Kiwifruit (Actinidia chinensis) R1R2R3-MYB transcription factor AcMYB3R enhances drought and salinity tolerance in Arabidopsis thaliana
ZHANG Ya-bin, TANG Wei , WANG Li-huan HU Ya-wen LlU Xian-wen, LlU Yong-sheng
1 Key Laboratory of Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University,Chengdu 610065, P.R.China
2 School of Food Science and Engineering, Hefei University of Technology, Hefei 230009, P.R.China
Abstract Kiwifruit is an important fruit crop that is highly sensitive to environmental stresses, such as drought, heat, cold, water logging and phytopathogens. Therefore it is indispensable to identify stress-responsive candidate genes in kiwifruit cultivars for the stress resistance improvement. Here we report the isolation and characterization of a novel kiwifruit R1R2R3-MYB homolog(AcMYB3R) whose expression was induced by drought, salinity and cold stress. In vitro assays showed that AcMYB3R is a nuclear protein with transcriptional activation activity by binding to the cis-element of the kiwifruit orthologue of G2/M phase-specific gene KNOLLE. The Arabidopsis transgenic plants overexpressing AcMYB3R showed drastically enhanced tolerance to drought and salt stress. The expressions of stress-responsive genes such as RD29A, RD29B, COR15A and RD22 were prominently up-regulated by ectopic expression of AcMYB3R. Our study provides a valuable piece of information for functional genomics studies of kiwifruit and molecular breeding in improving stress tolerance for crop production.
Keywords: kiwifruit, R1R2R3-MYB, drought, abiotic stress
1. lntroduction
Due to the sessile nature, plants are ever challenged by diverse environmental stresses such as drought, salt and/or low temperature. These stresses adversely affect plants growth and development, and consequently reduce crop yield (Yamaguchi-Shinozaki and Shinozaki 2006; Mittler and Blumwald 2010).
Many genes or their encoded proteins are essential for plants to cope with abiotic stress, especially transcription factors (Li Tet al. 2016; Huet al. 2017). In plants, a number of transcription factors (TF) have been demonstrated to be involved in regulating stress tolerance (Yamaguchi-Shinozaki and Shinozaki 2001; Songet al. 2016). The MYB transcription factor is one of the largest transcription factor families in plants (Duboset al. 2010). MYB proteins can be divided into different classes based on the number of MYB domain (one, two, three or four) (Rosinski and Atchley 1998; Duboset al. 2010). MYB3R (R1R2R3-MYB)containing three conserved MYB domains has been shown to play a pivotal role in response to abiotic stress in plants.Overexpression of a riceMYB3Rgene (OsMYB3R-2)significantly increased tolerance to cold, drought and salt stress in transgenicArabidopsis(Daiet al. 2007; Maet al.2009). A wheatMYB3Rgene,TaMYB3R1, was shown to participate in regulation of osmotic stress tolerance in transgenicArabidopsisby in fluencing expressions of both ABA-dependent and ABA-independent genes (Caiet al.2011, 2015). In addition, MYB3R proteins have been reported to play multifunctional roles in regulating the G2/M phase transition. In tobacco, NtmybA1 and NtmybA2 were demonstrated to activate the specific AACGG core motif sequence, the mitosis-specific activator (MSA) element,which is present in kinesin-like protein geneNACK1and B-type cyclin genes (Itoet al. 2001). InArabidopsis, both MYB3R1 and MYB3R4were shown to regulate cytokinesis through activation of specific G2/M phase geneKNOLLE(Hagaet al. 2007). These investigations demonstrated MYB3R proteins are important TFs involved in regulating abiotic stress tolerance in various plant species (Fenget al.2017).
Kiwifruit, well known as its high ascorbic acid content in fruit, usually encounters various abiotic environmental stresses, such as drought, heat, cold, water logging and phytopathogens, which severely impair its growth and development (Tanget al. 2016). Therefore, it is necessary to unearth novel genes related to abiotic stresses and apply them to improve stress resistance of kiwifruit varieties. In previous study,AcDHAR1andAcDHAR2have been isolated and characterized in kiwifruit, suggesting they play a critical role in the regeneration of L-ascorbic acid and response to environmental stimuli in kiwifruit such as salt stress (Liuet al. 2016).
Here we report the isolation and characterization of a kiwifruitMYB3Rhomolog (AcMYB3R; accession no.Ach00g380251) fromActinidia chinensiscv. Hongyang(Huanget al. 2013). Real-time PCR analysis showed expression ofAcMYB3Rwas activated by drought,salt and cold stresses. Subcellular localization assay indicated AcMYB3R is a nuclear-localized protein with transcriptional activation activity. GUS assay demonstrated the G2/M phase specific-geneAcKNOLLEwas activated byAcMYB3R. TransgenicArabidopsisoverexpressingAcMYB3Rexhibited a significantly enhanced tolerance to drought and/or salt stress.
2. Materials and methods
2.1. Plant materials
The tissue samples of root, stem, leaf, flower and fruit were collected from 5-year-oldA.chinensiscv. Hongyang plants regenerated from tissue culture in the farm at Hefei University of Technology, China. The sample tissues were immediately frozen in liquid nitrogen and stored at–80°C. Three biological replicates were conducted for each sampling.
TheArabidopsis thalianaecotype Columbia was employed in this study.Arabidopsisseeds were surfacesterilized for 1 min with 75% ethanol and subsequently 10 min with diluted (1:3, v/v) NaClO, followed by rinsing with sterile water five times. The plants were grown either in soil or Murashige and Skoog (MS) medium at 22°C under 16 h-light/8 h-dark cycle illumination. MS medium contains 0.5× MS salts, 2% (w/v) sucrose, and 0.7% (w/v) agar.
To determine the expression level ofAcMYB3Rin Hongyang root, 7-month-old plants regenerated from tissue culture were treated with 200 mmol L–1NaCl or 350 mmol L–1mannitol or 4°C. The treated roots were harvested at the indicated times (0, 1, 3, 6, 12 and 24 h) after treatments,then immediately frozen in liquid nitrogen and stored at–80°C. To analyze the expression patterns of drought stress-responsive genes inArabidopsis, 3-week-old wild type (WT) and transgenic plants were treated with 350 mmol L–1mannitol for 6 h, and intact plants were collected at the indicated time (0, 1, 3 and 6 h), then immediately frozen in liquid nitrogen and stored at –80°C.
2.2. lsolation and sequence analysis of the AcMYB3R from kiwifruit
Based on Kiwifruit Information Resource (Yueet al. 2015),gene-specific primers were designed and used for amplifying the cDNA ofAcMYB3R(accession no. Ach00g380251)by RT-PCR with PrimeStar DNA polymerase (TaKaRa,Japan). After sequencing, theAcMYB3Rgene fragment cut from pEASY-Blunt simple vector was cloned into theHindIII-XbaI site of a pHB vector to create the35S::AcMYB3Rrecombinant vector driven by CaMV 35S promoter. Gene-specific primers (AcMYB3R-F,5´-CCCAAGCTTATGATAGAGGTTAAGAGAGAG-3´;AcMYB3R-R, 5´-TGCTCTAGATTAAGGAGTCAAC ATCACATT-3´) were used forAcMYB3Ramplification, with underlined nucleotides as restriction enzyme cutting sites.
Alignment of amino acid sequences was conducted by DNAMAN v 8.0. AcMYB3R and its homologous sequences were collected from Kiwifruit Information Resource (http://bdg.hfut.edu.cn/kir/index.html) and NCBI (https://www.ncbi.nlm.nih.gov/) by BLASTp search tool. The R1-, R2- and R3-MYB domain sequences of these proteins were defined by the sequence search in SMART (http://smart.emblheidelberg.de/). The phylogenetic analysis was carried out using MEGA v. 5.0viathe neighbor-joining (NJ) method withp-distance under the standard parameters.
2.3. Arabidopsis transformation of AcMYB3R
The recombinant construct of35S::AcMYB3Rwas transformed into 5-week-oldArabidopsisplant usingAgrobacterium tumefaciensstrain GV3101 by floral dip method (Zhanget al. 2006). Transformed progenies were screened on 0.5× MS medium with 50 μg mL–1Basta(Sigma,USA). At T2 generation, qRT-PCR analysis was used to detect the transgene expression. Three independent T3 homozygous lines (#3, #5 and #6) were used for further studies.
2.4. Subcellular localization
The open reading frame (ORF) ofAcMYB3Rwithout the termination codon was ampli fied by PCR using gene-specific primers (5´-GGTACCATGATAGAGGTTAAGAGAGAGG-3´and 5´-AAGCTTAGGAGTCAACATCACATTT-3´), with the underlined nucleotides as theKpnI andHindIII restriction enzyme cutting site, respectively. The product was inserted into the expression vector pART27 containing the green fluorescent protein (GFP) coding sequence at the 5´ terminal of GFP gene under the control of CaMV 35S promoter to generate a fusion protein construct35S:AcMYB3R-GFP.The recombinant vectorA. tumefaciensstrain GV3101 harboring35S:AcMYB3R-GFP construct was in filtrated into tobacco (Nicotiana benthamiana) leaves. Two days after in filtration, the tobacco protoplasts were isolated and fixed in W5 buffer containing 0.5% (v/v) glutaraldehydefor 20 min and subsequently treated with 10 μg mL–14´,6-diamidino-2-phenylindole (DAPI) solution for 30 min (Liuet al. 2016). The GFP signal was detected by confocal microscope (FV1000;Olympus, Tokyo, Japan).
2.5. Transcription activation assay in yeast
Transcription activation assay was performed using the MATCHMAKER LexA Two-Hybrid System (Clontech,Japan). The ORF ofAcMYB3Rwas introduced into the pEG202 vector to createAcMYB3R-pEG202 construct.The construct was transformed into EGY48 strains by the lithium acetate-mediated method (Gietzet al. 1992).The positive transformants were selected on synthetic medium plates (SD medium) lacking Ura/His (SD/-Ura/-His) at 28°C, and 2–4 days later these yeast strains were tested on selective plate medium or by beta-galactosidase activity assay (LiY Xet al. 2016). The fragment ofAcMYB3Rwas ampli fied using the gene-specific primers(5´-GAATTCATGATAGAGGTTAAGAGAGAG-3´ and 5´-CTCGAGTTAAGGAGTCAACATCACATT-3´), with the underlined nucleotides as restriction enzyme cutting site.
2.6. Quantitative real-time PCR
Total RNA ofArabidopsisand kiwifruit samples was extracted using Trizol regent (TianGen, China) followed by treatment with RNase-free DNase I (Promega, USA) according to the manufacturers’ protocols. Then, 2 μg total RNA was used for synthesizing the first strand cDNA using PrimeScript™RT reagent Kit (Perfect Real Time) (TaKaRa, Japan).Quantitative real-time PCR (qRT-PCR) was performed on the ABI StepOne real-time PCR System (Applied Biosystem,USA). qRT-PCR was carried out in a total volume of 20 μL, containing 10 μL of SYBR mix (TransGene, China),0.4 μmol L–1of each primer, 6 μL of 1:20 diluted cDNA and 3.6 μL ddH2O. The reactions were performed with the program: 95°C for 10 min, 40 cycles at 95°C for 15 s, and 60°C for 60 s. Amplification ofAtActin2(accession no. AK063598)andAcActin2(accession no. Ach05g107181) were used as internal controls for calculating relative transcript levels inArabidopsisand kiwifruit, respectively. Relative expression levels were calculated by the 2–ΔΔCTmethod (Livak and Schmittgen 2001). Each sample was quanti fied in triplicate.Gene-specific primer pairs used for qRT-PCR were listed forAcMYB3R(5´-CAGGGACCTTCATCTCCTGA-3´ and 5´-CAAAAGAGGCTTCCCTGAGA-3´),AcActin2(5´-CT TACAGAGGCACCACTCAACC-3´ and 5´-ATAGAT GGGGACCGTGTGACT-3´),RD29A(5´-TGCACCAGGC GTAACAGG-3´ and 5´-ATCGGAAGA CGCGACAGGAA-3´),RD29B(5´-AGAGGTGGTGT AACGGGTAA-3´ and 5´-GG CTCAATG GGTTTGGTG-3´),COR15A(5´-TCAGTTCGTCG TCGTTTC-3´ and 5´-CATCTGCTAATGCCTCTTT-3´),RD22(5´-ACTTGGTAA ATATCACGTCAGGGCT-3´ and 5´-CTG AGGTGTTC TTGTGGCATACC-3´),AtActin2(5´-GCTCCTCTTAACCCAAAGGC-3´ and 5´-CACACCATCA CCAGAATCCAGC-3´), andAtKNOLLE(5´-CTAATCA GAAGAGTGAAAAAGATG-3´ and 5´-TTCGTAGAAG CCATCTCAAGATC-3´).
2.7. Drought and/or salt stress treatment in transgenic Arabidopsis
For root length measurement, seeds from WT and transgenic lines were germinated on 0.5× MS medium for 2 days, then transferred to fresh medium (supplement with or without 120 and 150 mmol L–1NaCl, 300 and 350 mmol L–1mannitol) for vertical growth. After growing 10 days, root length was measured. All experiments were replicated at least three times.
For testing salinity tolerance, 3-week-old WT and transgenic plants were irrigated by solutions containing different concentrations of salt for 12 days at 4-day intervals,if rst with 100 mmol L–1, then with 200 mmol L–1and finally with 300 mmol L–1NaCl, after which their survival rate was recorded.
For drought tolerance test, water was withheld for 14 days from 3-week-old WT and transgenic plants grown under suf ficient moisture, and subsequently all plants were rewatered. Their growth parameters were scored 7 days later.
2.8. GUS assay
To create β-glucuronidase (GUS) reporter constructs,about 2-kb promoter region ofAcKNOLLE(accession no. Ach23g121631) was ampli fied by PCR and cloned in front of the GUS gene in pBI121 binary vector (35S::GUS). Primers used for PCR were 5´-ccatgattacgccaagcttGAAATTGCCGGAAGAGGAC-3´ and 5´-actgaccacccggggatccCTTGAGTGCTCTGTTTCAGGA-3´.The resultant constructpAcKNOLLE:GUSwas transformed intoA.tumefaciensGV3101.Agrobacteriumcell cultures carrying appropriate constructs were syringe-in filtrated intoN.benthamianaleaves. After 2 days, the in filtrated leaves were collected and immediately stained with X-gal staining solutionviavacuum in filtration, then stained 1 day at 37°C in the dark. Stained leaves were placed in bleaching solution(ethanol:acetic acid:glycerol=3:1:1) and incubated at 37°C until the leaves were cleared of chlorophyll (Liuet al. 2012).GUS activity was analyzed using fluorometric assays according to previously described procedure (Jeffersonet al. 1987).
2.9. The determination of malondialdehyde (MDA)and proline content
Leaf tissues (about 500 mg) from stress-treatedArabidopsisseedlings was used for determining MDA and proline content as described previously (Gaoet al. 2014; Tanget al. 2014).Three replications were conducted for each of the wild type and transgenic plants.
2.10. DAB staining and H2O2 assay
Histochemical staining with 3,3´-diaminobenzidine (DAB)was used to analysisin situaccumulation of H2O2according to previously described procedure (Tanget al. 2014). Leaf tissues (500 mg) were used for determining H2O2content following the described method (Tanget al. 2014). All experiments were repeated at least three times.
2.11. Statistical analysis
At least three independent replicates were performed, and the average values were presented. Error bars represent SD(n>3). Statistical significance was determined by Student’st-test. Significant differences were indicated by asterisks (*,P<0.05;**,P<0.01). All data were analyzed by using SPSS statistical ver. 22.0 software package.
3. Results
3.1. Expression pattern of AcMYB3R
Using qRT-PCR analysis, we examined the expression pattern ofAcMYB3Rin various kiwifruit tissues. The results showed thatAcMYB3Rexpressed constitutively in all tissues tested, with significantly higher expression level in roots (Fig. 1-A). Moreover, using 7-month-old Hongyang seedlings regenerated from tissue culture, we found that expression ofAcMYB3Rin roots was significantly enhanced by adding stress treatments. As shown in Fig. 1-B and C, theAcMYB3Rwas initially expressed at relatively low levels and its expression abundance gradually increased until 24 h under salt or cold treatments. The expression ofAcMYB3Rwas also induced by mannitol, peaked at 6 h after the treatment and then gradually declined (Fig. 1-D). This result suggestsAcMYB3Ris probably involved in regulation of environmental stress tolerance in kiwifruit.
3.2. lsolation and sequence analysis of AcMYB3R
Based on Kiwifruit Information Resource (Yueet al. 2015),AcMYB3Rencodes a putative R1R2R3-MYB transcription factor with 1 779 bp nucleotides coding for 593 amino acids. Multiple sequence alignments of AcMYB3R and its homologous proteins revealed they possess three highly conserved MYB domains individually containing 49–50 amino acids (Fig. 2-A). Phylogenetic analysis showed thatAcMYB3Ris highly close to theMYB3Rgenes derived fromArabidopsis, tobacco and rice (Fig. 2-B), suggesting that MYB3R proteins with conversed structure may have the conserved functions in plants.
3.3. AcMYB3R is a nuclear protein with transcriptional activation activity
To examine the subcellular localization of AcMYB3R in plant cell, green fluorescent protein (GFP) was fused in-frame to its C-terminal. Transiently expressed AcMYB3R-GFP protein signal was detected exclusively in the nucleus,whereas the positive control showed GFP signal throughout the cytoplasm and nucleus (Fig. 2-C). This result suggested AcMYB3R is a nuclear-localized protein.
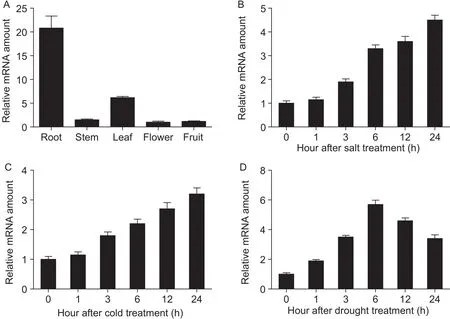
Fig. 1 AcMYB3R expression in kiwifruit (Actinidia chinensis cv. Hongyang). A, AcMYB3R expression in different tissues. The tissue samples were collected from 5-year-old Hongyang plants regenerated from tissue culture. Fruit samples were collected at 130 days after pollination. B, C and D, AcMYB3R expression in response to 200 mmol L–1 NaCl, 4°C cold stress and 300 mmol L–1 mannitol detected in Hongyang roots. The root samples were collected from 7-month-old plants regenerated from tissue culture.AcActin2 was used as an internal control. Data are mean±SD of three independent replicates.
To investigate whetherAcMYB3Rhas transcriptional activation activity,AcMYB3Rwas fused to pEG202 vector with a DNA-binding domain in the MATCHMAKER LexA Two-Hybrid System. The transformed yeasts were grown on the selective medium lacking Ura/His (SD/-Ura/-His)supplemented X-gal. As a result, both the positive control and the yeasts transformed with pEG202-AcMYB3Rshowed β-galactosidase activity, whereas the negative control exhibited no β-galactosidase activity (Fig. 2-D), indicating thatAcMYB3Rpossesses transcriptional activation activity.
3.4. AcMYB3R activates the type B cell cyclin gene AcKNOLLE
MYB3Rmember has been previously reported to play an essential role in regulating the G2/M phase transitionviabinding to MSA elements in the promoter of G2/M specificgeneKNOLLE(Hagaet al. 2007). Three MSA motifs were found in kiwifruit homolog (AcKNOLLE; accession no.Ach23g121631) of the cell cyclin geneKNOLLEbetween–236 to –152 bp in the upstream of its transcription start site (Fig. 3-A). To evaluate whetherAcMYB3Ris capable of activating theAcKNOLLE, we employedAgrobacteriummediated transient expression assays (Yanget al. 2000).Agrobacteriumcell cultures carryingpAcKNOLLE:GUSalone or combined with35S:AcMYB3Rwere syringein filtrated intoN.benthamianaleaves. Empty vector pBI121(35S:GUS) or construct35S:AcMYB3Rserved as positive or negative control, respectively. Using histochemical staining assay, strong GUS signal was detected in the positive control and co-transformed leaves with bothpAcKNOLLE:GUSand35S:AcMYB3R, by contrast no GUS signal was observed in the negative control andpAcKNOLLE::GUS-in filtrated leaves (Fig. 3-B). Their GUS activities were measured and shown in Fig. 3-C.To investigate ifAcMYB3Rcould ectopically activate the expression ofAtKNOLLEinArabidopsis, we generated transgenic plants overexpressingAcMYB3Rby using theAgrobacterium-mediated floral-dip method (Zhanget al.2006). Three independent homozygous transgenic lines (#3,#5, #6) were isolated according to the differentially elevated mRNA expression levels of the transgene (Fig. 3-D). As shown in Fig. 3-E, the expression ofAtKNOLLEwas significantly enhanced in transgenic lines as compared to WT. These results suggest that the promoter of theAcKNOLLEcan be recognized by AcMYB3R protein that acts as a transcriptional activator capable of activating the expression of G2/M phase-specific gene.
3.5. Overexpression of AcMYB3R increases tolerance to drought and salt in Arabidopsis
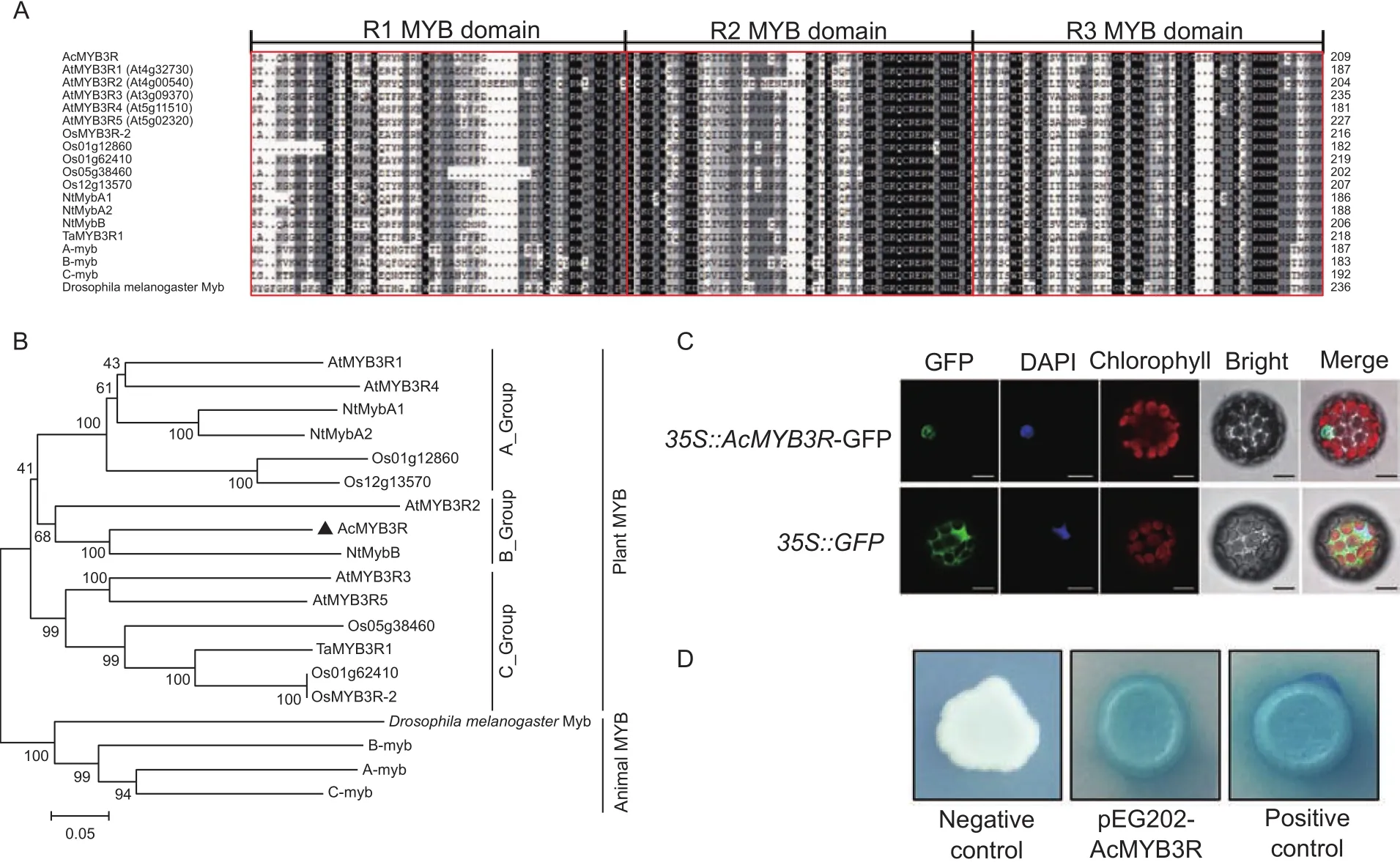
Fig. 2 Sequence alignment, phylogenetic tree, transcription activation and subcellular localization of AcMYB3R. A, multiple sequence alignment of amino acid sequences of AcMYB3R and its homologous proteins. The R1, R2 and R3 MYB domains are marked in red. B, the phylogenetic tree of MYB3R proteins. The tree was constructed with MEGA v. 5.0 via the neighbor-joining (NJ)method. GenBank accession numbers are included as AcMYB3R (Ach00g380251) from kiwifruit (Actinidia chinensis); AtMYB3R-1(At4g32730), AtMYB3R-2 (At4g00540), AtMYB3R-3 (At3g09370), AtMYB3R-4 (At5g11510) and AtMYB3R-5 (At5g02320) from Arabidopsis (Arabidopsis thaliana); NtmybA1 (AB056122.1), NtmybA2 (AB056123.1), and NtmybB (AB056124.1) from tobacco(Nicotiana tabacum); OsMYB3R-2 (BAD81765), Os01g12860 (LOC_Os01g12860.1), Os01g62410 (LOC_Os01g62410.1),Os05g38460 (LOC_Os05g38460.1) and Os12g13570 (LOC_Os12g13570.1) from rice (Oryza sativa); TaMYB3R1 (ADO32617.1)from wheat (Triticum aestivum); c-Myb (M15024), A-Myb (X13294), and B-Myb (X13293) from humans (Homo sapiens) and Drosophila melanogaster Myb (X05939). AcMYB3R is marked with triangle. C, subcellular localization of AcMYB3R-GFP fusion protein transiently expressed in Nicotiana benthamiana protoplasts. Transiently expressed AcMYB3R-GFP protein signal was observed in tobacco leaves protoplasts after Agrobacterium-mediated transformation. Bars=10 μm. D, transcription activation assay of AcMYB3R protein. The transformed yeasts were grown on the selective medium lacking Ura/His (SD/-Ura/-His) supplemented X-gal.
Mannitol was used to mimic drought stress condition for the transgenic study. There is no significant difference in growth and development observed between the transgenic and WT plants grown on 0.5× MS medium without adding mannitol (Fig. 4-A). By contrast, in the presence of mannitol(0, 300 or 350 mmol L–1, respectively), the seedling growth of WT was much more severely inhibited as compared to the transgenic lines (Fig. 4-A). We observed the root length of the transgenic lines overexpressingAcMYB3Rwas significantly longer than WT (Fig. 4-B). In addition, upon drought stress treatments and recovery experiment, the transgenic lines showed significantly higher survival rates than the WT plants (Fig. 4-C). Similar results were obtained by using NaCl stress treatments (0, 120 or 150 mmol L–1,respectively) (Fig. 4-D, E and F).
In addition, H2O2, MDA and proline contents were examined for WT and the transgenic plants under drought or salt treatments. The results showed that the contents of H2O2and MDA in transgenic plants were considerably decreased under drought and salt stress as compared to WT, while the proline content was significantly increased (Fig. 5-A–C).We also employed DAB staining to quantitatively detect H2O2in the treated plants (Tanget al. 2014). As shown in Fig. 5-D, transgenicArabidopsisoverexpressingAcMYB3Raccumulated higher levels of H2O2as compared to the WT leaves under drought and salt stress treatments.
3.6. AcMYB3R accelerates the expression of some stress-responsive genes under drought stress
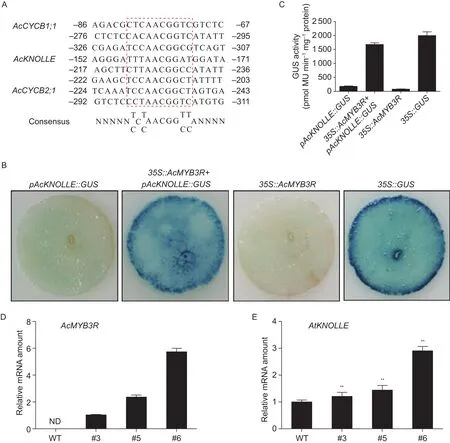
Fig. 3 β-Glucuronidase (GUS) activities under control of the interaction between AcMYB3R protein and AcKNOLLE promoter. A,the alignment of MSA-like sequences from promoters of type B cyclin genes including AcCYCB1;1 (Ach26g179151), AcKNOLLE(Ach23g121631), AcCYCB2;1 (Achn01g076081) in kiwifruit. The boxed sequences with red are highly homologous. The nucleotide positions are numbered from the transcription start site (ATG). B, GUS activity was detected using histochemical staining in the transiently expressed Nicotiana benthamiana leaves. Agrobacterium tumefaciens strain GV3101 harboring 35S:AcMYB3R or pAcKNOLLE::GUS was separately or simultaneously syringe-in filtrated into the abaxial surfaces of expanding N. benthamiana leaves. Construct 35S:GUS or 35S:AcMYB3R served as a positive or negative control, respectively. The AcKNOLLE promotor sequence from –2 000 to –1 containing three MSA motifs was fused to the GUS reporter gene to construct pAcKNOLLE:GUS vector.C, GUS activity driven by AcKNOLLE promoter detected by fluorometric assay and expressed as pmol 4-methylumbelliferone(MU) μg–1 protein min–1. D, expression level of AcMYB3R transgene in wild type (WT) and three independent transgenic lines (#3,#5 and #6) determined by real-time quantitative PCR. AtActin2 was used for an internal control. ND, not detected. E, expression level of AtKNOLLE in wild type (WT) and three independent transgenic lines (#3, #5 and #6) determined by real-time quantitative PCR. AtActin2 was used for an internal control. **, significant differences at P<0.01 by Student’s t-test. Data in Fig. 3-C–E are mean±SD from three independent experiment.
To further investigate the molecular mechanism ofAcMYB3R-mediated tolerance to drought stress, expression alteration of several stress-responsive genes was determined under drought stress by qRT-PCR. Under normal conditions, WT and transgenic plants showed similar expression patterns of the stress-responsive genes, while under drought stress,higher expression levels ofRD29A(Fig. 6-A),RD29B(Fig. 6-B),COR15(Fig. 6-C) andRD22(Fig. 6-D) were detected in transgenic plants as compared to the WT. This result suggested thatAcMYB3Ris able to accelerate stressresponsive genes expression in response to abiotic stress.
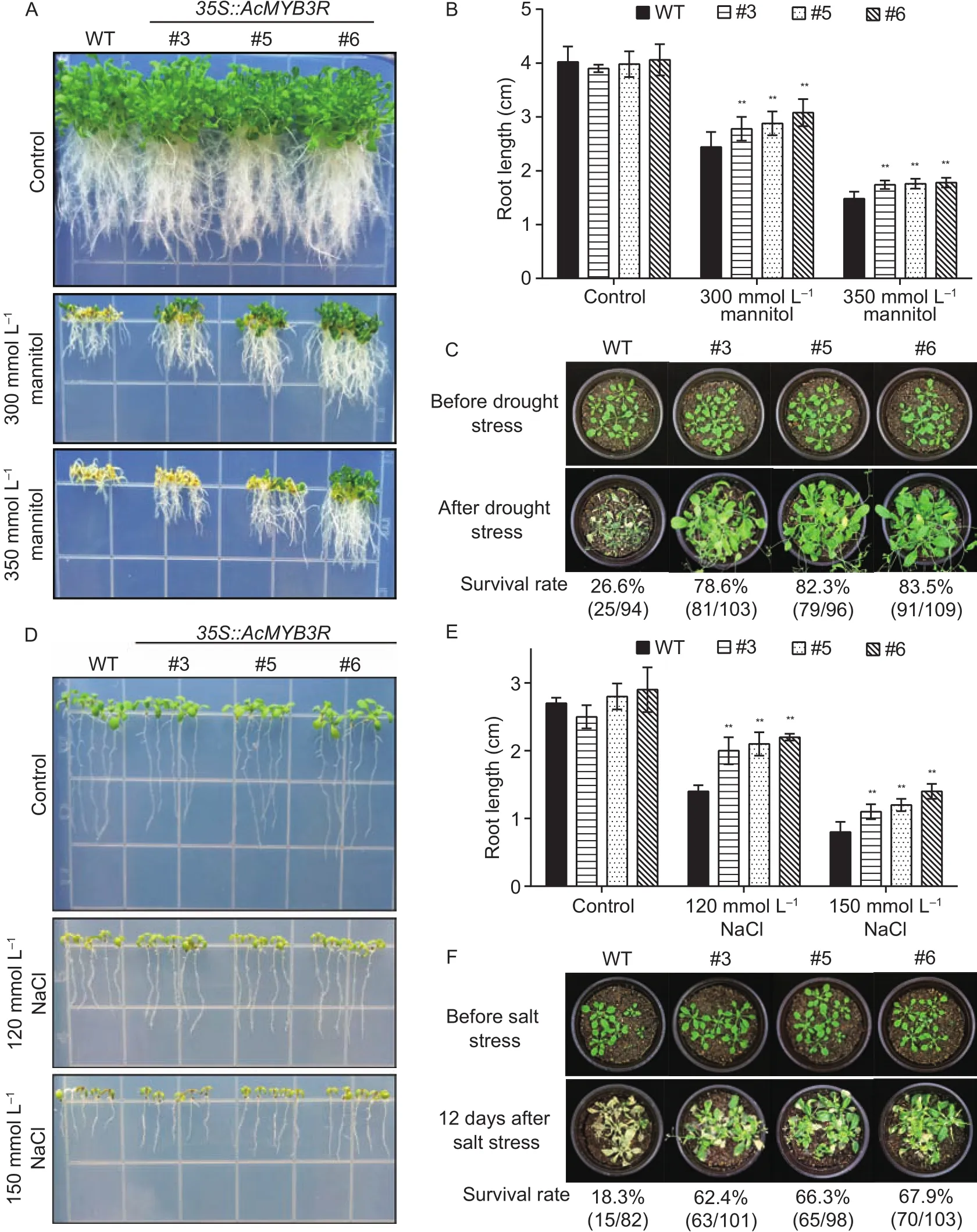
Fig. 4 Overexpression of AcMYB3R results in enhanced drought or salt tolerance. A and B, phenotypes and the root length of wild type (WT) and AcMYB3R-overexpressing lines (#3, #5 and #6) under mimic drought stress on 0.5× MS medium. C, phenotype of WT and AcMYB3R-overexpressing lines (#3, #5 and #6) before and after drought stress treatments in pots. D and E, phenotypes and the root length of WT and AcMYB3R-overexpressing lines (#3, #5 and #6) under salt stress on 0.5× MS medium. F, phenotype of WT and AcMYB3R-overexpressing lines (#3, #5 and #6) before and after NaCl treatment in pots. Data in Fig. 4-B and E are mean±SD from three independent experiment. **, significant differences at P<0.01 by Student’s t-test.
4. Discussion
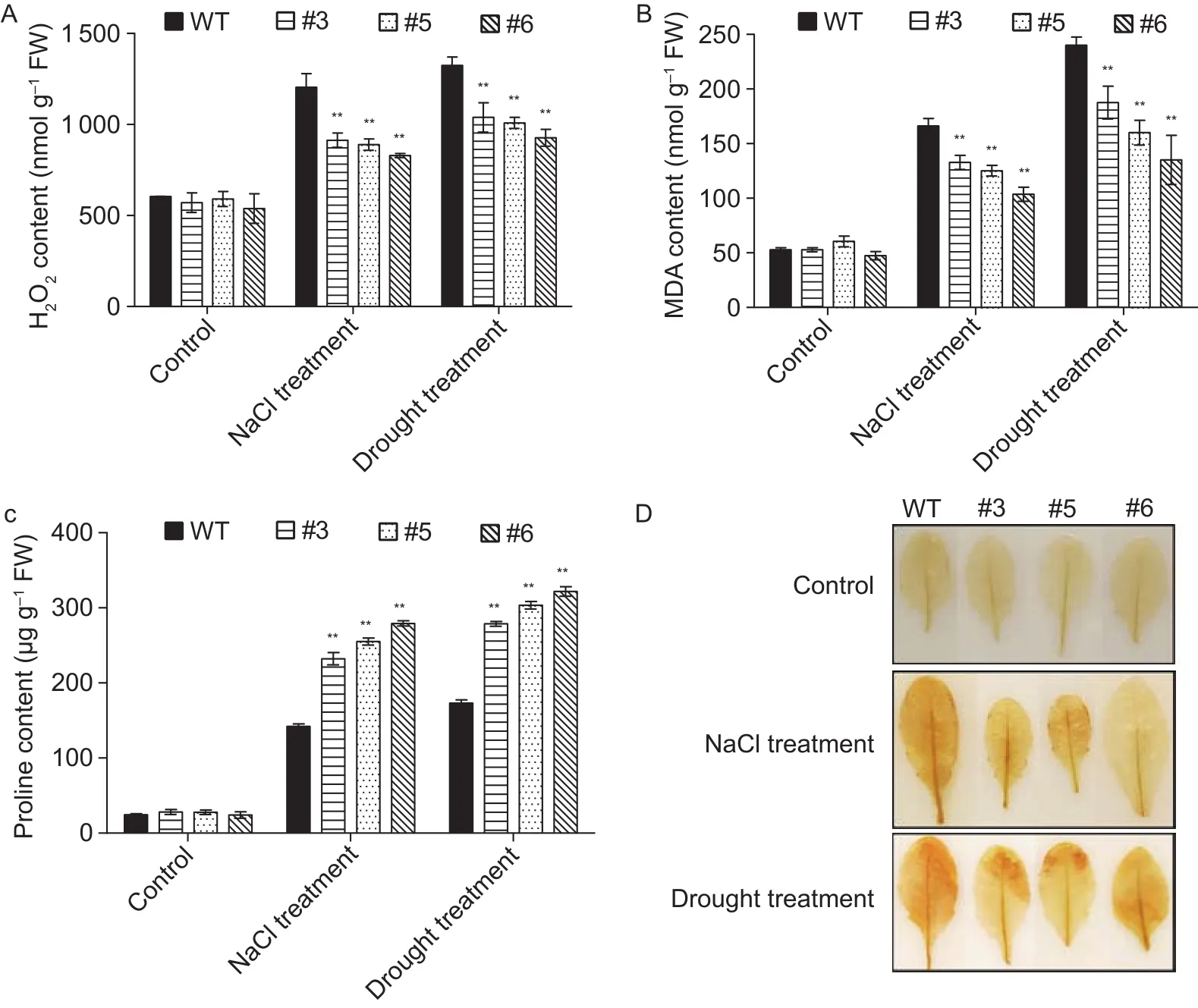
Fig. 5 H2O2 (A), malondialdehyde (MDA, B), and proline (C) contents in wild type (WT) and AcMYB3R-overexpressing lines (#3,#5 and #6) grown under drought or salt treatment. D, differential H2O2 production detected by diaminobenzidine (DAB) staining in the treated leaves of WT and AcMYB3R-overexpressing lines under salt or drought stress treatment. Data are mean±SD from three independent experiments. **, significant differences at P<0.01 by Student’s t-test.
Here we report the characterization of a kiwifruit(A. chinensis) MYB3R homologAcMYB3R. AcMYB3R protein was shown to belong to R1R2R3-MYB subfamily and contains three MYB domains with nuclear localization(Fig. 2-A–C). Its expression was detected in various tissues and enhanced by drought, salt or cold treatments (Fig. 1).This is consistent with the observations in several other MYB3R homologs. In rice, expression ofOsMYB3R-2was induced by cold, drought, and salt stress (Daiet al.2007). In wheat,TaMYB3R1expression was activated by exogenous phytohormone abscisic acid (ABA) and methyl jasmonate acid (MeJA), but not by salicylic acid (SA) or ethylene (ET) (Caiet al. 2011). In addition,TaMYB3R1expression level was gradually increased under drought,salt or cold stress (Caiet al. 2011). These results suggest the MYB3R proteins existed in different plant species might be the orthologous individuals functioning against abiotic stresses.
In plants,MYB3Rhas been reported to mediate the G2/M phase transition of the cell cycleviabinding to MSAcis-element. InArabidopsis,MYB3R1andMYB3R4were demonstrated to be transcriptional activators of cell cycle genes, whileMYB3R3andMYB3R5were shown to act as transcriptional repressors (Hagaet al. 200, 2011). In tobacco,NtmybA1andNtmybA2were shown to mediate the transition of G2/M phase, by contrastNtmybBacted as a negative transcriptional regulator (Itoet al. 2001). In rice,OsMYB3R-2was reported to be able to recognize the core sequence of MSA in type B cell cyclin genes,such asOsCycB1;1andOsKNOLLE, and to mediate their expressions (Maet al. 2009). In this study, we isolated the MSAcis-element from the kiwifruit homolog of the type B cell cyclinKNOLLEand showedAcMYB3Rwas capable of functioning as a R1R2R3-MYB protein and activating its expression (Fig. 3-B, C). MSAcis-element has been found in many G2/M phase genes (Fig. 3-A). Intriguingly,AtMYB3R1,AtMYB3R4,NtMybA1andNtMybA2, members from group A ofMYB3Rsubfamily, function as positive activators, whileAtMYB3R3andAtMYB3R5, the group C members, act in repressing the expression of cell cyclin genes. Our study showedAcMYB3Ris probably a positive regulator involved in modulating the G2/M phase of the cell cycle.
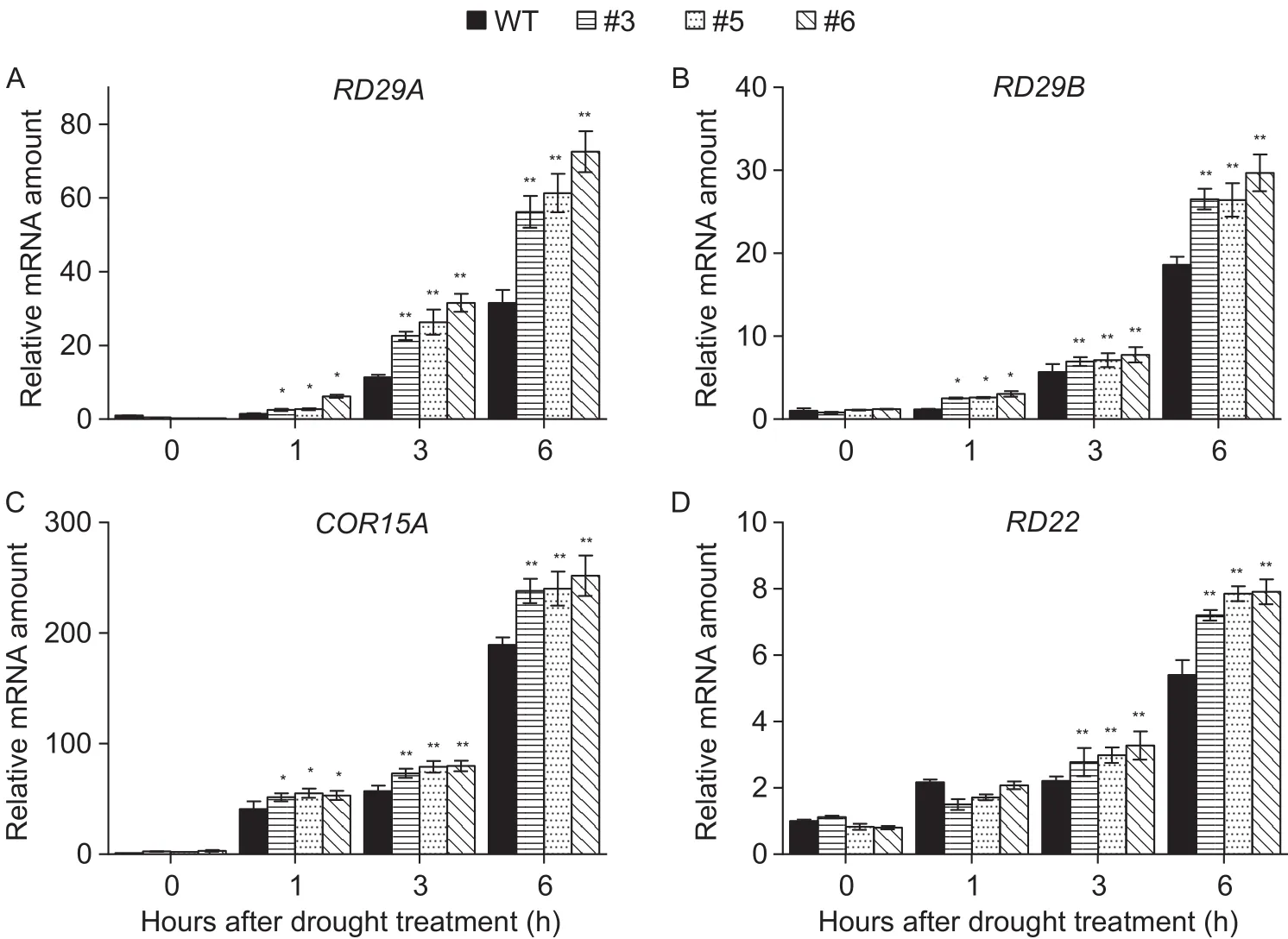
Fig. 6 Expression patterns of drought stress-responsive genes in wild type (WT) and 35S:AcMYB3R transgenic plants grown under drought treatment. 3-week-old WT and AcMYB3R-overexpressing lines were treated with 350 mmol L–1 mannitol for 6 h.The transcription levels of RD29A (A), RD29B (B), COR15A (C) and RD22 (D) were measured by qRT-PCR. AtActin2 was used for an internal control. All values are mean±SD from three independent experiments. * and **, significant differences at P<0.05 and P<0.01, respectively, by Student’s t-test.
The transcription factor is indispensable for plants to cope with abiotic stress by regulating the expressions of downstream genes involved in abiotic stress. The R2R3-MYB subfamily is the largest group of the plant MYB family and play multifunctional roles in plant growth, developmental,morphogenesis, secondary metabolism and responses to environmental stresses (Jin and Martin 1999; Allanet al.2008). In comparison with a large number of R2R3-MYB subfamily members that have been investigated, relatively a few of R1R2R3-MYB proteins were characterized in plant.Our transgenic studies demonstrated overexpression ofAcMYB3Rresulted in drastically increased tolerance to drought or salt stress inArabidopsis(Figs. 4 and 5) probably by accelerating cell cycle progress (Fig. 3) and activating the stress-responsive genes’ expressions (Fig. 6). Consistent with the observations in rice and wheat (Daiet al. 2007;Caiet al. 2015), our results suggestMYB3Rsubfamily is likely to play an important role in plants’ environmental adaption.
5. Conclusion
Here we isolated and characterized a novel 3R-MYB transcription factor fromActinidia chinensiscv. Hongyang(AcMYB3R). It is a nuclear protein with transcriptional activity. Its expression can be enhanced by cold,drought, and salt stress. TransgenicArabidopsisplants overexpressingAcMYB3Rshowed drastically enhanced tolerance to drought and salt stresses with elevated expression of the stress-responsive genes. Collectively,our study represents a valuable piece of information for functional genomics study of kiwifruit and molecular breeding for the crop improvement.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (31471157 and 31700266).
 Journal of Integrative Agriculture2019年2期
Journal of Integrative Agriculture2019年2期
- Journal of Integrative Agriculture的其它文章
- Digital mapping in agriculture and environment
- Maize production under risk: The simultaneous adoption of off-farm income diversification and agricultural credit to manage risk
- miR-34c inhibits proliferation and enhances apoptosis in immature porcine Sertoli cells by targeting the SMAD7 gene
- lnhibition of KU70 and KU80 by CRlSPR interference, not NgAgo interference, increases the ef ficiency of homologous recombination in pig fetal fibroblasts
- Protective roles of trehalose in Pleurotus pulmonarius during heat stress response
- Arbuscular mycorrhizal fungi combined with exogenous calcium improves the growth of peanut (Arachis hypogaea L.) seedlings under continuous cropping
