containing both 5 mm proximal and distal stumps at the injury site) were dissected, and fixed in Bouin's fixative overnight. The samples were dehydrated,and embedded in paraffin for longitudinal sectioning. The 4 μm thick slices were stained using the avidin-biotin staining technique. Briefly, slides were dewaxed, rehydrated, and boiled in a citrate buffer (0.1 M sodium citrate buffer, pH 6.0)for antigen retrieval. The slides were incubated in 2% H2O2to inactivate endogenous peroxidase at room temperature for 20 minutes. Next, the slides were incubated in blocking buffer (20% normal goat serum in phosphate buffered saline with 1% bovine serum albumin) for 30 minutes and incubated with primary antibody (anti-SUMO1, ab32058, rabbit,monoclonal antibodies, 1:250, Abcam, Cambridge, MA,USA; anti-SUMO2/3, ab3742, rabbit, polyclonal antibodies,1:800, Abcam) overnight at 4°C. After the slides had been washed with phosphate buffered saline, they were incubated with avidin-conjugated secondary antibodies (ab64264,mouse and rabbit, 1:500, Abcam) for 30 minutes at room temperature. An avidin-biotin complex solution was added for 30 minutes at room temperature. The antigens were visualized with 3.3′-diaminobenzidine (ab64264, Abcam).Finally, the slides were washed with double distilled water,air-dried, and fixed. Sections were analyzed with a standard light microscope focused on the injury site (Zeiss Axioskop(Carl Zeiss AG, Oberkochen, Germany) with a UC30 camera (Olympus Corp., Tokyo, Japan). The integrated optical density of each image was calculated using Image-Pro Plus 6.0 version software (Media Cybernetics, Inc., Rockville,MD, USA) as described (Feng and Yuan, 2015).
RT-qPCR
The sciatic nerves were harvested from four mice in each group, snap frozen and stored at -80°C until further use.The nerves were homogenized in TRIzol with a Potter Elvehjem homogenizer, and small fragments were further homogenized by sonication. The RNeasy Lipid Tissue kit(QIAGEN Co., Ltd., Shanghai, China) was used to extract the total RNA according to the manufacturer's protocol.Gel electrophoresis was used to verify the quality of the RNA. DNase treatment was performed with TURBO DNase(Invitrogen Life Technologies, Carlsbad, CA, USA). The Superscript III first strand synthesis system for RT-qPCR(Invitrogen Life Technologies) was used to produce cDNA.RT-qPCR was done with 10 ng cDNA in SYBR Green I mix and run on an ABI Prism 7900 HT Sequence Detection System (Applied Biosystems). All reactions were performed in triplicate. PrimerBank was used for the design and making of primers. Primer sequences are listed in Table 1. RT-qPCR data were normalized according to the method described by Vandesompele et al. (2002). With an improved version of the 2-ΔΔCtmethod (Hellemans et al., 2007), the raw data were processed and the relative quantities were normalized.
Western blot assay
At each designated time, sciatic nerves were harvested from four mice from each group. Total protein was extracted by a cell lysis solution. Western blot assay was performed using 4-15% sodium dodecyl sulfate polyacrylamide gel electrophoresis gels (Bio-Rad Laboratories, Inc.). The proteins were transferred onto polyvinylidene fluoride membranes (Bio-Rad Laboratories, Inc.) by electrophoresis. The membranes were blocked using Tris/HCl-buffered salt solution (0.1%Tween-20 and 5% skim milk powder), and then incubated with antibodies overnight at 4°C (anti-SUMO 1, ab133352,rabbit, monoclonal antibodies, 1:2000; Abcam, Cambridge,MA, USA; anti-SUMO 2/3, ab3742, rabbit, polyclonal antibodies, 1/5000; Abcam; anti-UBC9, ab227733, rabbit,polyclonal antibodies, 1:1000; Abcam). The membranes were washed five times in 0.1% Tris-buffered saline with Tween and then incubated with the secondary antibodies chicken anti-rabbit IgG conjugated to horseradish peroxidase (sc516087, 1:2000, chicken anti-rabbit, polyclonal antibodies; Santa Cruz Biotechnology, Inc.) for 1 hour. The Super Signal protein detection kit (Pierce, Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to detect the labeled proteins. Afterwards, the membranes were stripped and reprobed with an anti-GAPDH (ab8245, mouse, monoclonal antibodies, 1:2000; Abcam). Changes in the levels of SUMO conjugated proteins were evaluated using ImageJ software 1.37c (NIH, Bethesda, MD, USA). The high molecular weight area was cropped and analyzed in each lane.

Table 1 Primers for real-time quantitative polymerase chain reaction
Statistical analysis
Statistical analyses were performed using SPSS version 16.0 software (SPSS, Inc., Chicago, IL, USA). Data are expressed as the mean ± SD and were evaluated by one-way analysis of variance followed by Tukey's honestly significant difference post hoc test for multiple comparisons depending on the experimental design. A value of P < 0.05 was considered statistically significant.
Results
Immunopositivity for SUMO1 and SUMO2/3 after sciatic nerve injury
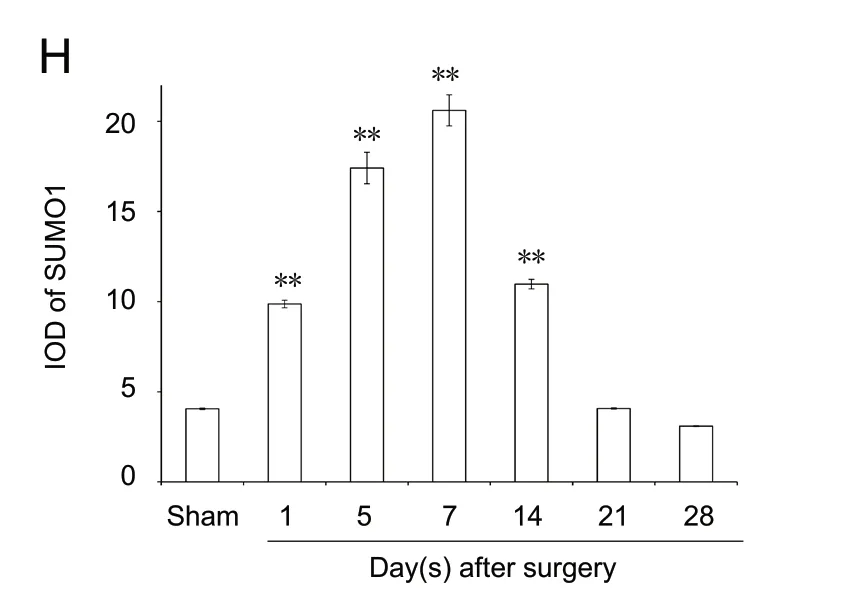
Immunopositivity for SUMO1 and SUMO2/3 was observed in all groups, but especially at the suture point of sciatic nerve(Figures 2A-G and 3A-G). SUMO1 expression significantly increased initially after injury in the experimental groups compared with the sham group at different time points (P< 0.01), but returned to control levels 21 and 28 days after injury. SUMO2/3 expression increased significantly at different time points in the experimental group compared with the sham group (P < 0.01). Both SUMO1 (0.206 ± 0.009) and SUMO2/3 (0.238 ± 0.003) peaked 7 days after injury in the experimental group (Figures 2H and 3H).
SUMO1, SUMO2, SUMO3 and UBC9 mRNA expression levels increase after sciatic nerve injury
The pattern of changes in mRNA expression levels of SUMO1, SUMO2, SUMO3 and UBC9 varied after injury(Figure 4A). Compared with the sham group, SUMO1,SUMO2, SUMO3 and UBC9 expression levels did not increase in the experimental group (P > 0.05; Figure 4B).
SUMO1, SUMO2/3 and UBC9 protein expression levels increase after sciatic nerve injury
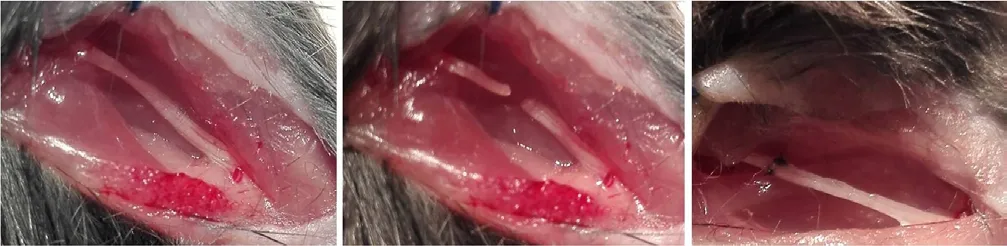
Figure 1 Suture model of sciatic nerve.(A) Sciatic nerve exposure; (B) sciatic nerve transection at the sciatic notch;(C) epineurial neurorrhaphy of the transected sciatic nerve.
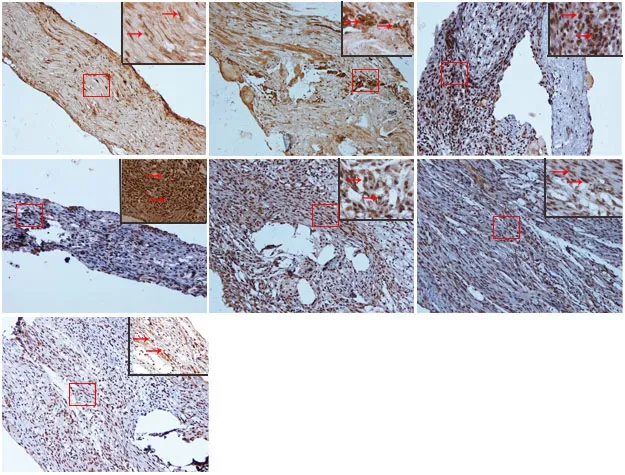
Figure 2 Immunohistochemical staining for SUMO1 in sciatic nerves.(A) SUMO1 immunopositive staining (arrows) in sciatic nerve of the sham group (1 cm sciatic nerves around the sciatic notch); (B-G) SUMO1 immunopositive staining (arrows) in the sciatic nerve (containing both 5 mm proximal and distal stumps at the injury site) at 1, 5, 7, 14, 21, and 28 days after sciatic nerve injury in the experimental group, respectively.SUMO1 immunopositive staining was observed in A-G, especially in the sciatic nerve suture point (the boxed areas) of Figure B-E. Using optical microscope, original magnification, 20×; insets are higher magnification of the boxed areas,original magnification, 100×. (H) Quantitative analysis of SUMO1 immunopositivity in each group. **P < 0.01, vs. sham group (mean ± SD, n = 4, one-way analysis of variance followed by Tukey's honestly significant difference post hoc test).IOD: Integrated optical density; SUMO: small ubiquitin-like modifier.
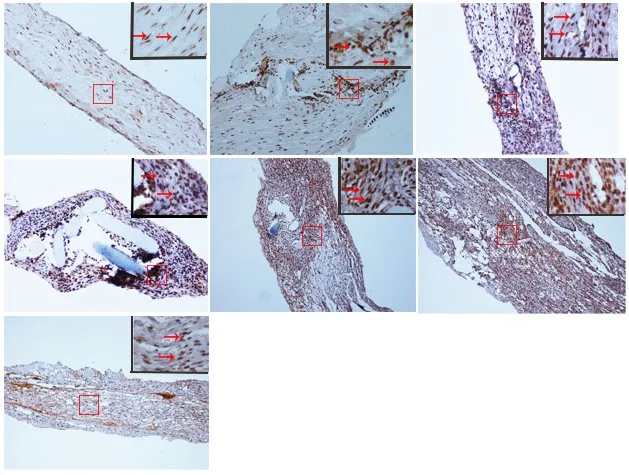
Figure 3 Immunohistochemical staining for SUMO2/3 in the sciatic nerve.(A) SUMO2/3 immunopositive staining (arrows) in the sciatic nerve of sham group (1 cm sciatic nerves around the sciatic notch); (B-G) SUMO2/3 immunopositive staining (arrows) in the sciatic nerve (containing both 5 mm proximal and distal stumps at the injury site) at 1, 5, 7, 14, 21 and 28 days after sciatic nerve injury in the experimental group, respectively.SUMO2/3 immunopositive staining was observed in A-G, especially at the sciatic nerve suture point (the boxed areas) of B-G. Using optical microscope, original magnification, 20×; insets are higher magnification of the boxed areas, original magnification, 100×. (H) Quantitative analysis of SUMO2/3 immunopositivity in each group. **P < 0.01, vs. sham group(mean ± SD, n = 4, one-way analysis of variance followed by Tukey's honestly significant difference post hoc test). IOD:Integrated optical density; SUMO: small ubiquitin-like modifier.
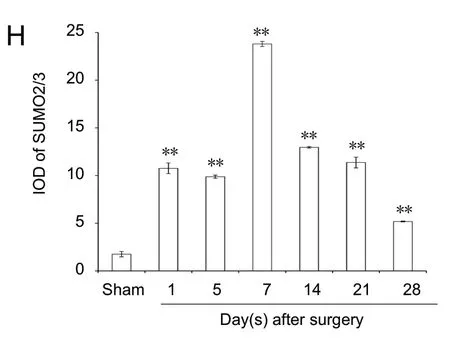

Figure 4 SUMO1, SUMO2, SUMO3, and UBC9 mRNA expression levels after sciatic nerve injury.(A) Real-time quantitative polymerase chain reaction analyses for SUMO1,SUMO2, SUMO3, and UBC9 of the sciatic nerve. (B) Quantification of expression of SUMO1, SUMO2, SUMO3, and UBC9 in each group: GAPDH was used as an internal control. UBC9: Ubiquitin-conjugating enzyme 9; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; SUMO:small ubiquitin-like modifier.
The results of western blot assay revealed increasing protein expression levels in both covalent and free states of SUMO1 and SUMO2/3 in the days after sciatic injury, especially in the covalent state (SUMO1 protein level in the covalent state: P < 0.01; SUMO1 protein level in the free state: P <0.01; SUMO2/3 protein level in the covalent state: P < 0.01;SUMO2/3 protein level in the free state: P < 0.01) compared with the sham group (Figures 5 and 6). The protein levels of UBC9 and the SUMO-conjugating enzyme were significantly higher at each time point after nerve injury, similar to the tendencies of SUMO1 and SUMO2/3 in the covalent states,than those in the sham group (P < 0.01; Figure 7).


Figure 5 SUMO1 protein expression in sciatic nerve.(A) Detection of SUMO1 levels in covalent and free states by western blot assay. (B)Quantification of expression of SUMO1 in each group, GAPDH was used as an internal control. *P < 0.05, **P < 0.01, vs. sham group (mean ± SD, n = 4, one-way analysis of variance followed by Tukey's honestly significant difference post hoc test). SUMO: Small ubiquitin-like modifier; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
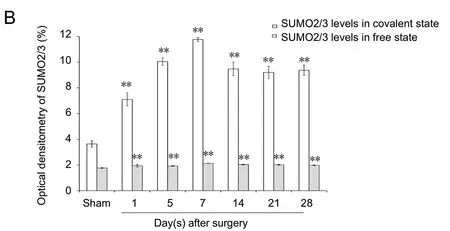
Figure 6 SUMO2/3 protein expression in sciatic nerve.(A) Detection of SUMO2/3 levels in covalent and free states by western blot assay. (B)Quantification of expression of SUMO2/3 in each group: GAPDH was used as an internal control. **P < 0.01, vs. sham group (mean ± SD, n = 4, one-way analysis of variance followed by Tukey's honestly significant difference post hoc test). SUMO: Small ubiquitin-like modifier; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; d: day(s).
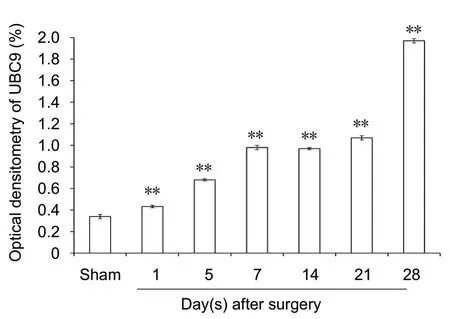
Figure 7 UBC9 protein expression in sciatic nerve.(A) Detection of UBC9 levels by western blot assay. (B) Quantification of expression of UBC9 in each group: GAPDH was used as an internal control. **P< 0.01, vs. sham group (mean± SD, n = 4, one-way analysis of variance followed by Tukey's honestly significant difference post hoc test). UBC9: Ubiquitin-conjugating enzyme; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
Discussion
Peripheral nerve injury is a common clinical problem that significantly affects the patients' quality of life and causes economic or social disability. Its treatment remains a great challenge in the clinic (Liu et al., 2015). Much research has focused on the functional recovery after peripheral nerve injury; however, recovery has generally been unsatisfactory with poor functional outcomes (Deumens et al., 2010; Paprottka et al., 2013; Liu et al., 2017b). There are many pathophysiological changes that occur following peripheral nerve injury, such as demyelination, degeneration, remyelination and regeneration (Scheschonka et al., 2007; Webber and Zochodne, 2010; Liu et al., 2018; Yang et al., 2018). Therefore,it is important to thoroughly understand the pathophysiological changes following peripheral nerve injury, which may provide an avenue for the treatment of peripheral nerve injury.
SUMOylation is a process when the SUMO protein covalently modifies acceptor lysine residues of a diverse range of target proteins to alter their functional properties (Eifler and Vertegaal, 2015; Iribarren et al., 2015; Citro and Chiocca,2017; Han et al., 2018). It plays a key role in many essential physiological processes, and in pathological conditions (Luo et al., 2013; Chang and Ding, 2018; Zhao, 2018). SUMOylation is increasingly attracting attention in neuroscience,because of its key roles in physiological neurological functions (Wilkinson et al., 2010; Henley et al., 2014, 2018). Many neurodegenerative diseases, such as Parkinson's disease, Alzheimer's disease, oxidative stress, osmotic stress and hypoxic stress, had been shown to be associated with SUMOylation(Martins et al., 2016; Anderson et al., 2017; Peters et al., 2017;Princz and Tavernarakis, 2017). Therefore, it is important to understand whether SUMOylation impacts peripheral nerve regeneration. In this study, the western blot assay results revealed an increase in both the covalent and free states of the protein levels of SUMO1 and SUMO2/3 after sciatic nerve injury, but especially in the covalent state. Furthermore,SUMO1 and SUMO2/3 immunopositivities were predominantly observed in the sciatic nerve, especially at the suture point, which is consistent with the results from the western blot assay. It suggested that SUMOs may have a role in promoting peripheral nerve regeneration.
Ubiquitin-conjugating enzyme 9 (UBC9) is the sole specific E2-type conjugating enzyme for SUMO proteins. Thus, SUMOylation is dependent upon UBC9 expression (Golebiowski et al., 2003; Moschos et al., 2010), making it an ideal target for the activation or blockage of the SUMO modification pathway. UBC9 expression in mammalian neurons has been investigated in several studies (Scheschonka et al., 2007; Henley et al., 2014, 2018). Although increasing attention is being paid to SUMOylation in the nervous system (Wang et al., 2007).In the present study, UBC9 protein levels were remarkably increased in the experimental group compared with the sham group at all time points after injury, showing similarities with changes in the covalent states of SUMO1 and SUMO2/3. This supports the role of UBC9 and SUMOs at the post transcriptional stage. Our results suggest that protein SUMOylation has a role in peripheral nerve regeneration.
Much research has suggested that neurodegenerative diseases are fundamentally associated with disorders of the RNA and it has been suggested that the mRNA expression might have a role in the pathological processes of degenerating neurons (Andreassi et al., 2018). In our study, SUMO1,SUMO2, SUMO3 and UBC9 mRNA expression levels did not increase after injury, comparing with sham group. However, the increased changes we observed in protein expression of UBC9 and SUMOs suggest that the post transcriptional protein SUMOylation is involved in peripheral nerve regeneration rather than at the transcriptional level.
This study had some limitations. Our results found that SUMOs and UBC9 were expressed differently in the injured sciatic nerve of mice. The sequence of changes requires further investigation to discover the potential neuroprotective role of SUMOylation after peripheral nerve injury.
In summary, SUMO1, SUMO2/3 and UBC9 had different patterns of expression after sciatic nerve injury, suggesting that SUMOs might have a neuroprotective role in peripheral nerve regeneration, especially in post-translational protein modification. Our results reveal a novel mechanism for possible therapy of peripheral nerve injury to improve the success rate of peripheral nerve regeneration and ultimately to improve patient rehabilitation.
Author contributions: Study design: DYZ and KY; experimental implementation: ZY, XZL, XFM and YXL; data analysis: ZY; paper writing:DYZ, KY and ZY. All authors approved the final version of the paper.
Conflicts of interest:The authors declare that there are no conflicts of interest associated with this manuscript.
Financial support: This study was supported by the National Natural Science Foundation of China (General Program), No. 316781246 (to DYZ);the Science and Technology Fund of Health Planning Commission of Binhai New Area of Tianjin of China, No. 2011BHKY008, 2012BWKZ002 (to KY); the National Key Research and Development Program of China, No.2016YFC1101600 (to KY); the National Key Research and Development Program of China, No. 2016YFC1101600 (to DYZ). The funding sources had no role in study conception and design, data analysis or interpretation, paper writing or deciding to submit this paper for publication.
Institutional review board statement: All animal protocols were approved by the Institutional Animal Care and Use Committee of Tianjin Fifth Central Hospital, China (approval number: TJWZXLL2018041) on November 8, 2018. All experimental procedures described here were in accordance with the National Institutes of Health (NIH) guidelines for the Care and Use of Laboratory Animals.
Copyright license agreement:The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review:Externally peer reviewed.
Open access statement:This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-Non-Commercial-ShareAlike 4.0 License, which allows others to remix, tweak,and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
- 中国神经再生研究(英文版)的其它文章
- Improvement of ataxia in a patient with cerebellar infarction by recovery of injured cortico-ponto-cerebellar tract and dentato-rubro-thalamic tract: a diffusion tensor tractography study
- Tandem pore TWIK-related potassium channels and neuroprotection
- Dendritic shrinkage after injury: a cellular killer or a necessity for axonal regeneration?
- Regenerative biomarkers for Duchenne muscular dystrophy
- Exploring the efficacy of natural products in alleviating Alzheimer's disease
- Role of macrophages in peripheral nerve injury and repair

