Multiple Roles of Berberine Bridge Enzyme Gene HbBBE1 in Response to Stress in Hevea brasiliensis Muell. Arg.
WANG Meng ZHNG Dong LU Yan-Xi XIAO Hua-Xing ZHENG Fu-Cong ZHANG Yu
(Institute of Tropical Agriculture and Forestry,Hainan University,Haikou 570228)
Abstract Berberine bridge enzyme(BBE,EC1.5.3.9) is a key enzyme in berberine biosynthesis. In this study, HbBBE1 gene was cloned in rubber tree by RT-PCR. The HbBBE1 promoter contained light, gibberellin, salicylic acid, wounding, fungal elicitor, defense, and stress responsive elements. The predicted protein of HbBBE1 contains a signal peptide, a trans membrane region, an FAD binding domain, and a BBE domain which is found specifically in the berberine bridge enzymes and berberine bridge-like enzymes. It shared the highest similarity with BBE protein from Manihot esculenta with 80.1% identical residues. Comparing expression among different tissues of rubber tree demonstrated that HbBBE1 has a particularly high expression level in the latex. HbBBE1 expression was significantly upregulated by biotic and abiotic stress, i.e. powdery mildew infection, drought stress, high intensity light irradiation, H2O2, and both tapping treatment in virgin trees and mechanical wounding in seedlings. Furthermore, HbBBE1 transcript was remarkably induced by application of exogenous hormones, including gibberellic acid(GA3), salicylic acid(SA), ethephon(ET), methyl jasmonate(MeJA), and abscisic acid(ABA). Among these treatments, HbBBE1 transcript was quickly induced by GA3 stimulation, and reached a maximum(6.3-fold) at 0.5 h. The increased expression of HbBBE1 was highly induced by ABA, ET, and H2O2 stimuli, and it accumulated dozens of times comparing that at the untreated control.To summarize, our study suggested that HbBBE1 played multiple roles in response to biotic and abiotic stress in rubber tree.
Key words Hevea brasiliensis;berberine bridge enzyme;HbBBE1;stress response
The berberine bridge enzyme(BBE) catalyzes the formation of the berberine bridgehead in benzophenanthridine alkaloids[1~2]. This is the first committed step in benzophenanthridine alkaloid biosynthesis. Some benzophenanthridine alkaloids, such as the antimicrobial alkaloid sanguinarine[3~4], are involved in the plant defense response to pathogen infection. The accumulation of sanguinarine in response to the addition of various elicitors has been found in cell-suspension cultures of California poppy(Eschscholziacalifornica)[5], and opium poppy(PapaversomniferumL.)[6]. Expression analysis by RNA blot hybridization in opium poppy plants showed thatBBEgenes are expressed in roots and stems of mature plants and in seedlings within 3 d after germination. Rapid and transientBBEmRNA accumulation also occurred after treatment with a fungal elicitor or with methyl jasmonate. However, sanguinarine was found only in roots, seedlings, and fungal elicitor-treated cell cultures[7]. These studies revealed that BBE play a role in plant defense response and alkaloid biosynthesis. Although the function of BBE in alkaloid biosynthesis has been well studied, its function in defense response of plant is rarely described.
Rubber tree(HeveabrasiliensisMuell. Arg.) is one of important sources of natural rubber production[8]. Biosynthesis of natural rubber, like other secondary metabolites, is affected by various plant hormones and stress[9~11]. For instance, rubber tree bark increases the production of latex between 1.5- to 2-fold under ethephon(an ethylene releaser) treatment[12~13]. Even though the exact mechanism of action of ethylene on the rubber tree is poorly understood, it is presumed that ethylene affect the genes involved in rubber biosynthesis directly or indirectly by modifying general nitrogen metabolism in the latex[12~14]. The bark of rubber tree is regular tapped for collecting latex, which can be regarded as a form of controlled wounding. So rubber tree regularly suffers from wounding stress during latex harvesting. It is still unknown that rubber tree adapt to these stress conditions and regulating natural rubber production.
We previously found that powdery mildew affected physiological response in rubber tree[15]. Based on this, a transcriptome sequencing dataset was obtained by RNA-Seq of rubber tree leaves after powdery mildew infection, and aBBEgene(named asHbBBE1) was obviously upregulated by powdery mildew infection(unpublished data). Important roles of BBE in the biosynthesis of secondary metabolites and defense response in other plants prompted us to study the functions of BBE in rubber tree. In this paper, we cloned theBBEgene in rubber tree and characterized the structure ofBBEin rubber tree. The tissue-specific expression ofBBE, and inducible expression ofBBEunder various stress and hormone treatments will be discussed in rubber tree.
1 Materials and methods
1.1 Plant materials and treatments
The rubber tree clones CATAS 7-33-97 and CATAS 8-79 planted at the experimental farm of Environment and Plant Protection College, Danzhou, Hainan Province, China. Samples of different tissues were harvested from 10-year-old healthy tapping trees of CATAS 7-33-97.
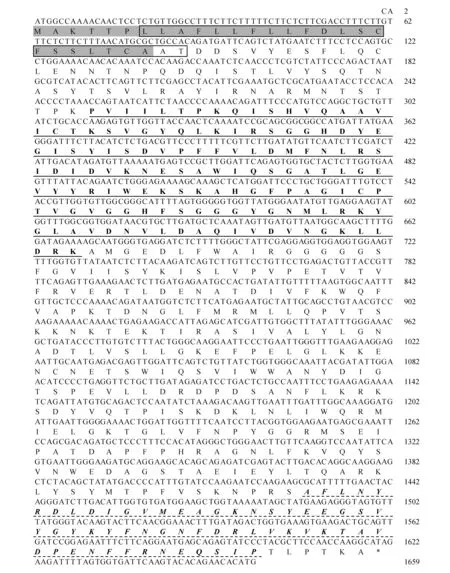
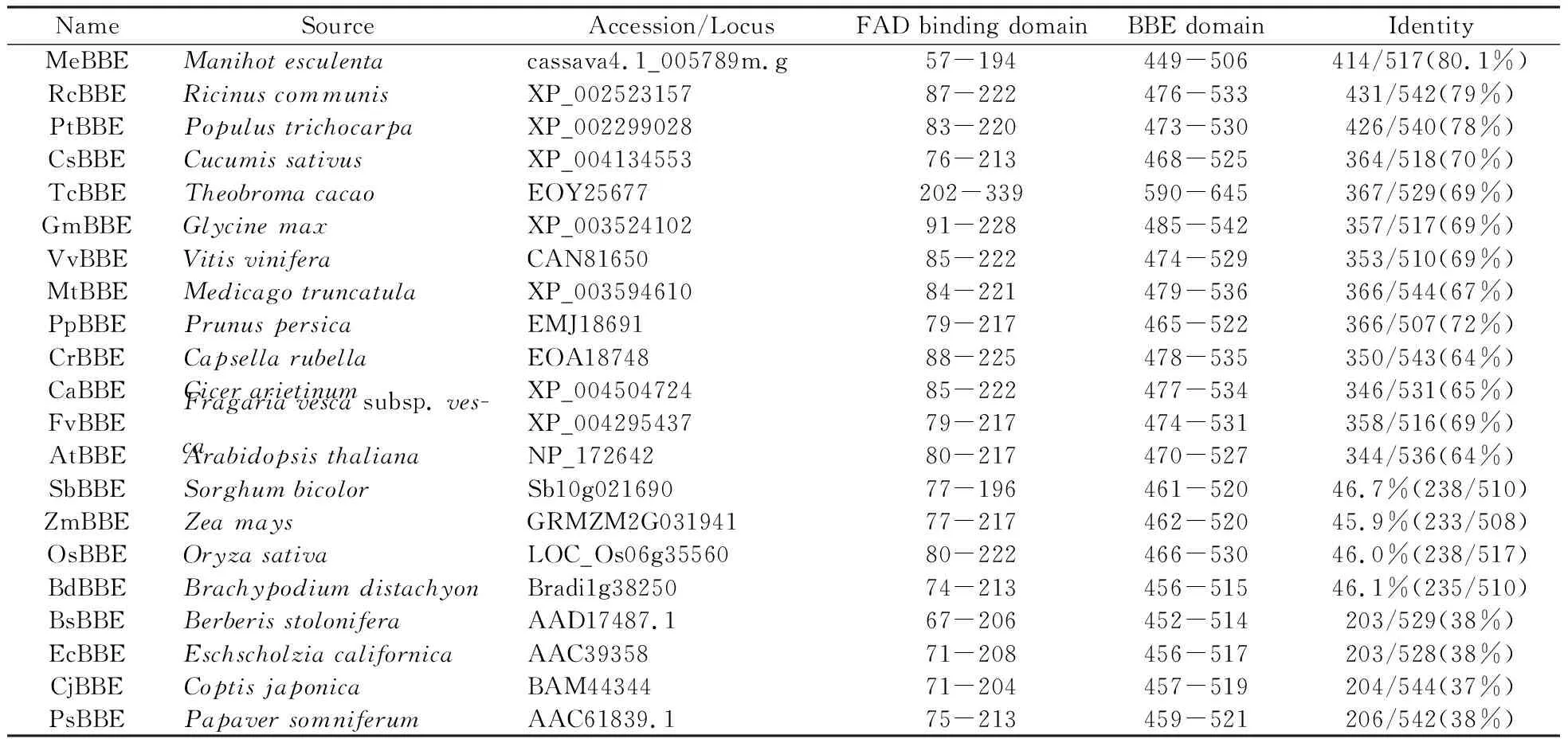
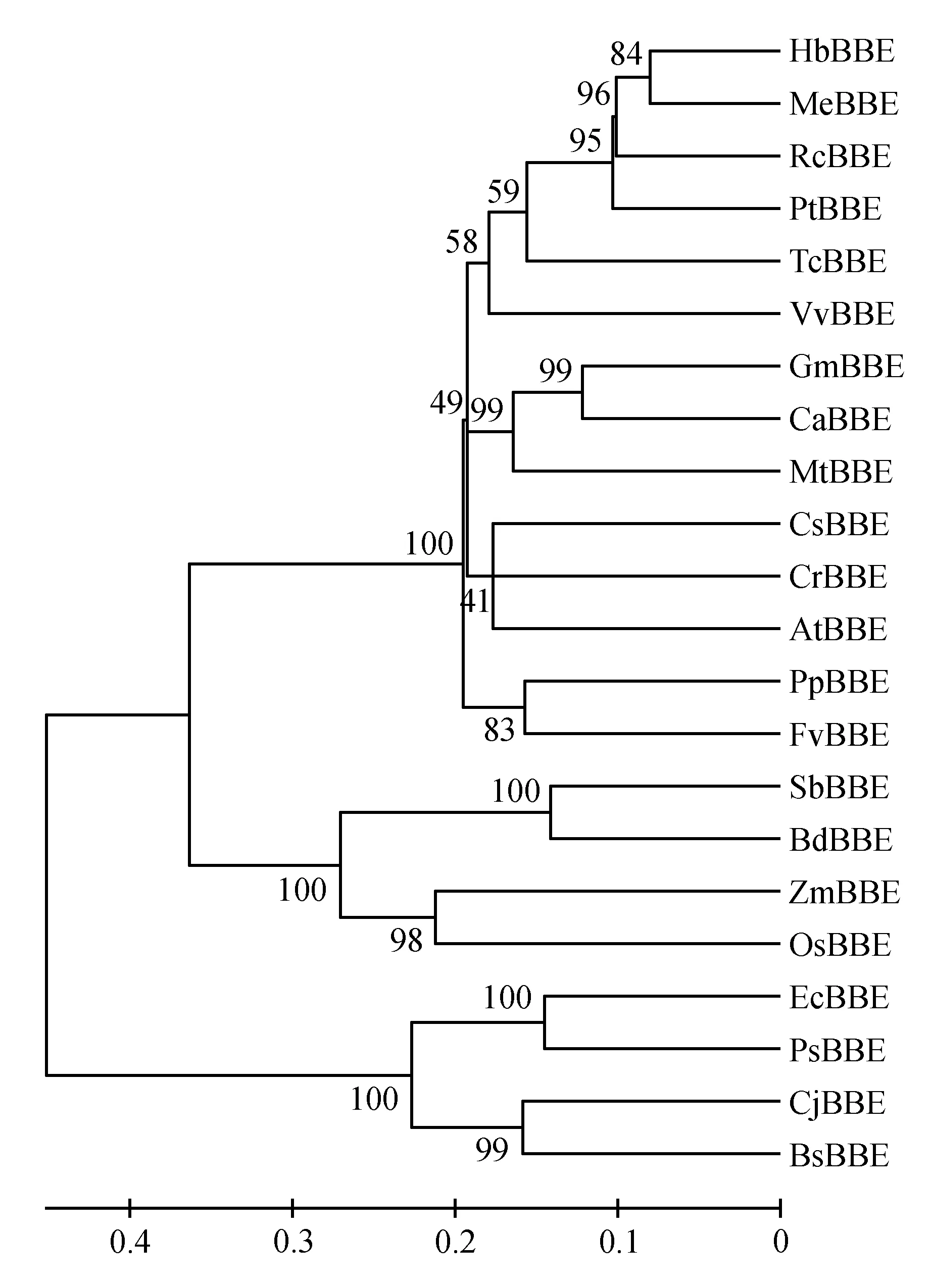
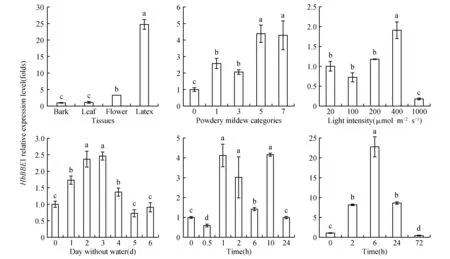
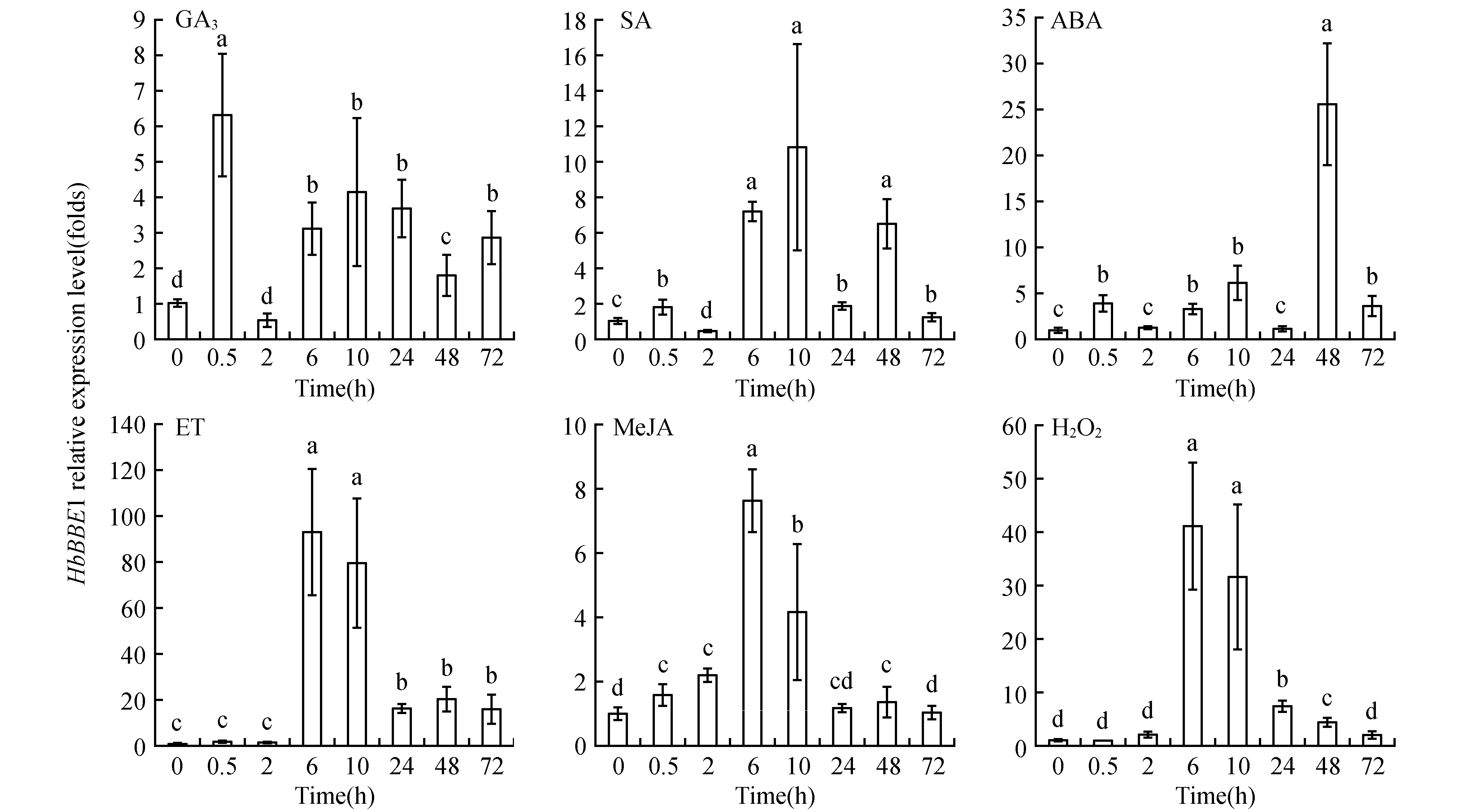
The grated budding seedlings of rubber tree clone CATAS 7-33-97 grown in the plastic pots were used for different treatments. For mechanical wounding treatment, it was performed according the methord of[16], and leaf sections were harvested at 0, 0.5, 1, 2, 6, 10 and 24 h. Powdery mildew(OidiumheveaeSteinmann) inoculation was according to the reported method[15]. Powdery mildew categories were classified as 0, 1, 3, 5, 7 and 9 grade according to leaf lesions area after powdery mildew infection. Seedling leaves with no pathogen infection were referred as level 0. The proportion of the lesions area in the whole leaf was defined as X, while 0 Total RNA was extracted from latex and leaves according to Wang’s study[18]. The quality and concentration of the extracted RNA checked by agarose gel electrophoresis and measured by a spectrophotometer(Thermo NanoDrop 2000 UV/Vis Spectrophotometer, USA). The sequence ofHeveaBBEwhich showed high similarity to BBE protein family was identified from theHeveatranscriptome assembly database, and it was used to design primers(HbBBE1F and HbBBE1R, Table 1) for cloningHeveaBBEby RT-PCR. Using cDNA of CATAS 8-79 as template, the full length cDNA ofHbBBE1 was isolated. The sequence ofHbBBE1 was confirmed by sequencing the PCR product. The PCR reaction was performed using Pyrobest® DNA polymerase(Takara) with the following thermal cycling parameters: 94℃ for 5 min, followed by 35 cycles of 94℃ for 30 s, 55℃ for 1 min and 72℃ for 2 min, and the final extension was performed at 72℃ for 10 min. Table 1 Sequences of the oligo nucleotides First strand cDNA synthesized from 2 μg of RNA with M-MuLV reverse transcriptase and random hexamer primers(Takara) according to the manufacturer’s instructions. The cDNA was diluted 1∶20 with nuclease-free water. Aliquots of the same cDNA sample were used for qRT-PCR withHbBBE1 specific primers(HbBBE1-QFand HbBBE1-QR), andActin(Actin-F and Actin-R) was used as a housekeeping gene. Reactions performed in a 25 μL volume containing 500 nmol·L-1(w/v) of each primer and 1×SYBER Green PCR master mix(Takara). qRT-PCR was performed using the Bio-Rad CFX96 system(Bio-Rad, Hercules, CA, USA). The reactions carried out as follows: 3 min at 95℃ for denaturation, 10 s at 94℃, 20 s at 60℃, and 30 s at 72℃ for amplification for 35 cycles. The relative abundance of transcripts was calculated according to the software instructions in Bio-Rad CFX96 Manager. For each sample, qRT-PCR was performed with three technical replicates on three biological replicates. The 2-ΔΔCTmethod was used to analyze relative transcript levels of target gene[20]. All data were analyzed with software IBM-SPSS 23.0(IBM Corporation, New York, USA). One-way ANOVA and Tukey test were used to assess the different level.P<0.05(probability level) was considered significant difference. Figures were drawn by OriginPro2015(Origin Lab Corporation, Massachusetts, USA). A 1 659 bp sequence was obtained by RT-PCR amplification. It contained a 1 620 bp ORF(open reading frame) encoding a putative berberine bridge enzyme(BBE) with 539 amino acids(Fig.1). Since this is the firstBBEgene cloned from rubber tree(H.brasiliensis), and it was designated asHbBBE1. The predicted molecular mass ofHbBBE1 was 60.2 kDa, with a theoretical isoelectric point(pI) of 7.53. Based on structural properties indicated by the NCBI Conserved Domain search(http:// www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?) and SMART(http://smart.embl-heidelberg.de/), HbBBE1 had an FAD binding domain(from the 84thto 223thamino acid) and a BBE domain(from the 476thto 533thamino acid)(Fig.1) which is found in the berberine bridge and berberine bridge-like enzymes. Furthermore, HbBBE1 might be a transmembrane protein containing an N-terminal hydrophobic signal peptide(from the 1stto 27thamino acid) and a transmembrane domain(TM, from the 7thto 29thamino acid), and there were 21 amino acids overlapping between them(Fig.1). Fig.1 The cDNA and deduced amino acid sequences of rubber tree HbBBE1 The deduced amino acid sequences are given below the cDNA sequences. The termination codon is marked by an asterisk(*), a signal peptide is indicated by shadow, a transmembrane domain is shown in the box, a FAD binding domain is indicated by a solid line, and a BBE domain is indicated by a dotted line. The genomic DNA sequence ofHbBBE1 was obtained by blast searching in the genome database ofH.brasiliensis(https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&PROG_DEF=blastn&BLAST_SPEC=Assembly&ASSEMBLY_NAME=GCA_001654055.1). Comparing the genomic sequence ofHbBBE1(GenBank accession number of the scaffold: LVXX01000028) with its corresponding cDNA sequence revealed that there was no intron present in the genomic sequence ofHbBBE1. Furthermore, a 2 000 bp genomic sequence at the upstream of the initiation codon(ATG) ofHbBBE1 had been extracted from the genome database, and used for prediction of motifs present in the promoter ofHbBBE1. According the online prediction tool Plant CARE(http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), 15 kinds of motifs and/or elements were found in the putative promoter ofHbBBE1, including light, gibberellin, salicylic acid, wounding, fungal elicitor, defense and stress responsive elements(Table 2). To reveal the relationship of HbBBE1 with other BBEs in plants, blastp was performed against the NCBI nr(non-redundant) protein database for identifying HbBBE1 orthologs from different species. Orthologs were identified from 17 plant species, and used for phylogenetic analysis(Table 3). These orthologs share conserved FAD binding and BBE domain by the NCBI Conserved Domain search(http:// www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?)(Table 3). HbBBE1 shared the highest similarity with BBE ofManihotesculenta(MeBBE) with 80.1% identical residues, and 79% withRicinuscommunis(RcBBE). However, it shared low similarity with BBEs with known function, i.e.,E.californica(EcBBE) andCoptisjaponica(CjBBE) with 38% and 37% identity, respectively[21~22](Table 3). Phylogenetic analysis of HbBBE1 orthologs from various species and BBEs with known function showed that these BBEs were divided into three groups with 100% bootstrap support value.Berberisstolonifera(BsBBE),P.somniferum(PsBBE), EcBBE, and CjBBE which function in berberine biosynthesis have been confirmed[21~22], were clustered in a separate clade. HbBBE1 and its orthologs were divided into two subgroups, one consisted with BBEs from the grass species(includingSorghumbicolor,Zeamays,OryzasativaandBrachypodiumdistachyon), the other consisted of HbBBE1 and BBEs from 13 dicots which shared more than 60% identity with HbBBE1(Table 3 and Fig.2). This result suggested that the functions of HbBBE1 and its orthologs may be different from the function of the four known BBEs(i.e. BsBBE, PsBBE, EcBBE, and CjBBE). The GenBank accession numbers used in building the phylogenetic tree are listed in Table 2. The scale bar indicates the number of amino acid exchanges for a given length of the tree. Table2Functionofdifferentmotifsinthe2000bpupstreamsequenceofHbBBE1analyzedbyPlantCARE Motif nameMatrix scoreFunction3-AF1 binding site10Light responsive element5UTR Py-rich stretch9Cis-acting element conferring high transcription levelsARE6Cis-acting regulatory element essential for the anaerobic inductionAT1-motif11Part of a light responsive moduleBox 46Part of a conserved DNA module involved in light responsivenessBox I7Light responsive elementBox-W16Fungal elicitor responsive elementGARE-motif7Gibberellin-responsive elementGap-box9Part of a light responsive elementHSE9Cis-acting element involved in heat stress responsivenessSkn-1_motif5Cis-acting regulatory element required for endosperm expressionTC-rich repeats9Cis-acting element involved in defense and stress responsivenessTCA-element9Cis-acting element involved in salicylic acid responsivenessWUN-motif9Wound-responsive elementCAAT-box5Common cis-acting element in promoter and enhancer regionsTATA-box5Core promoter element around -30 of transcription start Table 3 Details of HbBBE1 orthologs from different species Fig.2 Phylogenetic analysis of HbBBE1 and its orthologs from different species The expression analysis in different tissues demonstrated thatHbBBE1 was ubiquitously expressed in all tissues, but its expression in the latex was much higher than that in other tissues, i.e. bark, leaf, and flower(Fig.3). Studies in Papaveraceae and Fumariaceae species showed that benzophenanthridine alkaloids are accumulated in response to pathogenic attack and function as phytoalexins to prevent pathogen infection. Meanwhile, BBE is essential to the biosynthesis of benzophenanthridine alkaloids. So the regulation ofHbBBE1 transcript was investigated under powdery mildew infection. As shown in Fig.3, powdery mildew infection was significantly increased theHbBBE1 expression level, and it increased more than 4 folds at the severely infected categories 5 and 7 comparing to the not infected control. For different tissues analysis was performed in 10-year-old healthy tapping trees of CATAS 7-33-97, powdery mildew infection, different light intensities, drought stress and mechanical wounding treatments were performed in seedlings of CATAS 7-33-97, and tapping were performed in virgin trees(7 years old, untapped trees) of CATAS 7-33-97. Bars with different letters show significant differences at theP<0.05 level. Since light, plant hormones, wounding, fungal elicitor, defense and stress responsive elements were present in the putative promoter ofHbBBE, expression profiles ofHbBBE1 were investigated under multiple stresses condition and phytohormone treatments in rubber tree. Under different illumination intensities,HbBBE1 expression was upregulated as light intensity increasing, and reached a maximum at 400 μmol·m-2·s-1, whereas it was obviously decreased at the high light intensity of 1 000 μmol·m-2·s-1(Fig.3). Under drought stress,HbBBE1 transcript was continuously increased at the initial stage of drought stress(from 1 day without water(dww) to 3 dww), but it decreased from 4 dww and then returned to the untreated control(0 dww) level(Fig.3). Mechanical wounding treatment in the leaves of rubber tree seedlings demonstrated thatHbBBE1 expression was slightly decreased at 0.5 h, whereas it was increased from 1 h, and the highest expression was occurred at 1 and 10 h, more than 4-fold over that of the untreated control(0 h)(Fig.3). The bark of rubber tree is regular tapped for collecting latex, which can be regarded as a form of controlled wounding. SoHbBBE1 expression was analyzed in the latex of virgin trees upon tapping. The transcription ofHbBBE1 was remarkably induced by tapping, and increased nearly 23 folds at 6 h comparing with that at 0 h. and then fallen to the untreated level(Fig.3). The amount ofHbBBE1 transcript was significantly induced by application of exogenous hormones. As shown in Fig.4,HbBBE1 transcript was quickly induced by GA3stimulation, and reached a maximum at 0.5 h and increased 6.3-fold over that at 0 h. SA treatment was also upregulatedHbBBE1 expression, and the highest expression level was occurred at 10 h(Fig.4). As for ABA treatment, the highest expression level was occurred at 48 h, and increased 25.5-fold over that at 0 h(Fig.4). Ethephon(ET, releasing ethylene), is widely used in natural rubber production, which can effectively increase latex production in rubber tree. Application of exogenous ET in rubber tree seedlings caused a distinct increase inHbBBE1 expression. Its expression reached a maximum at 6 h after ET treatment, and increased 93 folds comparing with its expression at 0 h(Fig.4). JA is another hormone closely associated with rubber tree production.HbBBE1 expression was also induced by application of exogenous MeJA, and accumulated 7.6-fold at 6 h comparing that at 0 h(Fig.4). Interestingly, the expression ofHbBBE1 was significantly upregulated by H2O2treatment. The highest expression level was occurred at 6 h after treatment, and increased 41 folds comparing that at 0 h(Fig.4). All the treatments were performed in seedlings of CATAS 7-33-97. Bars with different letters show significant differences at theP<0.05 level. Fig.3 HbBBE1 expression in different tissues, and under biotic and abiotic stresses condition Fig.4 The transcriptional regulation of HbBBE1 expression in response to application of exogenous plant hormones and H2O2 The flavoprotein Berberine Bridge Enzyme(BBE) catalyzes the regioselective oxidative cyclization of(S)-reticuline to(S)-scoulerine in an alkaloid biosynthetic pathway. A large number of genes encoding for berberine bridge enzyme(BBE) enzymes were contained in plant genomes. Despite the widespread occurrence and abundance of this gene family in the plants, the function of the genes remain unexplored. Since BBE is an important enzyme in plant secondary metabolism product, a large number ofBBEgenes have been identified in plants by genome sequencing. The 28BBEgenes were identified in the model plantArabidopsis,BBEgenes from California poppy(E.californicaCham.)[21]and Opium poppy(P.somniferum)[7]have also been identified, but it is still not recognized in rubber tree Although there are many BBE genes present in plant genomes, not all of them had functions in plants. For example, there are three BBE homologs in Opium Poppy, butPsBBE2 andPsBBE3 are pseudogenes and onlyPsBBElis expressed and encodes a functional BBE protein[7]. In this study, blastn searching in the genome database ofH.brasiliensisrevealed that three scaffolds(Accession: LVXX01000028, LVXX01001103, and LVXX01000676) showed high similarity withHbBBE1, with 99%, 89%, and 92% identity, respectively. This suggested that there might be three BBE homologs present in the genome ofH.brasiliensis. Furthermore, structure prediction by NCBI Conserved Domain search and SMART indicated that HbBBE1 had an FAD binding domain and a BBE domain, which are the specific characteristics of BBE(Fig.1 and Table 3). Due to the attachment of FAD cofactor, theHbBBE1 can be set apart. Together withHbBBE1 actively expressed under multiple stimuli, these characteristics demonstrated that HbBBE1 is a member of BBE family. Rubber tree suffered from taping, disease and environmental stress[23~24]. In rubber tree, latex is very important secondary metabolite products, in isopentenyl diphosphate(IPP) biosynthesis pathway[9,25]. Its production is inevitably affected by phytohormone, pathogen infection, drought and low temperature stress, etc. Genes were induced in response to low temperature[26], powdery mildew infection[27], and ethephon stimulation[14]in rubber tree. Since important roles of BBE enzyme in plant secondary metabolism products and defense response have been reported in other plants, none BBE enzyme in rubber tree has been characterized. To deeply explore the function ofHbBBE1 in response to the plant stresses in rubber tree, we obtained the expression profiles ofHbBBE1 by using qRT-PCR technology. In this study, we found thatHbBBE1 has a particularly high expression level in the latex of rubber tree(Fig.3). Furthermore,HbBBE1 expression in the latex was significantly upregulated by tapping in virgin trees(7 years old, untapped trees) of CATAS 7-33-97(Fig.3). Interestingly,HbBBE1 transcript was remarkably induced by application of exogenous ET and MeJA, and wounding treatments in the leaves of rubber tree seedlings of CATAS 7-33-97(Fig.3-4). Previous studies revealed that latex of rubber tree contain many defense proteins such as beta-1,3-Glucanase[24], Hevamine[28~29]. These defense proteins in latex work together to protecting rubber tree bark from wounding(regular tapping) and other stress[30~31]. These results suggested that HbBBE1 has a role in latex production, which may serve as a defense protein to protecting the bark of rubber tree upon tapping. In addition,HbBBE1 expression was also upregulated by powdery mildew infection, drought stress, H2O2, ABA and SA treatments(Fig.3-4). ABA, SA, JA and ET are important phytohormone in regulating plant defense response against biotic and abiotic stresses[32~33]. These results indicated thatHbBBE1 involves in biotic and abiotic stress responses in rubber tree. Previous studies mainly focus on the BBE protein functions in biosynthesis of nicotine[34]and alkaloids[2,35]. Recently, berberine bridge enzyme-like enzymes was found playing a role in monolignol metabolism and lignin formation inArabidopsis[36]. These indicated BBE plays important roles in plants development. In this study,HbBBE1 expression was induced by high intensity light irradiation and GA3treatment(Fig.3-4), which indicated its role in rubber tree development. Taken together, our study concluded that the expressions ofHbBBE1 were significantly affected in responses to stresses in rubber tree. It still needs further study to figure out whetherHbBBE1 plays a role in stress response.1.2 Isolation of total RNA
1.3 Molecular cloning of BBE gene in rubber tree

1.4 cDNA synthesis and real-time RT-PCR(qRT-PCR)
1.5 Statistical Analysis
2 Results
2.1 Isolation of Hevea BBE gene and analyses of its gene structure and putative protein architecture
2.2 Phylogenetic analysis of HbBBE1 and its orthologs from different species

2.3 HbBBE1 involves in phytohormone, biotic and abiotic stress responses
3 Discussions

