Effects of probiotic on microf l oral structure of live feed used in larval breeding of turbotScophthalmus maximus*
JIANG Yan (姜燕) ZHANG Zheng (张正) WANG Yingeng (王印庚) JING Yayun (景亚运) LIAO Meijie (廖梅杰) RONG Xiaojun (荣小军) LI Bin (李彬)CHEN Guiping (陈贵平) ZHANG Hesen (张和森)
1Yellow Sea Fisheries Research Institute,Chinese Academy of Fishery Sciences,Laboratory for Marine Fisheries Science and Food Production Processes,Qingdao National Laboratory for Marine Science and Technology,Qingdao 266071,China
2Qingdao General Aquatic Co.Ltd.,Qingdao 266000,China
AbstractThe effects of an exogenous probiotic (Bacillus amyloliquefaciens) on microbial community structure ofBranchionus plicatilsandArtemia sinicawere evaluated in this study during turbot(Scophthalmus maximus) larval breeding. The analysis and comparison of the microf l oral composition of live feed with probiotic was conducted using the Illumina HiSeq PE250. The abundance of microbial species and diversity of microf l ora in live feed withB.amyloliquefacienswere higher than those in the control. The microf l oral composition was similar among the three replicate experimental groups ofB.plicatilscompared with the control after enrichment.Lactococcus,Pseudoalteromonas, andAlteromonaswere always dominant. Additionally, some other bacterial species became dominant during the enrichment process. The microbial community during nutrient enrichment ofA.sinicawas rather similar among the three control replicates. Relative abundance ofCobetiasp., the most dominant species, was 54%-65.2%. Similarity in the microbial community was still high after addingB.amyloliquefaciens. Furthermore,PseudoalteromonasandAlteromonasreplacedCobetiaas the dominant species, and the abundance ofCobetiadecreased to 4.3%-25.3%. Mean common ratios at the operational taxonomic unit level were 50%-60% between the twoB.plicatilsandA.sinicatreatments. Therefore, the microbial community structure changed after addingB.amyloliquefaciensduring nutrient enrichment ofB.plicatilsorA.sinicaand tended to stabilize.Additionally, the abundance ofVibrioin any kind of live feed was not significantly different from that in the control. These results will help improve the microf l ora ofB.plicatilsandA.sinicaand can be used to understand the multiple-level transfer role of probiotic species among probiotic products, microf l ora of live feed, and fi sh larvae.
Keyword: Branchionus plicatils;Artemia sinica; microf l oral structure;Bacillus amyloliquefaciens;Scophthalmus maximus; larval breeding
1 INTRODUCTION
Rotifers andArtemiaare important live feed when breeding marine species, such as fi sh(Carnevali et al., 2004; Bakke et al., 2013; Skjermo et al., 2015), shrimp (Silva et al., 2012; Jamali et al., 2015), and crab (Sulkin and Epifanio, 1975;Ruscoe et al., 2004), because of their natural,nutritional, and operational advantages. Studies have confi rmed that rotifers andArtemiaare optimal vectors to deliver nutrient substances,vaccines, and probiotics to improve the nutritional value of cultured animals and their response to disease after Specific enrichment (Gatesoupe, 1991,1994; Campbell et al., 1993; Immanuel et al., 2007;Palma et al., 2011).
Branchionusplicatilsis often used as fi rst feed for aquatic larvae and brings various bacteria that can directly affect the health of larvae. Suitable drugs are usually added during traditional nutrient enrichment to improve survival rate and reduce the chances of the host becoming infected by bacteria (Martínez-Díaz et al., 2003; Battaglene et al., 2006). Similarly, drugs are used to insure quality and quantity duringArtemianutrient enrichment (Defoirdt et al., 2007; Asok et al.,2012). However, some other drugs can affect the balance of the larval microbial community structure by interfering with propagation of normal microf l ora(Gatesoupe, 2002; Suga et al., 2011) and cause some undesired changes in pathogenic bacteria, such as bacterial resistance (Smith et al., 1994; Subasinghe,1997; Verschuere et al., 2000a; Huys et al., 2007;Allameh et al., 2016). Additionally, drug residues are a significant disadvantage for long-term use and could seriously affect marine food safety.
Chemical drug abuse is a serious food safety issue.However, probiotics can inhibit reproduction of pathogenic bacteria (Shiri Harzevili et al., 1998;Verschuere et al., 2000b), and eliminate residues,toxins, and side effects. Therefore, probiotics are an important tool to prevent and control disease and improve disease resistance, the immune response, and nutrient supply (Díaz-Rosales et al., 2006; Kim and Austin, 2006; Gatesoupe, 2008; Nayak, 2010; Wu et al., 2015). Studies have reported that live feed might prevent pathogenic bacteria from reproducing, such as short soaking with a high concentration ofBacillusbefore feeding, to increase the growth and survival rates of grouper larvae (Sun et al., 2013).
In other reports,Bacillusamyloliquefacienshas been suggested as a potential probiotic in aquaculture to protect aquatic animals from diseases caused byEdwardsiellatarda,Aeromonashydrophila,Vibrio parahaemolyticus, andV.harveyi(Cao et al., 2011;Das et al., 2013). Research on the microf l ora structure ofB.plicatilsandA.sinicahas not been conducted.Thus, the multiple-level transfer role of probiotic species among probiotic products, microf l ora of live feed, and fi sh larvae remains unclear. In the present study,B.amyloliquefacienswas added during processing ofB.plicatilsorA.sinicafor artifi cial nutrient enrichment. The changes in microbial community structure were fi rst investigated to analyze the effects of the probiotic on the microf l ora of live feed used for turbot larvae. The results will provide a theoretical guide for using probiotics in turbot larval breeding and farming.
2 MATERIAL AND METHOD
2.1 Preparation of probiotic strain
Bacillusamyloliquefacienswas stored in our lab and isolated from the intestinal tract of healthy turbot by Fan (2010) through morphological observations,hemolytic testing, physiological and biochemical testing, and molecular biological identifi cation.Bacillusamyloliquefaciensis effective at inhibiting the growth and reproduction of pathogenic bacteria in vitro, such asV.anguillarum,V.archariae, andV.scophthalmi(Fan, 2010). No pathological change or death occurred when the turbot were fed a diet that included 109cfu/gB.amyloliquefaciens.
Bacillusamyloliquefacienswas cultured in trypticase soy broth (TSB) medium at 30°C for 1 d.Salt (2%) was added to the TSB during culture of the probiotic strain. Single colonies were then selected from solid medium, added to one liquid culture, and placed in a vibrating culture box at 180 r/min for 10 h.TheB.amyloliquefaciensconcentration was 109cfu/mL, and the suspension was later used as a nutrient enhancement for live feed.
2.2 Introducing the probiotic and sampling
Branchionusplicatilsunderwent nutrient enrichment immediately after being purchased each day.B.amyloliquefacienswas added to the tank whereB.plicatilshad been cultured for 2 h. This process was continued for 6 h, and theB.plicatilswas collected as live feed for turbot larvae.Artemiasinicaeggs were bought and hatched as required each day.Nutrient enrichment was carried out after hatching and the probiotic was added at the same for 6 h. The initial concentration of probiotic was 106-107cfu/mL in the experimentalB.plicatilsandA.sinicatanks.Additionally, a broad-spectrum antibiotic (5 mg/L enrof l oxacin) was added to the control to insure survival and quality of the turbot larvae. All other nutrient additives to the experimental and control groups were the same.
Branchionusplicatilswas sampled on days 3, 7,and 13 of turbot larval development. These three samples were obtained in parallel. Similarly, three parallelA.sinicasamples were obtained on days 13,21, and 27. After enrichment, the samples in each group were gathered through fi ltered sterile gauze,washed three times in sterile seawater, and stored in liquid nitrogen. .
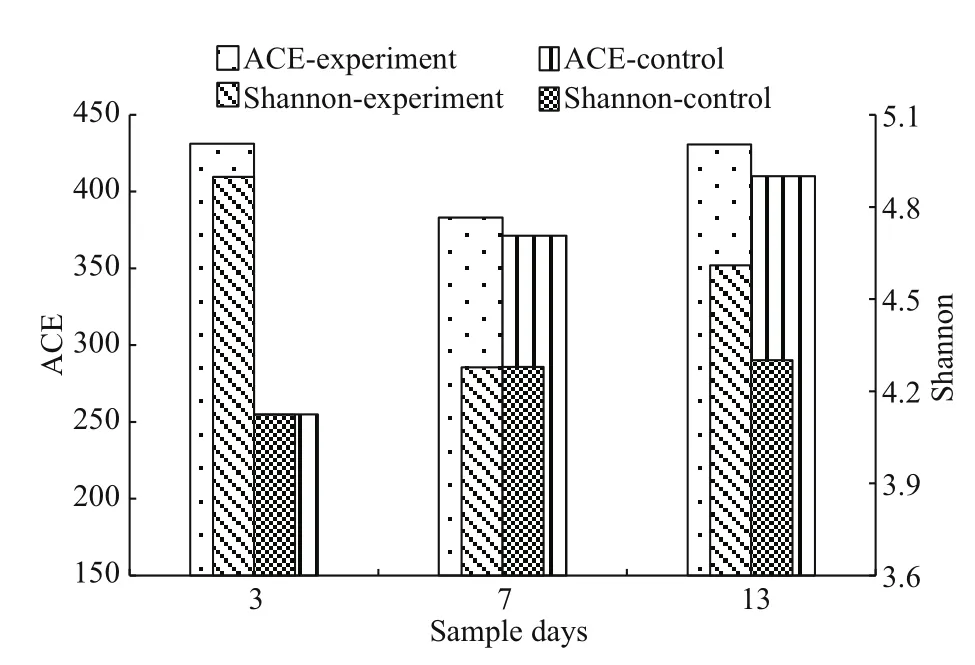
Fig.1 Abundance-based coverage estimator (ACE) and the Shannon diversity index of the B. plicatilis microf l ora
Suitable drugs are usually added during nutrient enrichment ofB.plicatilsandA.sinicato ensure their quality and quantity (Martínez-Díaz et al., 2003;Battaglene et al., 2006; Defoirdt et al., 2007; Asok et al., 2012). Enrof l oxacin was employed under the large-scale production condition for this artifi cial breeding trial in a turbot hatchery. Hence,B.plicatilsorA.sinicawith enrof l oxacin was considered the control.Bacillusamyloliquefacienswas added without the drug during enrichment as the experimental group to compare the pathogen inhibiting effect.Then, the effects ofB.amyloliquefaciensonB.plicatilsandA.sinicamicrof l oral structures were analyzed by comparing these two groups.
2.3 DNA extraction and sequencing
Total DNA of each sample was extracted with the E.Z.N.A.®Soil DNA Kit according to the manufacturer’s instructions. The V3 and V4 regions of 16S rDNA were amplified through polymerase chain reaction with primers: 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (3′-GGACTACNNGGGTATCTAAT-5′). High-throughput sequencing was accomplished with the Illumina HiSeq PE250 after the amplified DNA had been successfully detected via agarose gel electrophoresis.
2.4 Data analysis
Sequencing data were processed by splitting,splicing, fi ltering, and extracting before obtaining effective tags. Then, all effective tags of all samples were clustered into operational taxonomic units(OTUs) with Uparse v7.0.1001 (Caporaso et al.,2010) based on a sequencing identity of 97%. The OTU sequences were classified and analyzed with RDP Classifer 2.2 (Edgar, 2013) and GreenGene bank(Edgar et al., 2011) with a threshold of 0.8-1.0.

Fig.2 Abundance-based coverage estimator (ACE) and the Shannon diversity index of the A. sinica microf l ora
Student’st-test was used to examine significant differences between the experimental and control groups. AP-value<0.05 was considered significant.Values are given as mean±standard error.
3 RESULT
Raw data were obtained from six samples of eitherB.plicatilisorA.sinicausing the Illumina HiSeq PE250. After processing, 263 596 effective tags used in later analysis were gained inB.plicatilisand 260 446 inA.sinica. The samples averaged 43 933 effective tags inB.plicatilisand 43 408 inA.sinica.
3.1 Abundance-based coverage estimator (ACE)and the Shannon diversity index
ACE is used to estimate the abundance of a microbial species in ecology, whereas the Shannon diversity index expresses microf l oral diversity. The ACE and Shannon indices of the microf l ora inB.plicatilisare expressed in Fig.1. The ACE and Shannon indices in the experimental group were 383.09-431.10 and 4.28-4.90 and 254.84-410.08 and 4.12-4.30 in the control, respectively. Abundance of species and diversity in theB.plicatilisexperimental group were better than those in the control.
The changes in the ACE and Shannon indices are shown in Fig.2 between the control and experimental groups forA.sinica. The ACE and Shannon indices in the experimental group were 373.41-483.16 and 3.47-4.53 and 222.31-518.86 and 2.40-3.10 in the control, respectively. The ACEs were higher in the experimental group compared with the control, except on day 21. Changes in the Shannon index between the experimental groups forA.sinicaand the control were similar to those inB.plicatilis. Abundance of microbial species and diversity of microf l ora were all superior to those in the control.
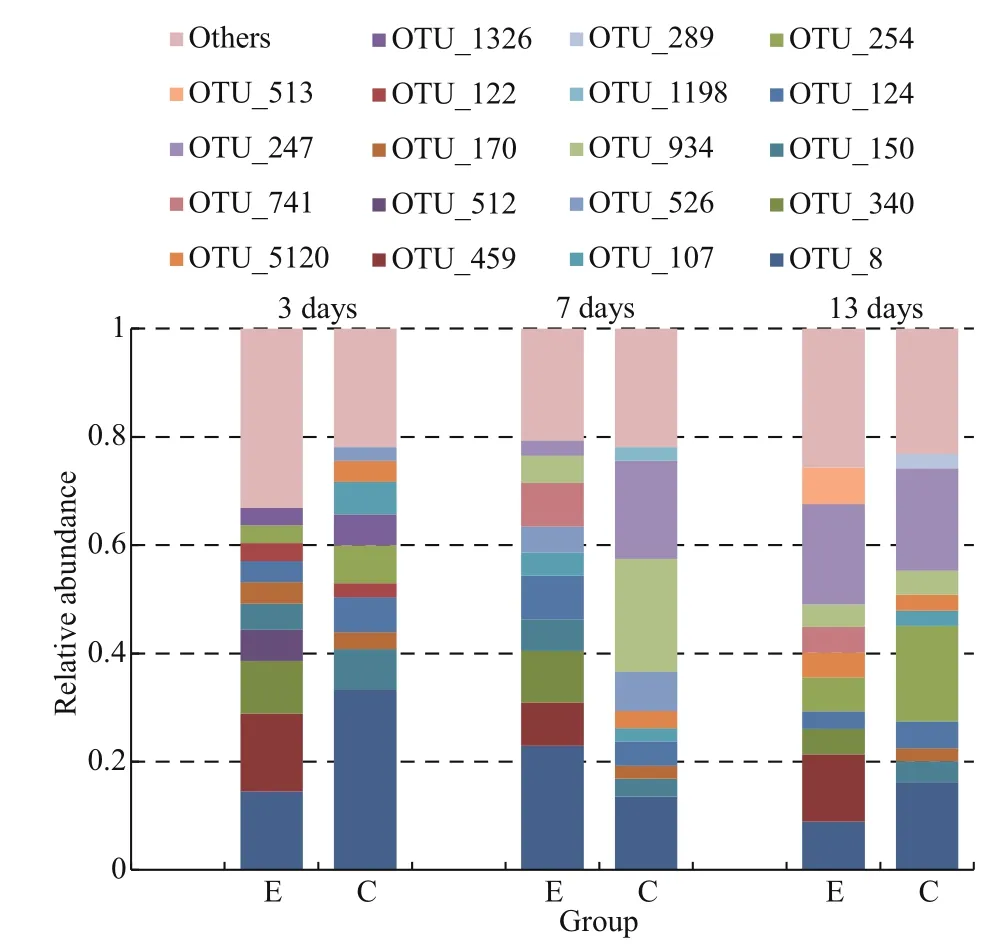
Fig.3 The microbial structure of B. plicatilis in the different treatmentsExperimental group represented by E, and control represented by C.The operational taxonomic units (OTUs) included in each bar are the top-ten OTUs in each sample.
3.2 Changes in the microf l oral composition of live feed
The top-ten OTUs of eachB.plicatilissample are represented by different colors in Fig.3. The sum of the relative abundances of OTU 8, OTU 150, OTU 254, OTU 124, and OTU 107 according to descending order was 0.603 in the control on day 3. The sum of the abundances of OTU 934, OTU 247, OTU 8, and OTU 526 was 0.598 on day 7, whereas the sum of OTU 247, OTU 254, OTU 8, OTU 124, and OTU 934 was 0.622 on day 13. Only OTU 8 was a common dominant species among the three control samples,and the microf l oral structure had no uniformity.However, the abundances of OTU 8, OTU 459, OTU 340, OTU 512, OTU 150, OTU 170, OTU 124, and OTU 122 were much higher in the experimental groups than other OTUs, and their sum was 0.604 on day 3. The sum of OTU 8, OTU 340, OTU 124,OTU741, OTU 459, OTU 150, and OTU 934 was 0.625 on day 7, and the sum of OTU 247, OTU 459,OTU 8, OTU 513, OTU 254, OTU340, and OTU 741 was 0.623 on day 13. OTU 8, OTU 459 and OTU 340 were always dominant in the three experimental groups compared with the control. Additionally,significant differences were observed in OTU 459 and OTU 340 between the control and experimental groups (P<0.05) (Fig.5).
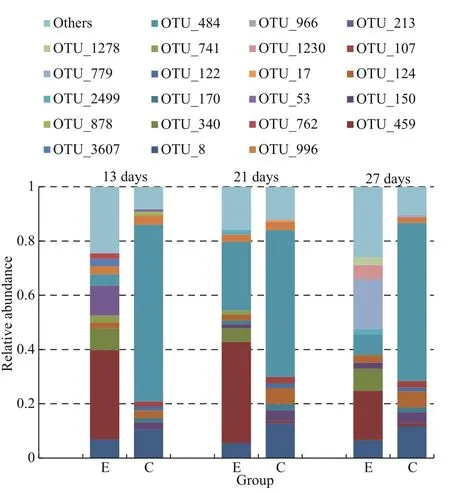
Fig.4 The microbial structure of A. sinica in the differenttreatmentsExperimental group represented by E and control represented by C.The operational taxonomic units (OTUs) included in each bar arethe top-ten OTUs in each sample.

Fig.5 Operational taxonomic units (OTUs) with significant differences in B. plicatilis between the control andexperimental groups Error bars represent standard errors of three replicates (P<0.05).
The distribution of the top-ten OTUs inA.sinicais presented in Fig.4. OTU 484 in the control was the fi rst dominant species with abundances of 0.540-0.652. OTU 8, OTU 124, and OTU 150 were according to descending order; the sums of these three were 0.160, 0.225, and 0.217 on days 13, 21,and 27, respectively. The microbial community composition was extremely similar in the control on the three sampling days. The abundances of OTU 484 and OTU 8 decreased in the experimental groups compared with the control, and the sums of these two OTUs were only 0.112, 0.308, and 0.143 on days 13,21, and 27, respectively. The abundances of OTU 459 and OTU 340 increased significantly in the experimental groups, and the sums of these two OTUs were 0.409, 0.423, and 0.263 on days 13, 21, and 27,respectively. OTU 459, OTU 340, OTU 484, and OTU 8 were always dominant in the three experimental groups. OTU 340, OTU 459, OTU 17, OTU 150,OTU 8, and OTU 484 were significantly different between the control and experimental groups (P<0.05)(Fig.6). In sum, clear differences in microbial community structure were observed between the control and experimental groups.
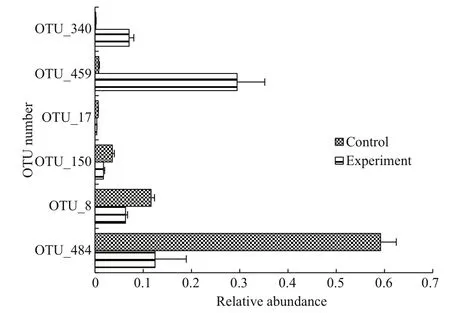
Fig.6 Operational taxonomic units (OTUs) with significant differences in A. sinica between the control andexperimental groups Error bars represent standard errors of three replicates (P<0.05).
3.3 Bate diversity
Similarity of theB.plicatilismicrobial community structure among the tested groups is shown in Fig.7 through the distance between two points. Each point on the principal components analysis (PCA) plot represents a relative sample, in which PC1 and PC2 are the top-two principal components in terms of OTU level. The percentages of PC1 and PC2 were 44.8% and 27.1%, respectively. Any one of the three samples in the experimental or control groups was relatively far from the others. Distances between C3 and E3, C7 and E7, and C13 and E13 were greater,suggesting that the microf l ora composition had obviously changed between the control and experimental groups.
The percentage of PC1 was 88% and much higher than PC2 (9.3%); therefore, PC1 was the deciding factor when analyzing the distance (Fig.8). The distance between any two of the three samples in the experimental or control groups was close to the PC1 axis. However, E13, E21, and E27 were farther from C13, C21, and C27, respectively. For the same reason,A.sinicamicrobial community composition were distinctly different in the experimental groups from those in the control.
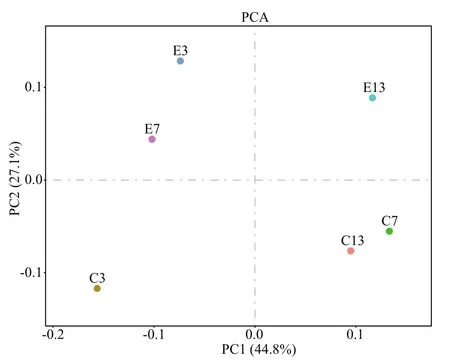
Fig.7 Principal components analysis (PCA) based on the operational taxonomic unit (OTU) level in B. plicatilisC3, C7, and C13 were three parallel samples in the control taken on days 3, 7, and 13 during turbot larval development, whereas E3, E7,and E13 correspond to the experimental groups.
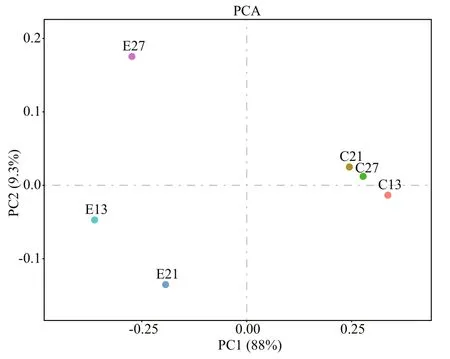
Fig.8 Principal components analysis (PCA) based on the operational taxonomic unit (OTU) level in A. sinicaC13, C21 and C27 were three parallel samples in the control taken on days 13, 21, and 27 during turbot larval development, whereas E13, E21, and E27 correspond to the experimental groups.
3.4 Microf l ora composition analysis
Analyses of the same OTUs inB.plicatiliswere carried out to determine the composition of the microbial community between the control and experimental groups (Fig.9). The average number of observed OTUs in the experimental group was higher than the control. The experimental groups with 56.31% OTUs were the same as those in the control based on average.
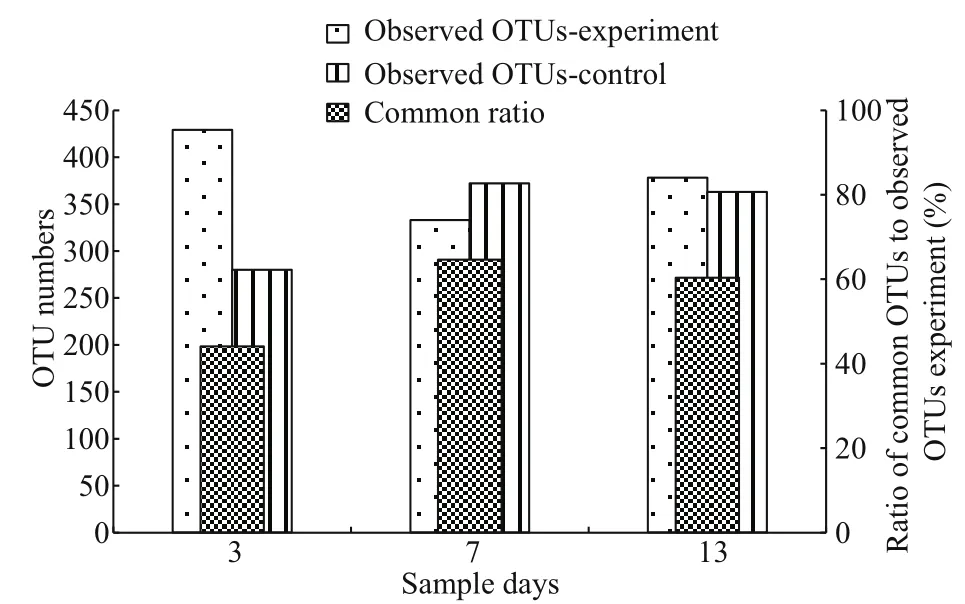
Fig.9 Branchionus plicatilis microf l oral similarity analysis between the two different treatments based on the operational taxonomic unit (OTU) level
A total of 356 OTUs was observed in theA.sinicaexperimental groups, which was higher than the 348 in the control (Fig.10). The average common ratio was 52.49% between the control and experimental groups.
3.5 Dominant OTUs
The major OTUs (abundance>0.01) inB.plicatilisand the relative classifi cation in GreenGene bank are shown in Table 1. Firmicutes, Proteobacteria,Cyanobacteria, and Bacteroidetes were the main taxa but most of the OTUs belonged to Firmicutes and Proteobacteria. Bacilli and Gammaproteobacteria were dominant at the class level. Furthermore,Lactococcus,Salinivibrio,Pseudoalteromonas, andPseudomonaswere the major genera. Additionally,Vibriocontaining OTU741 was not different between the two groups (P>0.05) inB.plicatilis(Table 1,Fig.5).
The main OTUs (abundance>0.01) inA.sinicawere classified according to GreenGene bank (Table 2). Firmicutes and Proteobacteria were the two main phyla with dominant OTUs, andCobetia,Pseudoalteromonas, andLactococcuswere the main genera. Similarly, no difference inVibriowas observed between the control andA.sinicaexperimental groups(Table 2 and Fig.6;P>0.05).
4 DISCUSSION
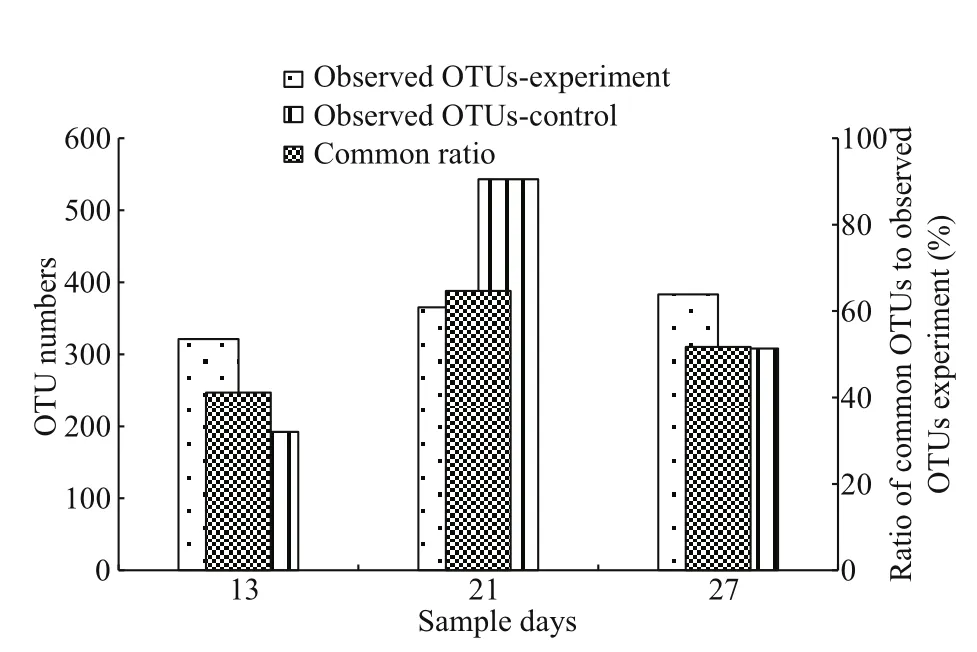
Fig.10 Artemia sinica microf l oral similarity analysis between the two different treatments based on the operational taxonomic unit (OTU) level
High-throughput sequencing has been developed from toxicological and pharmacological studies and clinical testing to reveal results on nutrition, drug resistance of bacteria and the safety of animal products(Chambers and Gong, 2011; Diaz-Sanchez et al.,2013; Fang et al., 2015). The applications of highthroughput sequencing extend into various large fi elds with the development of bioinformatics. Based on bioinformatics analysis, the use of high-throughput sequencing in aquaculture and microf l oral research has been increasing in recent years (Wang et al., 2014;Zhang et al., 2014). Traditional bacterial culture methods are limited when studying microbial community structure and do not fully reflect the total number of bacteria and species composition in an aquaculture system. Only a few bacteria, such asAeromonas,Pseudomnas, andVibrio, can be detected through traditional cultural methods (Planas et al.,2006; Shi et al., 2015). However, high-throughput sequencing is able detect all microbes. In this study,Lactococcus,Exiguobacterium,Solibacillus,Salinivibrio,Marinomonas,Cupriavidus,Tenacibaculum,Zobellia, andCobetiawere identified by highthroughput sequencing, and the diversity of species in live feed was realized and compared with the traditional cultural method. Hence, the results fully and accurately reflected the microbial community structure in the aquaculture system.
The ACE and Shannon diversity indices ofB.plicatilisandA.sinicawere higher in the experimental groups compared with the control,suggesting that addingB.amyloliquefaciensduring the enrichment process increased microbial species abundance and diversity. The results show that the microf l ora ofB.plicatiliswas different on each sampling day, although the hatching technique was the same. Therefore, obvious differences were detected in the dominant OTUs among the three replicates of the control, whereas the experimental groups revealed similar microf l ora among the three replicates. The reason might be that the reproductive capacity of more microbial species was inhibited byB.amyloliquefaciens.B.amyloliquefaciensbecame the dominant species inB.plicatiliswith nocompetition, such as OTU 459 and OTU 340. These results all indicate thatB.amyloliquefacienshad better effects on the adaptability and unifi cation of microbial community structure compared with drugs.TheA.sinicamicrobial community composition among the three replicates in the control was close because larvae came from the same egg batch and hatching technique. Similar microbial structures were obtained among the three replicates of the experimental groups. After addingB.amyloliquefaciens, OTU 459 and OTU 340 grew rapidly and became dominant species, but replication of OTU 484, OTU 150, OTU 8, and OTU 17 was significantly inhibited. It is possible that survival or competition among adhesive sites occurred between these two OTUs.B.amyloliquefaciensis thought to be the only factor that changed and unified theA.sinicamicrof l oral structure. Common ratios between the experimental and control groups forB.plicatilisandA.sinicawere<60%, suggesting thatB.amyloliquefacienschanged the microbial community composition. A previous study reported that probiotics improve rotifer microf l ora through the initial method (Gianelli et al.,1997). Similarly, microbial community structure of the intestine of cultured fi sh changes after adding a probiotic (Bergh et al., 1994; Huys et al., 2001). The reaction of microf l ora in the turbot larval intestinal tract toB.amyloliquefacienswill be analyzed and reported in another study.

Table 1 classified information of dominant operational taxonomic units (OTUs) in B. plicatilis
Vibriois regarded as a major pathogenic bacteria in aquaculture. However,V.alginolyticusandV.anguillarumhave been detected in healthyB.plicatilisandArtemia(Munro et al., 1995; Villamil et al., 2003). Several studies have also suggested that vibrios are common bacteria during live feed nutrient enrichment and turbot larval rearing (Gatesoupe,1990; Villamil et al., 2003). In this study, althoughVibrio(OTU 741) was the major microbe in the twolive feeds and abundance in the experimental groups was higher than the control, there were no significant differences between them, indicating that the inhibitory effects ofB.amyloliquefaciensand the antibiotic on vibrios were quite similar. Some other studies have also reported thatLactobacilluspentosusandLactobacilluscaseiprotectArtemiafrom the pathogenic effects ofV.alginolyticus(Lamari et al.,2014; Garcés et al., 2015) andLactococcuslactisinhibitsV.anguillaruminB.plicatils(Shiri Harzevili et al., 1998). Hence, it is possible thatB.amyloliquefaciensreplaced the antibiotic and prevented pathogenic bacteria from reproducing duringB.plicatilisandA.sinicanutrient enrichment,which agrees with other studies (Defoirdt et al., 2007;Ahmed et al., 2015).

Table 2 classified information of dominant operational taxonomic units (OTUs) in A. sinica
Lactococcuswas a dominant genus inB.plicatilisorA.sinica, and noLactococcussp. are pathogenic,exceptL.garvieae(Hoshina et al., 1958; Chen et al.,2001, 2002). However,L.garvieaehas been found in the intestinal tract of fi sh without any apparent disease symptoms (Cai et al., 1998). Therefore,Lactococcusprobably did not adversely affect the health of these two live feeds. Additionally,Pseudomonas,Pseudoalteromonas, andAlteromonaswere the other major species in this study, and their abundances were all higher in the experimental groups than in the control. However, some representative species in these genera are pathogenic or are conditioned pathogens in aquaculture (Ferguson et al., 2004;Garnier et al., 2007; Hjelm et al., 2004; Magi et al.,2009). Probiotics promote the growth of some microbes by inhibiting survival of their competitors,which was suggested by the changes in relative abundances in this study. However, one probiotic will not protect all live feed from infections by all pathogenic bacteria. Various probiotics work with each other. Therefore, selecting a probiotic formula with a focus on disease control and prevention is important.
5 CONCLUSION
In summary,B.amyloliquefacienswas added during nutrient enrichment and clearly affectedB.plicatilisandA.sinicamicrobial community structures. Additionally, the inhibiting effect ofB.amyloliquefacienson vibrios was almost similar to a broad-spectrum antibiotic, suggesting thatB.amyloliquefacienscould take the place of an antibiotic during hatching ofB.plicatilisandA.sinica.
BranchionusplicatilsandA.sinicaare two of the most important and common live feeds for fi sh,shrimp, and crab seedlings in many countries.Introducing a probiotic through live feed can play the role of disease control and prevention, which may reduce antibiotic abuse, as Specific pathogens are antagonized by application of certain probiotic species. In this case, good husbandry and environmentally friendly aquaculture can help. All of these aspects will be a focus in the future.
6 DATA AVAILABILITY STATEMENT
The datasets generated and analyzed during the current study are available from the corresponding author.
 Journal of Oceanology and Limnology2018年3期
Journal of Oceanology and Limnology2018年3期
- Journal of Oceanology and Limnology的其它文章
- Editorial Statement
- The post-larval and juvenile fi sh assemblage in the Sukhothai Floodplain, Thailand*
- Comparison ofintestinal microbiota and activities of digestive and immune-related enzymes of sea cucumberApostichopus japonicusin two habitats*
- Otolith shape analysis for stock discrimination of twoCollichthysgenus croaker (Pieces: Sciaenidae,) from the northern Chinese coast*
- The impact of spatial autocorrelation on CPUE standardization between two different fi sheries*
- Stocking density affects the growth performance and metabolism of Amur sturgeon by regulating expression of genes in the GH/IGF axis*
