The landscape of molecular mechanisms for salt tolerance in wheat
*
aThe Key Laboratory of Plant Cell Engineering and Germplasm Innovation,Ministry of Education,School of Life Sciences,Shandong University,Jinan 250100,Shandong,China
bState Key Laboratory of Soil and Sustainable Agriculture,Institute of Soil Science,Chinese Academy of Sciences,Nanjing 210008,Jiangsu,China
1.Introduction
Bread wheat(Triticum aestivum L.),one of the most important staple crops globally,provides most of the calories for approximately 30%of the world population[1].Increasing attention is being given to the mechanisms of abiotic stress response due to greater awareness of the threats of climate change,and loss of arable land during urbanization,and environmental degradation caused by pollution[2].Since more than 800 Mha(6%)of arable land are affected by salinity worldwide[3],soil salinity is a major constraint upon wheat grain yield[4].
A direct consequence of soil salinity is the over-accumulation of intracellular sodium(Na+),resulting in serious ionic toxicity,especially in the leaves with direct inhibitory effects on photosynthesis.Moreover,salt stress can cause osmotic and oxidative stress,further disturbing metabolic processes and leading to DNA damage and even cell death[3].Therefore,understanding the mechanisms of response and adaptation to salt stress and then improving the salinity tolerance of crops are critical tasks for breeders and researchers.
Although some mechanisms,such as osmotic adjustment,tissue tolerance processes,and K+retention,have been elaborated in other crops[5],these are greater challenges for bread wheat due to its large,complicated and hexaploid genome[6].Nevertheless,the mechanisms underlying salinity tolerance in wheat,including leaf Na+exclusion mediated by high-affinity K+transporters(HKTs)and reactive oxygen species (ROS)detoxification,have been addressed in long-term and subtle ways[5].Multiple components involved in crosstalk of salinity response with other environmental or developmental signals were identified.Notably,in line with the continuous releases of wheat whole genome information[7]and recently established wheat mutant libraries[8],more versatile approaches will be available for salt tolerance improvement.Therefore,this review will provide an outline of the mechanisms of wheat salinity tolerance,and present an outlook on prospective key research on this topic.
2.HKT-type transporters confer wheat salinity tolerance by promoting sodium exclusion
It has long been known that tetraploid wheat is less salt tolerant than bread wheat[9,10],and that a major factor behind this difference is that bread wheat is able to maintain a higher ratio of potassium concentration to sodium concentration in the leaves[11].This trait was shown to be governed by Kna1 on chromosome 4D[12].A genetic analysis,based on a population derived from a cross between a standard durum wheat genotype and a line containing introgressions from the A genome diploid ancestral wheat relative Triticum monococcum showing high Na+exclusion ability,revealed that two loci,Nax1 and Nax2,were involved in excluding sodium ions[13].
Class 1 HKT genes are involved in regulating transport of Na+in higherplants[14].Several HKT1genes,includingHKT1;1/2-like,HKT1;3-like,HKT1;4-like,and HKT1;5-like,have been identified and mapped to wheat homoeologous chromosome groups2,6,2,and 4,respectively[15].Among these,Nax1 in chromosome arm 2AL co-segregated with sodium transporter gene HKT1;4-A2,which was shown to control Na+unloading from xylem in roots and sheaths and therefore was proposed as the functional candidate[16].Nax2 was mapped to the distal region of chromosome 5AL that is homoeologous to a region on chromosome 4DL containing Kna1.Based on synteny and phylogeny analysis with Nax2,TmHKT1;5-A was proposed to be the candidate of Nax2[17].In addition,field trials in saline soils demonstrated that the presence of TmHKT1;5-A significantly reduced leaf sodium content and increased durum wheat grain yield by 25%compared to lines without the Nax2 locus[18].Furthermore,decreased expression of TaHKT1;5-D,which is homoeologous to TmHKT1;5-A and underlies Kna1 locus in bread wheat,caused by target-specific RNA interference-induced silencing(RNAi),led to an accumulation of Na+in leaves[19],strongly suggesting that TaHKT1;5-D should be the candidate gene of Kna1.
Na+exclusion mediated by HKT genes in leaves has been recognized as a major mechanism in salinity tolerance of wheat.However,some fundamental issues need to be further addressed.One is how these HKT genes respond to salt stress in wheat.For example,TaHKT1;5-D exhibited a transcriptional reprogramming from constitutive high basal expression in diploid Aegilops tauschii to salt-induced expression in a newly synthetic allohexaploid wheat[20],whilst Byrt et al.[19]discovered no detectable difference in TaHKT1;5-D expression when hexaploid wheat cv.Bobwhite was challenged by salt stress.Additionally,a reduction inTaHKT1;5-Dtranscripts was revealed after salt treatment in both hexaploid wheat cv.JN177 and its introgression line SR3[21].These contradictory results bring about an interesting question of whether the response of TaHKT1;5-D to salinity is accession-dependent(that is,is there an association between the response mode and tolerance to salt stress?),or tissue-specific(as TaHKT1;5-D was previously implied to be predominantly functional within the stele,particularly within xylem parenchyma and pericycle cells adjacent to the xylem vessels[19]).
Another question is how these wheat HKT genes are regulated.The sole HKT gene in Arabidopsis,AtHKT1,is regulated by small RNA and DNA methylation[22].Moreover,DNA methylation also participated in the response of TaHKT1;5s to salt stress in wheat cv.JN177 and SR3[21].Intriguingly,the transcript levels of TaHKT1;5-B1 and TaHKT1;5-B2 were extremely low compared with that of TaHKT1;5-D[19].Epigenetics plays an important role in the dosage effect of homeologous transcription[7].Therefore,the contribution of epigenetics to the lower expressions of TaHKT1;5-B1 and TaHKT1;5-B2 should be further studied.Moreover,transcription factors,such as AtABI4[23]and OsMYBc[24],were shown to regulate HKT genes in plants,offering more candidate targets for enhancing salinity tolerance.However,an up-stream regulator(s)of wheat HKT genes is still unidentified possibly due to the complexity of the hexaploid wheat genome.
3.ROS homeostasis involved in salinity tolerance of a somatic hybrid introgression line
Wild relatives and related species often carry specific traits with potential for improvement of common wheat[25].For example,tall wheat grass(Thinopyrum ponticum),a species that normally grows in barren areas,exhibits tolerance to abiotic stress[26]and is therefore a valuable genetic resource for wheat improvement.However,hybrids between potentially beneficial species and common wheat may be restricted by the “recombination barrier”[27].Asymmetric somatic hybridization is a viable alternative to introgression,especially where inter-specific crosses are not possible[28].Utilizing this approach,the salinity-tolerant bread wheat cultivar Shanrong No.3(SR3)was generated as a derivative of a somatic hybrid between bread wheat and tall wheatgrass[29].This novel cultivar is not only an elite line for breeding,but also a valuable genetic resource to uncover mechanisms underlying salt tolerance.
Transcriptomic,proteomic and metabolomic comparisons of SR3 with its wild type cv.Jinan 177(JN177)wheat parent suggested that reactive oxygen species homeostasis was the major biochemical basis for the salt tolerance of cv.SR3[30].A mapping analysis localized a tolerance QTL on chromosome arm 5AL,at a position containing TaSRO1,a gene encoding a poly(ADP ribose)polymerase(PARP)domain protein.PARP proteins have been implicated in modulation of redox homeostasis.Sequence variation between the TaSRO1 alleles present in cv.SR3 and cv.JN177 was predicted to affect PARP catalytic activity that is significant for DNA repair under oxidative stress.The transgenic constitutive expression of the allele from cv.JN17,a sensitive cultivar,enhanced the levels of salinity and ROS tolerance,while RNAi-induced knock--down of the gene in cv.SR3 compromised the level of tolerance.Thus TaSRO1 was considered to be a strong candidate for the salt tolerance QTL in cv.SR3[31].
ADP-ribosylation is a kind of protein modification involved in signal transduction,DNA repair,and stress response[32].PARP-like genes are present in many eukaryotes,and the PARP catalytic domain is the major ADP ribosylation factor in mammalian cells[33].In Arabidopsis,AtRCD1 and six AtSRO genes(similar to RCD One)belong to the PARP subfamily,but intriguingly,none of these members exhibited PARP catalytic activity,even though they were implicated in stress response[34].You et al.showed that OsSRO1c had dual roles in drought and oxidative stress tolerance in rice by modulating stomatal closure and H2O2accumulation,but also had no PARP catalytic activity[35].In contrast,wheat TaSRO1 is unique in being the first SRO1 protein found to possess PARP catalytic activity in plants, and the higher PARP catalytic activity of the TaSRO1 allele in SR3 accounted for the stronger DNA repair capability under stress conditions.This at least partly contributed to the vigorous growth and stress tolerance of SR3[31].Moreover,TaSRO1 contained an RST (for RCD-SRO-TAF4) domain that functions in protein-protein interactions[34].It is meaningful to further identify TaSRO1-interacting proteins,and examine whether TaSRO1 ADP-ribosylates the interacting proteins and therefore contributes to the superior capacity of ROS homeostasis maintenance in SR3.
Somatic hybridization introduces a minimum of exogenous chromatin into a recipient genome, but causes genomic shock that induces high frequencies of both point mutations and indels(insertions and deletions)in coding sequences,and is thus capable of generating elite alleles[36].Genetic analysis indicated the remarkable salinity tolerance of SR3 by modulation of ROS homeostasis that was accomplished by a polygene effect.A zinc finger transcription factor,TaCHP,was activated in SR3 with much higher transcript abundance than in JN177[37].TaCHP facilitated salinity tolerance in wheat through improved leaf peroxidase(POD)activity to enhance ROS scavenging ability.Another example is the wheat oxophytodienoate reductase gene TaOPR1,whose expression was induced in roots by salt treatment with higher induction in SR3 than in JN177[38].TaOPR1 enhanced salt tolerance by triggering transcription of ROS homeostasis associated genes,consequently reducing malondialdehyde and ROS levels in an ABA-pathway dependent manner[38].
“Genomic shock”during the process of somatic hybridization also causes massive epigenetic reprogramming[39].A topic of increasing interest is the role of epigenetic variation in controlling gene expression.Observed differences in transcript abundances of TaFLS1,TaWRSI1,and TaTIP2;2 between JN177 and SR3 that could not be explained by differences in either the promoter or the coding sequences,were shown to vary with respect to DNA methylation level[21].In animals,the status of DNA methylation is affected by the level of ROS content[40].It is essential to determine whether the divergence of ROS accumulation and ROS homeostasis maintenance between SR3 and JN177 is associated with DNA methylation,and its effect on expression patterns of salt-stress responsive genes.
4.Genes involved in crosstalk between salinity response and other environmental or developmental signals in wheat
When plants are confronted with high salinity,complex physiological responses such as phytohormone signaling pathways and developmental signals are triggered to cope with or adapt to the stress[41].Therefore,it is essential to identify the node(s)linking salinity response and other environmental or developmental signals.An attempt to do this in wheat was firstly performed by looking at phytohormones,as most phytohormones are regulatory factors of both developmental processes and stress response.For example,the wheat gene TaAOC1,encoding an allene oxide cyclase involved in jasmonic acid(JA)synthesis,was induced by high salinity[42].Constitutive expression of TaAOC1 in both wheat and Arabidopsis restricted root growth,but enhanced salt tolerance and JA content.The evidence indicates JA was involved in the orchestration of salt stress response and developmental processes.Moreover,TaAOC1 and TaOPR1 encode two key enzymes of the α-linolenic acid metabolic pathway,catalyzing JA synthesis and OPRI branches,respectively.In line with the data of TaAOC1 and TaOPR1[37,41],we determined that these two branches provide salt tolerance via both the JA-and ABA-dependent pathways to promote expression of MYC2,a crucial component of the abiotic stress response-signaling pathway.These findings firstly indicate that different branches of a metabolic pathway participate in a single process but controlled by different mechanisms.Importantly,variation in TaAOC1 and TaOPR1 alleles could be exploitable in molecular breeding.
Another example is TaBASS2 that transports pyruvic acid from the cytoplasm into the chloroplasts,where it can be used as the precursor of ABA and other compounds.Overexpres-sion of TaBASS2 improved salinity tolerance and reactive oxygen species scavenging in wheat and Arabidops is through repression of ABI4 expression,indicating that ABA signaling and plastid retrograde signaling pathways were involved in the performance of TaBASS2[43].
Light is a basic factor that positively affects the growth and development of plants.TaGBF1,a blue light-specific responsive G-box binding factor,was induced after exposure to salt[44].TaGBF1 caused salt sensitivity and promoted blue light mediated photomorphogenesis,showing that it was a common component of the blue light-and salt stress-responsive signaling pathways.Interestingly,genetic analysis suggested that the role of TaGBF1 in response to salt relied on ABI5,a key component of the ABA signaling pathway,rather than light.
In summary,only fragmentary information has been mined on crosstalk between response to salinity and other environmental or developmental stimuli in wheat.Along with the enrichment of genomic data and other omics data forming a network(see next section),more key components embedded in the machinery will be dissected for wheat improvement.
5.New trends in functional genomic studies of salinity tolerance in wheat
5.1.Omics networks
Along with the recent advances in wheat whole genome sequencing,a new epoch for wheat research is emerging[7].Rapidly increasing information on genomics and other omics approaches,including transcriptomics, proteomics, epigenomics,metabolomics and phenomics, will accelerate the rate of gene discovery in wheat.
In earlier studies,subtraction hybridization[37]and cDNA microarrays[45]between salt-susceptible and tolerant wheat lines were performed to identify the molecular basis in salinity tolerance.For example, TaCHP,which was expressed at extremely low levels in JN177 but at high levels in SR3 was isolated by subtraction hybridization between JN177 and SR3[37].However,there were numerous omissions in prediction of candidate genes because of the low throughput and low resolution of the approach,especially in the absence of a whole genome sequence for wheat.More recently,high-throughput transcriptome sequencing profiles using wheat cultivarswith contrasting levels of salt tolerance enabled global gene expression reprogramming involving 36,804 genes following salt stress[46].Moreover,as the assembly and annotation of the transcripts were based on information from wheat genome survey sequences,the resolution was sufficiently high to permit expression partitioning of homologs and tandem duplications contributing to the variation in salt tolerance.
The rapidly improving technical capacity of next generation sequencing(NGS)and genomic enrichment information for wheat will also enable identification of the role of epigenomics in salinity tolerance.A preliminary DNA methylome analysis of salt stress differences between SR3 and JN177 revealed that multiple salt stress responsive genes were regulated by DNA methylation[21].Recently,genome-wide DNA methylation was measured in wheat under different temperature conditions,using the whole genome sequence to distinguish sub-genome-specific methylation[47].A similar high-resolution DNA methylome analysis following salinity treatment has not yet been reported.Furthermore,bulks of small RNAs in response to salt stress were discovered through miRNome analysis[48,49].However,functional validations of these candidate small RNAs and their putative targeted genes were rarely performed and need further study.The first functional noncoding mi RNA screened from high-resolution omics data was involved in wheat β-diketone wax metabolism[50].Such studies will facilitate the functional study of wheat small RNAs in response to salt stress.
The rapid accumulation of omics data from multiple tissues and temporal developmental time-courses,and various stress conditions in wheat has encouraged the building of a pan-omics database[51].That omics network will greatly promote the exploitation of functional genes,and give us a more comprehensive understanding ofsalinity tolerance.Moreover,co-/multi-regulatory genetic bases of salinity tolerance together with other environmental or developmental stimuli will be easily identified from the network intersections.
5.2.Salt-resistant germ plasm
Specific germplasms,including the diploid ancestral wheat relative T.monococcum and the somatic hybrid introgression,were fundamental in elaborating two major mechanisms(ionic and ROS homeostasis)of wheat salinity tolerance.More elegant systems need to be applied to generate novel salt-resistant germplasms for gene discovery and breeding.A further example is the salt-tolerant wheat germplasm RH8706049,a mutant derived from anther culture,EMS induction and selection for salt tolerance,from which several salt-responsive genes were identified.Preliminary functional analyses of these genes were made in Arabidopsis[52,53].Further analyses of the genes in transgenic wheat are needed.Moreover,multiple approaches such as that in SR3 using multiple omics,population genetics and salt-tolerant QTL analysis are essential for further investigation of RH8706049.
Another recent trend in functional genomics is the establishment of comprehensive EMS mutant libraries of tetraploid wheat and hexaploid wheat[8].Mutant sites were sequenced and cataloged using the next-generation sequenc-ing and exome capture platform.Based on phenotype screening,novel genes involving salinity tolerance will be easily identified from these potentially informative libraries.Moreover,selected mutants can be used to validate the functions of salt stress responsive genes,hopefully avoiding the tedious process of wheat transformation.
6.Conclusions
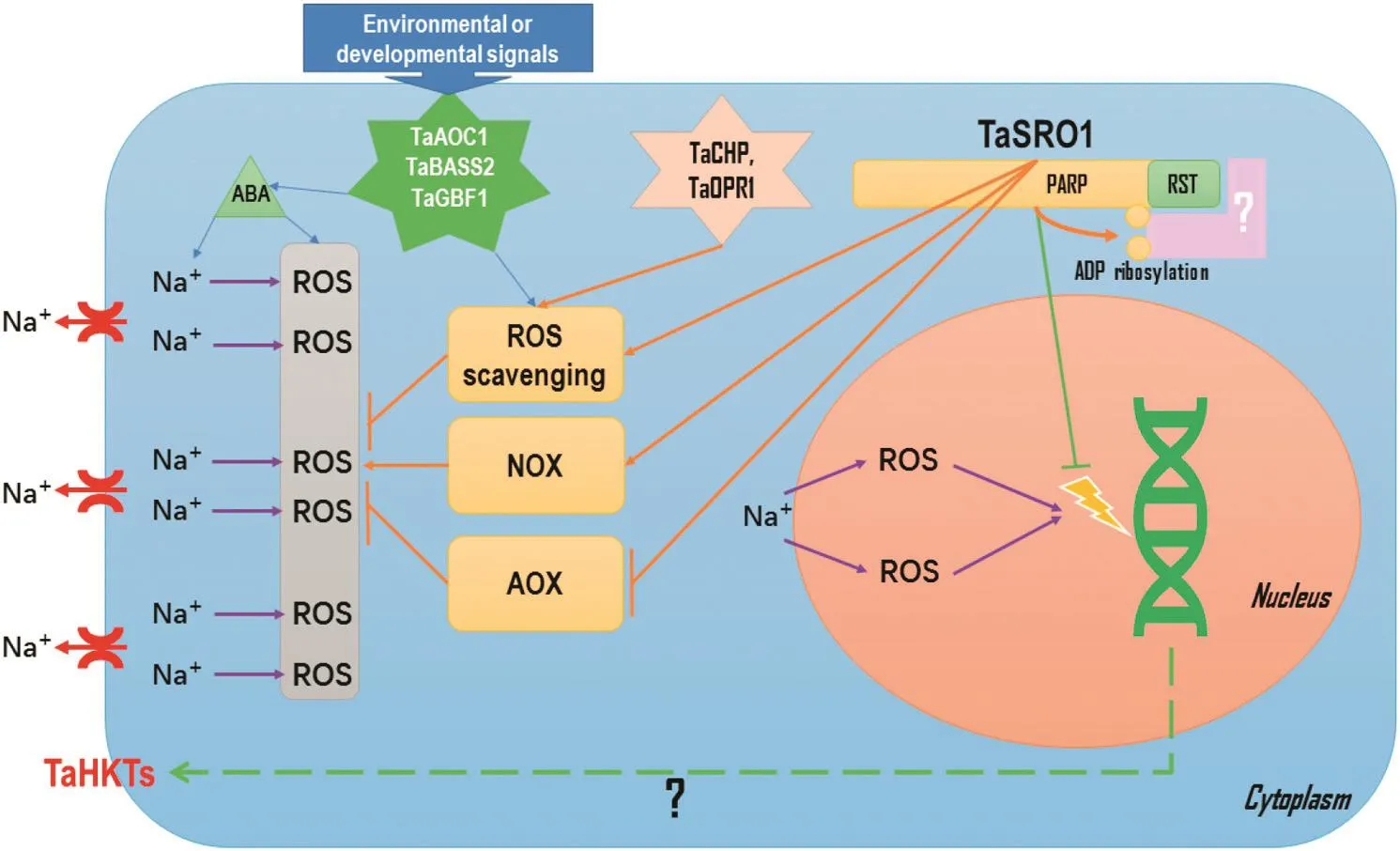
Fig.1–Pathways involved in wheat salinity tolerance.ROS,reactive oxygen species;NOX,NADPH oxidase;AOX,alternative oxidase;ABA,abscisic acid;wheat genes(Ta),including TaAOC1,an allene oxide cyclase gene;TaBASS2,a pyruvic acid transporter gene;TaGBF1,a G-box binding factor gene;TaCHP,a zinc finger transcription factor gene;TaOPR1,an oxophytodienoate reductase gene;TaSRO1,a similar to RCD One gene;TaHKTs,high-affinity potassium transporter genes.
Despite the complexity of the wheat genome,two major pathways,namely HKT genes that mediate Na+exclusion and the SRO gene that regulates ROS homeostasis,are pivotal in wheat salinity tolerance(Fig.1).The issues of regulation of HKT genes and the interaction protein from the SRO gene need to be further addressed.The discovery of the two pathways was greatly assisted by the use of specific germplasms(such as the diploid ancestral wheat relative T.monococcum and the somatic hybrid introgression line SR3).With the emergence of NGS and wheat whole genome information,functional genomics studies of salinity tolerance will be accelerated in wheat.Recent advances in the development of wheat EMS mutant libraries and genome editing will hasten validation and exploitation of salt stress responsive genes.More genes conferring salinity tolerance are likely to be identified and used in wheat improvement.
This study was supported by the National Key Research and Development Project(2016YFD0101004),NationalNatural Science Foundation of China(31430060,31601306),and China Postdoctoral Science Foundation(2016M601161).
R E F E R E N C E S
[1]Food and Agriculture Organization of the United Nations,www.fao.org/worldfoodsituation/csdb/en/.
[2]International Wheat Genome Sequencing Consortium(IWGSC),A chromosome-based draft sequence of the hexaploid bread wheat(Triticum aestivum)genome,Science 345(2014)1251788.
[3]R.Munns,M.Tester,Mechanisms of salinity tolerance,Annu.Rev.Plant Biol.59(2008)651–681.
[4]D.B.Lobell,W.Schlenker,J.Costa-Roberts,Climate trends and global crop production since 1980,Science 333(2011)616–620.
[5]R.Munns,M.Gilliham,Salinity tolerance of crops–what is the cost?New Phytol.208(2015)668–673.
[6]T.Marcussen,S.R.Sandve,L.Heier,M.Spannagl,M.Pfeifer,K.S.Jakobsen,B.B.Wulff,B.Steuernagel,K.F.Mayer,O.A.Olsen,Ancient hybridizations among the ancestral genomes of bread wheat,Science 345(2014)1250092.
[7]M.Wang,S.B.Wang,G.M.Xia,From genome to gene:a new epoch for wheat research?Trends Plant Sci.20(2015)380–387.
[8]K.V.Krasileva,H.A.Vasquez-Gross,T.Howell,P.Bailey,F.Paraiso,L.Clissold,J.Simmonds,R.H.Ramirez-Gonzalez,X.D.Wang,P.Borrill,C.Foskerc,S.Aylingc,A.L.Phillips,C.Uauy,J.Dubcovsky,Uncovering hidden variation in polyploid wheat,Proc.Natl.Acad.Sci.U.S.A.114(2017)201619268.
[9]L.Francois,E.Maas,T.Donovan,V.Youngs,Effect of salinity on grain yield and quality,vegetative growth,and germination of semi-dwarf and durum wheat,Agron.J.78(1986)1053–1058.
[10]H.Rawson,R.Richards,R.Munns,An examination of selection criteria for salt tolerance in wheat,barley and triticale genotypes,Aust.J.Agric.Res.39(1988)759–772.
[11]J.Gorham,R.G.W.Jones,A.Bristol,Partial characterization of the trait for enhanced K+–Na+discrimination in the D genome of wheat,Planta 180(1990)590–597.
[12]J.Dubcovsky,G.Santa Maria,E.Epstein,M.C.Luo,J.Dvořák,Mapping of the K+/Na+discrimination locus Kna1 in wheat,Theor.Appl.Genet.92(1996)448–454.
[13]R.Munns,R.Hare,R.James,G.Rebetzke,Genetic variation for improving the salt tolerance of durum wheat,Aust.J.Agric.Res.51(2000)69–74.
[14]T.Horie,F.Hauser,J.I.Schroeder,HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants,Trends Plant Sci.14(2009)660–668.
[15]S.B.Huang,W.Spielmeyer,E.S.Lagudah,R.Munns,Comparative mapping of HKT genes in wheat,barley,and rice,key determinants of Na+transport,and salt tolerance,J.Exp.Bot.59(2008)927–937.
[16]S.B.Huang,W.Spielmeyer,E.S.Lagudah,R.A.James,J.D.Platten,E.S.Dennis,R.Munns,A sodium transporter(HKT7)is a candidate for Nax1,a gene for salt tolerance in durum wheat,Plant Physiol.142(2006)1718–1727.
[17]C.S.Byrt,J.D.Platten,W.Spielmeyer,R.A.James,E.S.Lagudah,E.S.Dennis,M.Tester,R.Munns,HKT1;5-like cation transporters linked to Na+exclusion loci in wheat,Nax2 and Kna1,Plant Physiol.143(2007)1918–1928.
[18]R.Munns,R.A.James,B.Xu,A.Athman,S.J.Conn,C.Jordans,C.S.Byrt,R.A.Hare,S.D.Tyerman,M.Tester,M.Gilliham,Wheat grain yield on saline soils is improved by an ancestral Na+transporter gene,Nat.Biotechnol.30(2012)360–364.
[19]C.S.Byrt,B.Xu,M.Krishnan,D.J.Lightfoot,A.Athman,A.K.Jacobs,N.S.Watson-Haigh,D.Plett,R.Munns,M.Tester,M.Gilliham,The Na+transporter,TaHKT1;5–D,limits shoot Na+accumulation in bread wheat,Plant J.80(2014)516–526.
[20]C.W.Yang,L.Zhao,H.K.Zhang,Z.Z.Yang,H.Wang,S.S.Wen,C.Y.Zhang,S.Rustgi,D.von Wettstein,B.Liu,Evolution of physiological responses to salt stress in hexaploid wheat,Proc.Natl.Acad.Sci.U.S.A.111(2014)11882–11887.
[21]M.Wang,L.M.Qin,C.Xie,W.Li,J.R.Yuan,L.N.Kong,W.L.Yu,G.M.Xia,S.W.Liu,Induced and constitutive DNA methylation in a salinity tolerant wheat introgression line,Plant Cell Physiol.(2014)1354–1365.
[22]D.Baek,J.Jiang,J.S.Chung,B.Wang,J.Chen,Z.Xin,H.Shi,Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance,Plant Cell Physiol.52(2011)149–161.
[23]D.Shkolnik-Inbar,G.Adler,D.Bar-Zvi,ABI4 downregulates expression of the sodium transporter HKT1;1 in Arabidopsis roots and affects salt tolerance,Plant J.73(2013)993–1005.
[24]R.Wang,W.Jing,L.Y.Xiao,Y.K.Jin,L.Shen,W.H.Zhang,The rice high-affinity potassium transporter1;1 is involved in salt tolerance and regulated by an MYB-type transcription factor,Plant Physiol.168(2015)1076–1090.
[25]T.Cox,Deepening the wheat gene pool,J.Crop.Prod.1(1997)1–25.
[26]T.D.Colmer,T.J.Flowers,R.Munns,Use of wild relatives to improve salt tolerance in wheat,J.Exp.Bot.57(2006)1059–1078.
[27]C.Feuillet,P.Langridge,R.Waugh,Cereal breeding takes a walk on the wild side,Trends Genet.24(2008)24–32.
[28]G.M.Xia,Progress of chromosome engineering mediated by asymmetric somatic hybridization,J.Genet.Genomics 36(2009)547–556.
[29]G.M.Xia,F.N.Xiang,A.F.Zhou,H.Wang,H.M.Chen,Asymmetric somatic hybridization between wheat(Triticum aestivum L.)and Agropyron elongatum(Host)Nevishi,Theor.Appl.Genet.107(2003)299–305.
[30]Z.Y.Peng,M.C.Wang,F.Li,H.J.Lv,C.L.Li,G.M.Xia,A proteomic study of the response to salinity and drought stress in an introgression strain of bread wheat,Mol.Cell.Proteomics 8(2009)2676–2686.
[31]S.T.Liu,S.W.Liu,M.Wang,T.D.Wei,C.Meng,M.Wang,G.M.Xia,A wheat SIMILAR TO RCD-ONE gene enhances seedling growth and abiotic stress resistance by modulating redox homeostasis and maintaining genomic integrity,Plant Cell 26(2014)164–180.
[32]R.Gupte,Z.Y.Liu,W.L.Kraus,PARPs and ADP-ribosylation:recent advances linking molecular functions to biological outcomes,Genes Dev.31(2017)101–126.
[33]W.L.Kraus,PARPs and ADP-ribosylation:50 years…and counting,Mol.Cell 58(2015)902–910.
[34]S.Kangasjärvi,J.Kangasjärvi,Towards understanding extracellular ROS sensory and signaling systems in plants,Adv.Bot.2014(2014)538946.
[35]J.You,W.Zong,X.K.Li,J.Ning,H.H.Hu,X.H.Li,J.H.Xiao,L.Z.Xiong,The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice,J.Exp.Bot.64(2012)569–583.
[36]M.C.Wang,C.Liu,T.Xing,Y.X.Wang,G.M.Xia,Asymmetric somatic hybridization induces point mutations and indels in wheat,BMC Genomics 16(2015)807.
[37]C.L.Li,J.Lv,X.Zhao,X.H.Ai,X.L.Zhu,M.C.Wang,S.Y.Zhao,G.M.Xia,TaCHP:a wheat zinc finger protein gene downregulated by abscisic acid and salinity stress plays a positive role in stress tolerance,Plant Physiol.154(2010)211–221.
[38]W.Dong,M.C.Wang,F.Xu,T.Y.Quan,K.Q.Peng,L.T.Xiao,G.M.Xia,Wheat oxophytodienoate reductase gene TaOPR1 confers salinity tolerance via enhancement of abscisic acid signaling and reactive oxygen species scavenging,Plant Physiol.161(2013)1217–1228.
[39]S.W.Liu,F.Li,L.N.Kong,Y.Sun,L.M.Qin,S.Y.Chen,H.F.Cui,Y.H.Huang,G.M.Xia,Genetic and epigenetic changes in somatic hybrid introgression lines between wheat and tall wheat grass,Genetics 199(2015)1035–1045.
[40]Q.H.Wu,X.H.Ni,ROS-mediated DNA methylation pattern alterations in carcinogenesis,Curr.Drug Targets 16(2015)13–19.
[41]D.Golldack,C.Li,H.Mohan,N.Probst,Tolerance to drought and salt stress in plants:unraveling the signaling networks,Front.Plant Sci.5(2014)151.
[42]Y.Zhao,W.Dong,N.B.Zhang,X.H.Ai,M.C.Wang,Z.G.Huang,L.T.Xiao,G.M.Xia,A wheat allene oxide cyclase gene enhances salinity tolerance via jasmonate signaling,Plant Physiol.164(2014)1068–1076.
[43]Y.Zhao,X.H.Ai,M.C.Wang,L.T.Xiao,G.M.Xia,A putative pyruvate transporter TaBASS2 positively regulates salinity tolerance in wheat via modulation of ABI4 expression,BMC Plant Biol.16(2016)109.
[44]Y.Sun,W.Xu,Y.B.Jia,M.C.Wang,G.M.Xia,The wheat TaGBF1 gene is involved in the blue-light response and salt tolerance,Plant J.84(2015)1219–1230.
[45]C.Liu,S.Li,M.C.Wang,G.M.Xia,A transcriptomic analysis reveals the nature of salinity tolerance of a wheat introgression line,Plant Mol.Biol.78(2012)159–169.
[46]Y.M.Zhang,Z.S.Liu,A.A.Khan,Q.Lin,Y.Han,P.Mu,Y.G.Liu,H.S.Zhang,L.Y.Li,X.H.Meng,Z.F.Ni,M.M.Xin,Expression partitioning of homeologs and tandem duplications contribute to salt tolerance in wheat(Triticum aestivum L.),Sci Rep 6(2016)21476.
[47]L.J.Gardiner,M.Quinton-Tulloch,L.Olohan,J.Price,N.Hall,A.Hall,A genome-wide survey of DNA methylation in hexaploid wheat,Genome Biol.16(2015)273.
[48]H.Eren,M.Pekmezci,S.Okay,M.Turktas,B.Inal,E.Ilhan,M.Atak,M.Erayman,T.Unver,Hexaploid wheat(Triticum aestivum)root miRNome analysis in response to salt stress,Ann.Appl.Biol.167(2015)208–216.
[49]B.Wang,Y.F.Sun,N.Song,J.P.Wei,X.J.Wang,H.Feng,Z.Y.Yin,Z.S.Kang,Micro RNAs involving in cold,wounding and salt stresses in Triticum aestivum L.Plant Physiol.Biochem.80(2014)90–96.
[50]D.Q.Huang,J.A.Feurtado,M.A.Smith,L.K.Flatman,C.Koh,A.J.Cutler,Long noncoding mi RNA gene represses wheat βdiketone waxes,Proc.Natl.Acad.Sci.U.S.A.114(2017)E3149–E3158.
[51]C.Uauy,Wheat genomics comes of age,Curr.Opin.Plant Biol.36(2017)142–148.
[52]Z.X.Gao,X.L.He,B.C.Zhao,C.J.Zhou,Y.Z.Liang,R.C.Ge,Y.S.Shen,Z.J.Huang,Overexpressing a putative aquaporin gene from wheat,TaNIP,enhances salt tolerance in transgenic Arabidopsis,Plant Cell Physiol.51(2010)767–775.
[53]X.Huang,Y.Zhang,B.Jiao,G.P.Chen,S.H.Huang,F.Guo,Y.Z.Shen,Z.J.Huang,B.C.Zhao,Overexpression of the wheat salt tolerance-related gene TaSC enhances salt tolerance in Arabidopsis,J.Exp.Bot.63(2012)5463–5473.
- The Crop Journal的其它文章
- A journey to understand wheat Fusarium head blight resistance in the Chinese wheat landrace Wangshuibai
- Genetic improvement of heat tolerance in wheat:Recent progress in understanding the underlying molecular mechanisms
- Wheat genome editing expedited by efficient transformation techniques: Progress and perspectives
- Corrigendum to “A new method for evaluating the drought tolerance of upland rice cultivars”[Crop J. 5 (2017) 488–489]
- Mapping stripe rust resistance genes by BSR-Seq:YrMM58 and YrHY1 on chromosome 2AS in Chinese wheat lines Mengmai 58 and Huaiyang 1 are Yr17
- Wheat breeding in the hometown of Chinese Spring

