邻苯二甲酸酯降解细菌的多样性、降解机理及环境应用
韩永和,何睿文,李超,向萍,罗军,崔昕毅
南京大学环境学院 污染控制与资源化研究国家重点实验室,南京210046
邻苯二甲酸酯降解细菌的多样性、降解机理及环境应用
韩永和,何睿文,李超,向萍,罗军,崔昕毅*
南京大学环境学院 污染控制与资源化研究国家重点实验室,南京210046
邻苯二甲酸酯(phthalic acid esters, PAEs)是一类对人体内分泌系统有干扰作用的持续性有机污染物(persistent organic pollutants, POPs)。PAEs在环境介质如水体、底泥和土壤中长期赋存会对生物体产生毒害效应,其分布广、浓度高和难降解等特点是限制有效环境治理的主要因素。作为环境的重要组成部分,微生物对污染物有很强的适应能力和高效的降解能力,这为PAEs的生物修复提供了可能。与物理化学修复法相比,微生物修复技术具有可控性强、修复面广和灵活性高等优势。本文综述了已报道的大部分PAEs降解细菌的种类及其代谢机制,并分析了其在PAEs污染水体和土壤修复中的应用现状与前景,以期为PAEs环境行为与生物修复研究提供参考。
内分泌干扰物;持久性有机污染物;邻苯二甲酸酯;微生物;生物降解;健康风险
Received 7 November 2015 accepted 22 December 2015
邻苯二甲酸酯(phthalic acid esters, PAEs)是一类以人工合成为主要来源的环境内分泌干扰物,因其在塑料制品、医疗用品、家电和玩具等材料中的广泛使用,可在土壤、水体和大气中积累并直接或间接地影响人类健康[1-2]。作为最常见的单体塑化剂,环境中PAEs主要包括邻苯二甲酸二(2-乙基)己酯(di-(2-ethylhexyl) phthalate, DEHP)、邻苯二甲酸一丁酯(monobutyl phthalate, MBP)、邻苯二甲酸二丁酯(dibutyl phthalate, DBP)、邻苯二甲酸丁苄酯(butyl benzyle phthalate, BBP)和邻苯二甲酸二乙酯(diethyl phthlate, DEP)等。国际癌症研究机构(IARC)统计表明,塑料成品中PAEs含量通常为10%~60%[3]。其中,小分子量PAEs(DBP、邻苯二甲酸二甲酯(dimethyl phthlate, DMP)和DEP)主要添加至化妆品和个人护理产品中,起到香水保香和抑挥发及指甲油的抑脆裂作用,而大分子PAEs如BBP和DEHP是工业塑化制品如聚氯乙烯(polyvinylchloride, PVC)的主要原材料[1, 3]。在我国,90%以上PAEs被用于PVC的生产[2]。PAEs在合成材料中往往以非化学共价键的形式存在,可通过风化作用等向环境释放[1]。目前PAEs在市政固体废弃物,室内降尘,底泥和废水及垃圾渗滤液中被频繁检出;同时,PAEs在土壤微生物、植物和动物体内的富集及生物链传递引发的人体健康风险也备受关注[1]。
我国塑料制品使用泛滥,由此引发的PAEs污染问题日趋严重[2, 4]。在农业活动中,PAEs污染问题尤为突出。He等[2]认为,农业中塑料薄膜的使用、市政固体废物的排放、农药的使用和废水灌溉是我国农田PAEs污染的主要来源。对山东平原某蔬菜大棚PAEs污染的调查表明,37个采样点共检出16种PAEs,浓度均值为1.939~35.442 mg·kg-1,其中DBP、DMP、DEP和DEHP较高[4]。PAEs对农田生态系统的危害不仅体现在对土壤微生物和酶活性的抑制作用,还体现在对农作物产量和质量方面的影响[5]。
环境中PAEs的治理技术通常包含3类:物理-化学治理、生物治理和氧化降解[1-2]。其中,物理-化学治理的吸附法和絮凝-聚沉法、生物治理的微生物降解和植物修复以及光/氧化降解等工艺效果显著[1-2]。然而,物理-化学或光/氧化工艺无法在PAEs污染土壤修复中大规模应用,还有可能导致二次污染。环境中PAEs的降解与分布在很大程度上取决于微生物的群落与功能差异,理解微生物介导的PAEs代谢机制可为PAEs环境归趋研究与PAEs修复提供理论基础[5]。本文以环境中最常见的PAEs(包括DBP、MBP、BBP、DEP和DEHP)为研究对象,综述了目前国内外报道的PAEs降解菌及其多样性、PAEs降解机理及PAEs降解菌的环境应用,拟从微生物学、分子生物学、生态学和环境化学等角度阐述PAEs降解菌在环境中的特性及其意义。
1 PAEs降解菌多样性
微生物是环境的重要组成部分,其在地球上的出现可追溯至早太古代(~3.2 Ga)的河口沉积物中。极强的生命力、多样性和适应能力造就了环境中丰富多彩的微生物世界。PAEs作为一种人工合成塑化剂其在环境中出现的年代并不久远,但微生物很快对PAEs产生了适应能力,主要体现在对多种PAEs的有效降解[5]。环境中PAEs难以通过水解或光降解去除,研究认为微生物才是PAEs降解的主要参与者[1]。根据微生物分类学,PAEs降解菌以好氧细菌和部分真菌为主,具备厌氧降解功能的细菌较少[6]。另一方面,目前对PAEs降解菌群落结构与功能的认识还非常有限,关注PAEs降解的微生物群落效应是今后研究的重点内容。
2008年以前发现的PAEs降解细菌有节细菌Arthrobacter keyseri 12B(旧名为Micrococcus sp.),假单胞菌Pseudomonas fluorescens、P. putida、P. acidovorans、P. testosterone和P. pseodoalcaligenes,以及红球菌Rhodococcus rubropertinctus等[6]。虽然PAEs降解菌报道广泛,对其进行详细的遗传学分类较少。Liang等[6]统计表明,PAEs降解细菌主要分布在α、β和γ变形杆菌门(Proteobacteria),厚壁菌门(Firmicutes,低G+C含量),放线菌门(Actinobacteria,高G+C含量),拟杆菌门(Bacteroidetes)和绿菌门(Chlorobi)等5个门类的29个属。奇异球菌-栖热菌门(Deinococcus-Thermus)的2个菌株在2010年[7]和2014年[8]首次得到报道。目前PAEs降解细菌已扩充到了36个属(表1和图1A),有超过80个PAEs降解菌株得到了详细的研究与报道。
进化分析显示,具备PAEs降解能力的细菌集中在γ-Proteobacteria和Acinobacteria,其次是α和β-Proteobacteria,Firmicutes最少(图1A)。其中,Sphingomonas、Comamonas、Pseudomonas、Arthobacter和Rodococcus是PAEs降解菌的主要属[5]。这些微生物多数能以PAEs为唯一碳源和能源物质进行生长繁殖,且有近一半菌株具备降解多种PAEs的能力(表1)。例如,Wu等[9]研究表明,农杆菌Agrobacterium sp. JDC-49可同时降解DBP、DEP、DMP、邻苯二甲酸二正辛酯(di-n-octyl phthalate, DOP)、邻苯二甲酸二异壬酯(di-isononyl phthalate, DINP)和邻苯二甲酸(phthalic acid, PA)等7种PAEs。类似的多功能菌株还包括Diaphorobacter sp. QH-6[10],Rhodococcus sp. JDC-11[11]和鞘氨醇单胞菌Sphingobium sp. SM42[12]等。
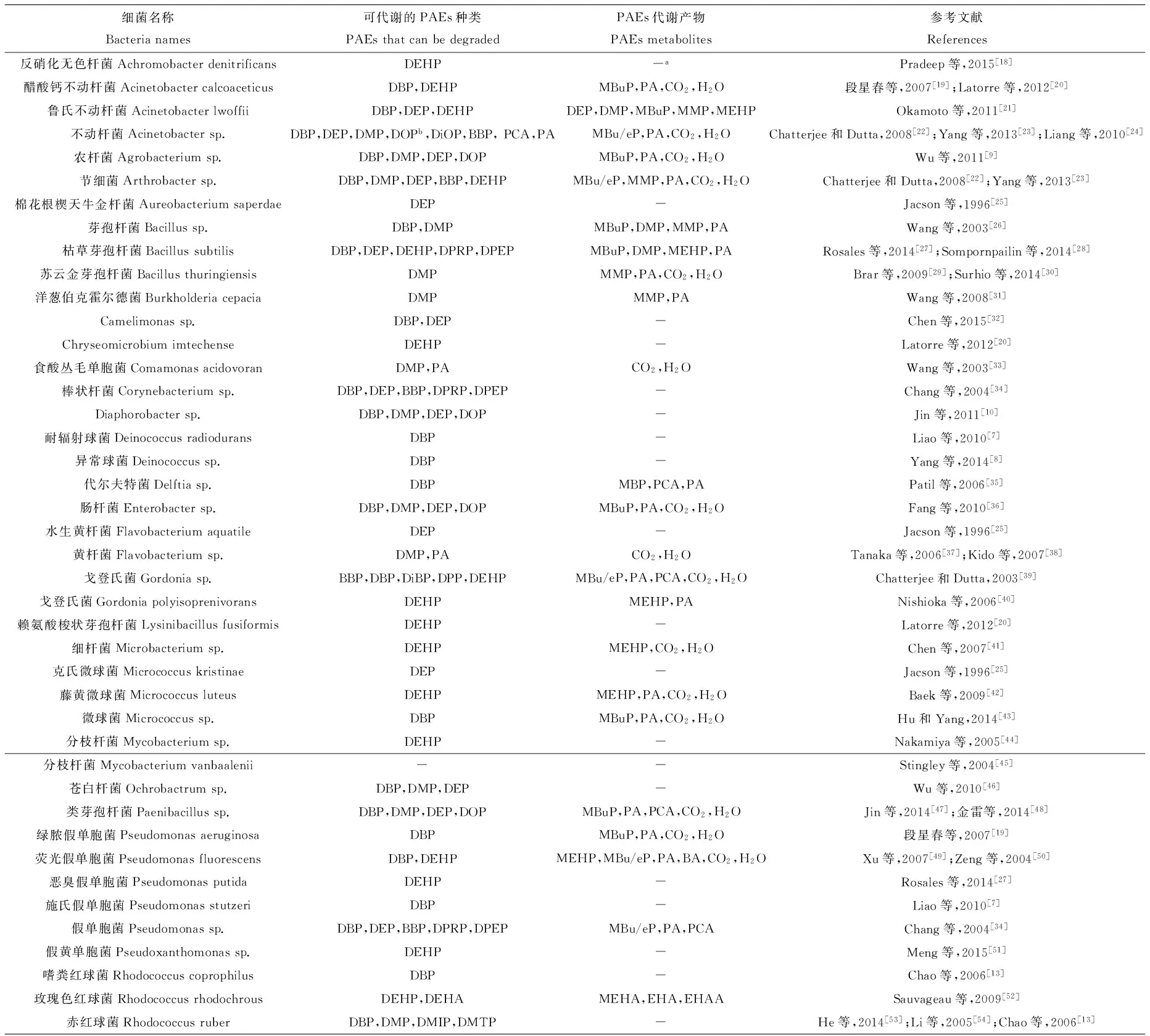
表1 部分已报道的PAEs降解细菌及其相关信息
注:DEHP=邻苯二甲酸二(2-乙基己)酯,DBP=邻苯二甲酸二丁酯,MBuP=邻苯二甲酸单丁酯,PA=邻苯二甲酸,DEP=邻苯二甲酸二乙酯,DMP=邻苯二甲酸二甲酯,MMP=单甲基邻苯二甲酸,MEHP=邻苯二甲酸单-2-乙基己酯,BBP=邻苯二甲酸丁苄酯,DOP=邻苯二甲酸二辛酯,DiOP=邻苯二甲酸二异辛酯,PCA=原儿茶酸,MBeP=邻苯二甲酸单苄酯,DPRP=邻苯二甲酸丙酯,DPEP=脱氧叶红初卟啉,DiBP=邻苯二甲酸二异丁酯,DPP=邻苯二甲酸二丙酯,MEHP=邻苯二甲酸单(2-乙基己基)酯,BA=苯甲酸,DEHA=己二酸二(2-乙基己)酯,MEHA=单乙基己基酯,EHA=乙基己醇,EHAA=乙基己酸,DMIP=间苯二甲酸二甲酯,DMTP=对苯二甲酸二甲酯,TA=对苯二甲酸。a未提及,bDOP:DEHP的别称。
Note:DEHP = di-2-ethylhexyl phthalate, DBP = di-butyl phthalate, MBuP = mono-n-butyl phthalate, PA = phthalic acid, DEP = diethyl phthalate, DMP = dimethyl phthalate, MMP = monomethyl phthalate, DPP = dipropyl phthalate, MEHP = mono-2-ethylhexyl phthalate, BBP = butyl benzyl phthalate, DOP = dioctyl phthalate, DiOP = di-iso-octyl phthalate, PCA = protocatechuic acid, MBeP = mono-benzyl phthalate, DPRP = dipropyl phthalate, DPEP = deoxophylloerythroetioporphyrins, DiBP = di-iso-butyl phthalate, MEHP = mono-2-ethylhexyl phthalate, BA = benzoic acid, DEHA = di-2-ethylhexyl adipate, MEHA = mono-ethylhexyl adipate, EHA= 2-ethylhexanal, EHAA = 2-ethylhexanoic acid, DMIP = dimethyl isophthalate, DMTP = dimethyl terephthalate, TA = terephthalic acid.anot mentioned,bDOP: also named DEHP.
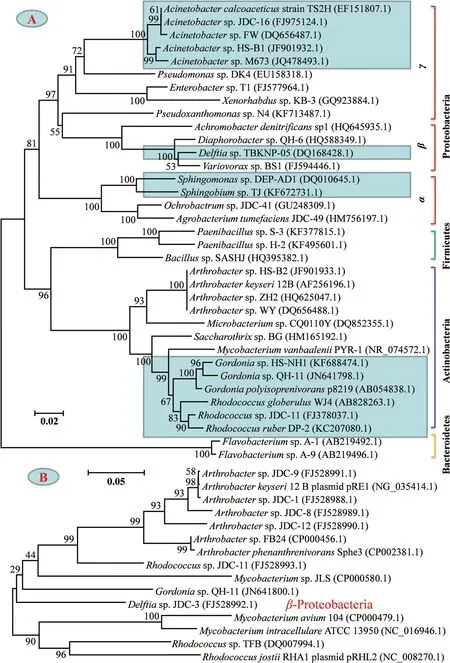
图1 基于16S rRNA(A)和邻苯二甲酸酯双加氧酶基因(B)序列同源性构建的细菌系统发育树 注:图示文献报道且在NCBI数据库同时提交了序列的邻苯二甲酸酯降解菌的基因信息。图1A中蓝色框示该属细菌的邻苯二甲酸酯双加氧酶基因已被提交到NCBI数据库。Fig. 1 Rooted phylogenetic trees based on 16S rRNA (A) and phthalate dioxygenase gene (B) sequences Note: Figures show the gene information of PAEs-degrading bacteria those have been reported in the literature and whose sequences have been submitted to NCBI database. Blue boxes in Fig. 1A indicate that the phthalate dioxygenase genes in these bacteria have been submitted to NCBI database.
然而,目前对PAEs降解细菌的研究还比较基础。在43株已报道且在NCBI数据库提交了16S rRNA信息的细菌中只有35条序列的片段长度1200 bp,据此构建的系统进化树如图1A所示。对应PAEs降解菌的16S rRNA多样性,只有Acinetobacter、Delftia、Sphingomonas、Rhdococcus和Gordonia等5个属的10多个菌株其邻苯二甲酸酯双加氧酶基因(phthalate dioxygenase gene, PDOG)得到了克隆分析(图1B)。除Delftia外,其他细菌均属于Acinobacteria(图1B),而R. coprophilus G9和Mycobacterium vanbaalenii PYR-1的PDOG片段长度只有420 bp和267 bp[13]。这说明,对于PAEs降解菌的分子生物学研究,未来还有很多工作值得开展。
2 PAEs对细菌群落的影响
环境中微生物资源丰富,但可培养微生物所占比例低于1%。如前所述,PAEs在环境中的广泛分布会对微生物群落结构和酶活性产生影响,相应地,PAEs降解菌会对PAEs的环境归趋产生直接或间接的作用[5]。
聚合酶链反应-变性梯度凝胶电泳(polymease chain reaction-denaturing gradient gel electrophoresis, PCR-DGGE)分析显示,往土壤中添加DEP浓度高达10 mg·kg-1时,土壤微生物的种类降低到了个位数,主要包括Sphingomonas spp.、Pseudomonas spp.和Actinomycetes spp.[5]。Chen等[14]研究表明,DMP、DEP和DOP都会显著降低土壤脲酶(urease)活性。有趣的是,即使DMP或DEP添加浓度高达500 mg·kg-1,脲酶活性也只降低了32%和31%。显然,微生物对PAEs的抗性和降解缓和了污染物毒性[14]。类似地,Cartwright等[15]发现,DEP低至1 mg·kg-1即可在1 d内显著降低土壤可培养细菌总数(47%,BIOLOG法),这可能与PAEs诱发的细菌细胞膜流动性增加有关。不同的是,DEHP并未对土壤微生物造成任何影响。事实上,土壤微生物数量变化与DEP的有效降解并无显著相关性(降解半衰期为0.75 d);相反,DEHP在70 d后只减少了10%,说明微生物的数量并不能反映土壤微生物对PAEs的降解活性[15]。因此,PAEs诱导效应可能主要体现在对群落结构的调整以增加PAEs代谢菌群丰度,这与Kapane等[5]的结论相符。对土壤多种酶活性的检测结果表明,添加DBP虽然降低了脲酶、纤维素酶(cellulose)和β-葡萄糖苷酶(β-glucosidase)活性,却增强了脱氢酶(dehydrogenase)、催化酶(catalase)、蛋白酶(protease)和磷酸酶(phosphatase)活性[16]。这种现象与微生物总数降低却依然能够有效降解PAEs是一致的,即具备PAEs降解功能的细菌在污染物诱导下成为了优势菌群。例如,Wang等[17]发现,随着DMP浓度从5 mg·kg-1升至20 mg·kg-1,微生物群落分类单元(operational taxonomic units, OTUs)随之降低,但DMP降解效率却随之升高,说明优势菌的存在是DMP降解的主要贡献者。
虽然环境中PAEs的微生物行为已有诸多报道,现有研究并未很好地跟踪微生物群落变化的基本规律。这一方面与环境体系的复杂性有关,同时也与PAEs种类的多样性及微生物降解PAEs的特异性及互作效用密切相关(见下文)。由于对PAEs降解菌种类及功能认识的不足(表1和图1),PAEs降解基因的多样性及其环境意义仍是今后研究的重点内容。
3 微生物降解PAEs的特异性和降解效率
研究指出,某些降解菌可同时代谢多种PAEs,但已报道的80多个菌株中超过50%只能代谢1种底物(表1)。此外,具备多种PAEs代谢功能的微生物也存在底物特异性,这种现象早于2003年已在Arthrobacter sp.和S. paucimobilis中被观察到。研究发现,Arthrobacter sp.可迅速将DMP降解为邻苯二甲酸单甲酯(monomethyl phthalate, MMP)和PA,此后PA被继续代谢为CO2和H2O;然而,该菌不能继续代谢MMP,却由S. paucimobilis完成这个过程[59]。最近研究也发现,Camelimonas sp. M11对DBP、DEP、邻苯二甲酸二丙酯(dipropyl phthalate, DPP)和邻苯二甲酸二戊酯(dipentyl phthalate, DNPP)有很强的降解能力,但不能降解DMP[32]。Vega等[59]认为,DBP被Arthrobacter sp.和S. paucimobilis代谢存在DBP→PA和DBP→MMP两种机制,并提出环境微生物协同完成复合污染物代谢的假设(见下文)。
鉴于微生物对PAEs的碳源利用特性,理论上它们具备较强的PAEs耐受能力和降解能力。一般而言,PAEs侧链越短,微生物降解效率也越高。然而,PAEs降解酶活性不受侧链长度影响,而与侧链产生的空间位阻有关[6]。Liang等[24]发现,不动杆菌Acinetobacter sp. JDC-16在20 h内(pH=8、35oC)可将500 mg·L-1DEP完全降解。动力学拟合结果表明,该菌在500 mg·L-1DEP暴露下只需6.9 h即可达到稳定期,DEP降解常数Rm高达54 mg·L-1·h-1[24]。多数PAEs降解菌对DBP的降解能力也很强。例如,Diaphorobacter sp. QH-6对500 mg·L-1底物的降解半衰期只需5.2 h[10],而R. ruber DP-2和Enterobacter sp. T5对高达1 200和1 500 mg·L-1DBP的降解半衰期也只需近30 h[36, 53]。相较而言,细菌对结构较复杂、分子量较大的BBP降解能力较差。例如,在最优pH条件下,P. fluorescence B-1对10 mg·L-1BBP代谢速率低至0.03 mg·L-1·h-1[49];Acinetobacter sp. FW略高,为2.1 mg·L-1·h-1[22]。类似地,虽然P. fluorescence FS1在60 d内可将200 mg·L-1DEHP中的75%有效降解,但降解半衰期高达17 d[63]。有趣的是,B. subtilis No. 66对DEHP有较强的降解能力,但几乎不能降解DBP或只能降解DEP、脱氧叶红初卟啉(deoxophylloerythroetioporphyrins, DPEP)和邻苯二甲酸丙酯(dipropyl phthalate, DPRP)的一小部分[64]。
值得注意的是,在污染物结构特定的情况下,微生物对PAEs的降解能力往往取决于微生物本身[23]。例如,Yang等[23]发现,Acinetobacter sp. HS-B1和Arthrobacter sp. HS-B2对起始浓度为500 mg·L-1BBP的24 h降解率分别达28%和59%。当培养基中添加1%LB时,HS-B1和HS-B2对BBP的去除率提高到了40%和75%,说明微生物对PAEs的代谢不仅是碳源利用,还可能存在纯粹的酶学解毒过程[23]。此外,Gordonia sp. MTCC 4818和P8219对PAEs的高效降解也备受关注,其对80 μmol BBP的90 h降解率和1 300 mg·L-1DEHP的45 h降解率可达100%[39-40]。
4 微生物介导的PAEs降解机理
如表1所示,多数微生物可以PAEs为唯一碳源和能源物质进行代谢生长。然而,微生物无法直接利用长侧链或带苯环等复杂结构的有机物大分子,将长碳侧链缩短、将双侧链降解成单侧链、将苯环开环并进一步代谢成CO2和H2O是微生物代谢PAEs的主要步骤[6]。虽然目前报道的PAEs降解酶基因信息很少(图1B),通过代谢产物分析技术如气相色谱-质谱联用(Gas Chromatograph-Mass Spectrometer, GC-MS)和液相色谱-质谱联用(Liquid Chromatography-Mass Spectrometer, LC-MS)等可推测几种比较经典的微生物代谢路径。
4.1 PAEs侧链的降解
侧链代谢是PAEs微生物降解的首要步骤,包括β-氧化作用(β-oxidation)、转酯化(trans-esterificaiton)或去烷基化作用(de-alkylation)和脱脂化作用(de-esterificaiton)[6]。对于侧链双酯基碳数大于2的PAEs,β-氧化介导的长链降解是一种很重要的代谢机制,该过程涉及双链乙烷基的同时脱落。Amir等[65]研究表明,在堆肥中添加DBP后,底泥中检测到了DEP和DMP,说明DBP→DEP的代谢过程属于β-氧化(图2)。然而,目前关于PAEs的β-氧化作用并未在纯菌体系中得到研究。对于PAEs单侧链的降解,则以转酯化或脱脂化作用为主。如图2所示,DEP→EMP→DMP属于单侧链的烷基脱落,这种侧链酯基发生变化的过程又称为转酯基作用。该路径最早由Cartwright等[66]于2000年提出,与β-氧化作用类似,其后续研究并未得到充分的开展。目前报道的唯一一种具备转酯化作用的微生物是A. lwoffii R-3[21]。脱脂化作用是研究最透彻的PAEs降解路径,其与侧链烷基变化无关,而是将PAEs一侧或双侧酯基水解,产物为带苯环的酸类物质。例如,某些微生物可将DEHP一侧酯基脱落形成MEHP,MEHP酯基可被进一步水解成PA(图2)。这类微生物有G. polyisoprenivorans p8219[40]、Microbacterium sp. CQ0110Y[41]、P. fluorescens FS1[63]和M. luteus等[42](表1)。类似地,DBP→MBuP,BBP→MBzP,MBzP、DEP、DMP和MMP→PA都属于微生物介导的脱脂化降解,相关微生物包括Gordonia sp. MTCC 4818[39]、Atrhrobacter sp. WY[22]、Paenibacillus sp. S-3/H-2[47-48]和P. fluorescens B-1[49]等(表1)。研究发现,脱脂化作用在厌氧和好氧条件下都可发生,该过程由酯酶介导完成[40, 59-60]。
如图2所示,PA是PAEs的代谢“中转站”,PA的进一步代谢与环境中含氧量有关。在好氧条件和3,4或4,5-邻苯二甲酸酯双加氧酶作用下,PA可被降解成3,4或4,5-双羟基邻苯二甲酸酯,进而形成PCA[6]。在厌氧条件下,PA将被转化成苯甲酸(benzoic acid, BAc),其中一部分BAc可转化成PCA并进入下一步代谢[6](图2)。由此可见,除PA外,PCA是PAEs降解的另一重要“中转站”。
需要指出的是,BBP的微生物降解产物还包括苯甲醇(benzyl alcohol, BAl)和1-丁醇(1-butanol)。与PA厌氧代谢类似,BAl可在相同条件下被代谢成BAc,从而进入下一个代谢路径(图2)。在Acinetobacter sp. FW和Gordonia sp. MTCC 4818等菌株中,1-丁醇的代谢主要由β-氧化作用完成[22, 39]。β-氧化作用的产物CO2和H2O将进入三羧酸循环(TCA cycle),为微生物生长提供碳源物质(图2)。
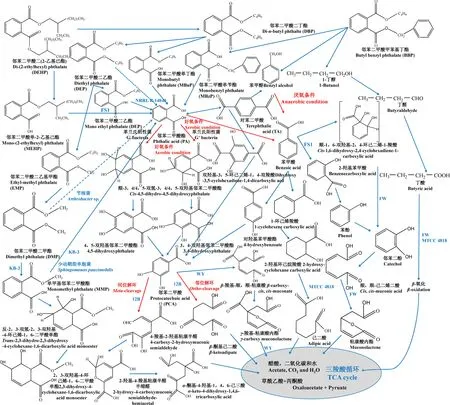
图2 微生物降解邻苯二甲酸酯的可能路径 注:实线和虚线分别表示PAEs的主要和次要代谢路径。Fig. 2 Possible pathways involved in microbes-mediated PAEs degradation Note: Solid and dotted lines indicate the major and minor pathways of PAEs degradation respectively.
4.2 PAEs苯环的降解
苯环降解是实现PAEs碳源利用的重要步骤,该过程以PA、PCA和BAc为主要代谢中间产物。在好氧条件下,革兰氏阳性菌可将PA转变成顺-3,4-双氢-3,4-双羟基邻苯二甲酸酯(cis-3,4-dihydro-3,4-dihydroxyphthalate),而革兰氏阴性细菌则在苯环的4,5位发生双氧化,产生顺-4,5-双氢-4,5-双羟基邻苯二甲酸酯[6](图2)。例如,R. ruber DP-2同时含有酚水解酶基因(phenol hydroxylase gene, pheu)和3,4-邻苯二甲酸酯双加氧酶基因(pht),其对600~1 200 mg·L-1DBP的降解时间只需15.8~27.8 h[53]。DBP经进一步还原和脱羧基作用,可被转化成另一种重要的代谢中间产物PCA。若PCA发生邻位(ortho-)或间位(meta-)解环,可分别产生β-酮基己二酸(β-ketoadipate)和4-羧基-2-双羟基粘康半醛(4-carboxy-2-hydroxymuconic semialdehyde)。经一系列代谢后,产生的草酰乙酸(oxaloacetate)和丙酮酸(pyruvate)将进入TAC循环[6](图2)。Eaton和Ribbons[67]认为,Micrococcus sp. 12B对DBP的降解属于这一代谢路径。以PCA为代谢中间产物,PAEs的完全降解还可通过β-羧基-顺,顺-粘康酸(β-carboxy-cis, cis-muconate)→γ-羧基-粘康酸内酯(γ-carboxy muconolactone)→TCA路径实现,这种代谢机制已在Arthrobacter sp. WY中得到了详细研究[22]。
除PCA外,BAc是PAEs代谢的另一“二级中转站”。如前所述,有部分PA在好氧条件下可通过BAc路径实现完全降解,而BAl→BAc转化发生在厌氧条件下[6](图2)。除BAc→PCA代谢路径外,目前还发现了另外3种典型的降解机制。第1种是BAc→1-环己烯羧酸(1-cyclohexene carboxylic acid)→2-羟基环己烷羧酸(2-hydroxy-cyclohexane carboxylic acid)→己二酸(adipic acid)路径。例如,Chatterjee和Dutta[39]报道显示,Gordonia sp. MTCC 4818不能将PA转化成PCA,但可实现BAc的己二酸代谢。第2种是BAc→2-羟基苯甲酸(benzenecarboxylic acid)→苯酚(phenol)→邻苯二酚(catechol)→顺,顺-己二烯二酸(cis, cis-muconic acid)→粘康酸内酯(muconolactone)路径。P. fluorescence FS1是该降解路径的代表菌株[50, 63]。在某些情况下,微生物还可将BAc转化成不稳定的中间产物如顺-1,6-双羟基-2,4-环己二烯-1-羧酸(cis-1,6-dihydroxy-2,4-cyclohenadiene-1-carboxylic acid),此后进入类似于路径二的代谢过程(图2)。Chatterjee和Dutta[22]验证了Acinetobacter sp. FW中该代谢路径的存在。
4.3 PAEs的协同代谢机制
微生物介导的PAEs降解是一个复杂系统,不同微生物或同一微生物在不同条件下,其代谢路径都可能存在较大差异(图2)。某些微生物代谢PAEs的能力很强。例如,P. fluorescence FS1可将DEHP进行逐级降解,产生终产物CO2和H2O[50, 63]。具备完全降解功能的微生物还包括Flavobacterium sp. A-1/A-9[37-38]、B. thuringiensis HD-1[30]和Variovorax sp. BS1[62]等(表1)。通常而言,具备产生CO2和H2O的菌株属于PAEs完全代谢菌。然而,环境中某些微生物不具备完全代谢PAEs的能力,该过程往往需要多种微生物的协同作用。例如,Vega和Bastide[59]研究发现,Arthrobacter sp.可将DMP代谢成MMP和PA,但该菌不能进一步代谢MMP。当该菌与筛自同一土壤的微生物S. paucimobilis (MMP降解菌)混合培养后,DMP和MMP都未在培养基中被检测到,说明二者共存时可协同代谢PAEs。由此可见,实际环境中PAEs的降解往往是多种微生物共同作用的结果[22]。
5 PAEs降解菌的环境应用
5.1 微生物修复PAEs污染水体/底泥
固定化技术是实现微生物负载及污染物环境治理的常规手段。为提高PAEs降解菌的活性持留,微生物固定化技术在PAEs废水中的应用也逐渐得到了推广[1]。在固定化微生物治理PAEs的实践中,清华大学王建龙教授课题组做出了突出贡献。1995年,作者率先尝试以聚乙烯醇(polyvinyl alcohol, PVA)固定化基质包埋微生物对DBP的降解研究[69]。结果表明,菌株A固定化小球可在40 h内将100 mg·L-1DBP完全降解,且固定化细胞的降解活性比游离细胞更高。目前已报道的固定化菌还包括B. subtilis[28],Bacillus sp.[26]、Micrococcus sp.[43]和Variovorax sp. BS1[62]等。为实现PAEs污染水体的有效治理,混合包埋不同功能的PAEs降解菌是今后研究的主要方向。
我国部分地区的底泥PAEs浓度高达1 250 mg·kg-1[2]。考虑到微生物固定化技术在底泥修复中应用的局限性,越来越多研究将重点转向土著微生物的筛选与回用。如表1所示,目前已得到深入研究的PAEs降解菌超过34种来源于底泥或沉积物。利用活性污泥中微生物群落的群感效应,可实现废水中PAEs的有效治理[1, 6]。Chang等[70]发现,好氧条件下,微生物对底泥中50 mg·L-1DEP、DBP、BBP和DEHP的降解半衰期只需2.7、1.8、2.1和3.8 d。底泥中PAEs在厌氧条件下的降解速率也很高。例如,10 mg·L-1DMP和DBP的降解半衰期低至1.0和1.4 d,但DOP的降解半衰期比较长,为19.4 d[71]。以上结论说明PAEs的微生物降解与其结构密切相关[1, 6]。因此,在长链PAEs浓度较高的废水中,有必要采取微生物-物理/化学(如高级氧化技术)相结合的手段实现其有效治理[1]。此外,研究发现,当往底泥中添加壬基酚(nonylphenol)或多环芳烃(polycyclic aromatic hydrocarbons, PAHs)时,Sphigomonas sp. DK4和Corynebacterium sp. O18对PAEs的降解能力会受到显著抑制[34]。鉴于PAEs污染废水可能还包含其他有毒有害重(类)金属和有机污染物,筛选具备多种污染物抗性的菌株及其生理生化研究是工程应用的基础。
5.2 微生物修复PAEs污染土壤
土壤微生物已被证明具备降解多种PAEs的能力(表1),利用这些微生物回土修复是一种理想的方法。Wang等[55]从DEHP污染土壤筛得一株高效降解菌Rhodococcus sp. WJ4,在液体培养基中该菌对200 mg·L-1DEHP的去除率高达96.4%。土壤修复实验表明,WJ4对1 g·kg-1DEHP污染土壤的21 d修复率达55%,显示出了较好的应用潜力。然而,类似的PAEs污染土壤的微生物修复实例未见报道。现有研究表明,生物泥浆和固相反应器(bioslurry and solid-phase bioractors, BSSB)修复污染土壤是比较可行的技术,目前已得到了一些推广与应用[2, 6]。例如,利用BSSB修复技术,Di Gennaro等[72]将5.51 mg·g-1DEHP降至0.63 mg·g-1,76 d内总去除率高达89%。这种方法的可行性取决于微生物培养条件的可控程度及营养物质的供给效率[72]。若土壤中添加堆肥的粒径很小,也有助于提高PAEs的降解效率[2]。
由此可见,将微生物进行回土修复或在泥浆相中进行强化修复都是可行的。Liang等[6]认为,生物泥浆法修复PAEs污染土壤的优势主要体现在1) 营养物质、末端电子受体和底物的均匀分布;2) 提高了微生物与污染物的有效接触。不足的是,生物泥浆法需要反复挖掘土壤,因此限制了其修复能力和实际应用[2]。
6 结论与展望
多数PAEs都具有内分泌干扰作用且广泛存在于各种环境介质中,实施PAEs的土壤和水体修复是保证人体健康的重要举措。微生物对多种PAEs都具备较强的降解能力。目前已有6个门类、30多个属的80多个PAEs降解菌株得到了详细研究,通过GC-MS和LC-MS等检测技术或代谢路径的模型拟合[54]提出了PAEs代谢的微生物学机制。然而,已报道的PAEs微生物降解酶基因数量有限,这限制了从生物化学和分子生物学水平认识PAEs代谢的机理,同时也不利于环境中PAEs降解菌的群落结构分析和功能研究。虽然PAEs污染水体(底泥)的微生物修复已取得一定成效,土壤修复仍是当前PAEs污染治理的主要内容。
基于PAEs微生物代谢的重要性及当前研究的不足,今后的工作可从以下几方面展开:1) 在现有基础上继续筛选PAEs降解菌并研究其降解能力;2) 深入阐释现有PAEs降解菌代谢PAEs的分子生物学机制,推动对新获菌株的全面研究;3) 利用高通量分析技术等实现对环境中PAEs降解菌多样性的充分认识;4) 继续探讨PAEs降解菌在土壤和水体污染治理中的应用。
[1] Abdel daiem M M, Rivera-Utrilla J, Ocampo-Pérez R, et al. Environmental impact of phthalic acid esters and their removal from water and sediments by different technologies-A review [J]. Journal of Environmental Management, 2012, 109: 164-178
[2] He L, Gielen G, Bolan N S, et al. Contamination and remediation of phthalic acid esters in agricultural soils in China: A review [J]. Agronomy for Sustainable Development, 2015, 35(2): 519-534
[3] IARC. Monographs on the Evaluation of Carcinogenic Risks to Humans. Industrial Chemicals [S]. Lyon, France: International Agency for Research on Cancer, 2000
[4] Liu X, Shi J, Bo T, et al. Occurrence of phthalic acid esters in source waters: A nationwide survey in China during the period of 2009-2012 [J]. Environmental Pollution, 2014, 184: 262-270
[5] Kapanen A, Stephen J R, Bruggemann J, et al. Diethyl phthalate in compost: ecotoxicological effects and response of the microbial community [J]. Chemosphere, 2007, 67(11): 2201-2209
[6] Liang D W, Zhang T, Fang H H, et al. Phthalates biodegradation in the environment [J]. Applied Microbiology and Biotechnology, 2008, 80(2): 183-198
[7] Liao C S, Chen L C, Chen B S, et al. Bioremediation of endocrine disruptor di-n-butyl phthalate ester by Deinococcus radiodurans and Pseudomonas stutzeri [J]. Chemosphere, 2010, 78(3): 342-346
[8] Yang C F, Wang C C, Chen C H. Di-n-butyl phthalate removal by strain Deinococcus sp. R5 in batch reactors [J]. International Biodeterioration & Biodegradation, 2014, 95: 55-60
[9] Wu X, Wang Y, Liang R, et al. Biodegradation of an endocrine-disrupting chemical di-n-butyl phthalate by newly isolated Agrobacterium sp. and the biochemical pathway [J]. Process Biochemistry, 2011, 46(5): 1090-1094
[10] Jin D, Wang P, Bai Z, et al. Biodegradation of di-n-butyl phthalate by a newly isolated Diaphorobacter sp. strain QH-6 [J]. African Journal of Microbiology Research, 2011, 5: 1322-1328
[11] Jin D C, Liang R X, Dai Q Y, et al. Biodegradation of di-n-butyl phthalate by Rhodococcus sp. JDC-11 and molecular detection of 3,4-phthalate dioxygenase gene [J]. Journal of Microbiology and Biotechnology, 2010, 20(10): 1440-1445
[12] Whangsuk W, Sungkeeree P, Nakasiri M, et al. Two endocrine disrupting dibutyl phthalate degrading esterases and their compensatory gene expression in Sphingobium sp. SM42 [J]. International Biodeterioration & Biodegradation, 2015, 99: 45-54
[13] Chao W, Lin C, Shiung I, et al. Degradation of di-butyl-phthalate by soil bacteria [J]. Chemosphere, 2006, 63(8): 1377-1383
[14] Chen H, Zhuang R, Yao J, et al. A comparative study on the impact of phthalate esters on soil microbial activity [J]. Bulletin of Environmental Contamination and Toxicology, 2013, 91(2): 217-223
[15] Cartwright C D, Thompson I P, Burns R G. Degradation and impact of phthalate plasticizers on soil microbial communities [J]. Environmental Toxicology and Chemistry, 2000, 19(5): 1253-1261
[16] Zhou Q, Wu Z, Cheng S, et al. Enzymatic activities in constructed wetlands and di-n-butyl phthalate (DBP) biodegradation [J]. Soil Biology and Biochemistry, 2005, 37(8): 1454-1459
[17] Wang Z G, Hu Y L, Xu W H, et al. Impacts of dimethyl phthalate on the bacterial community and functions in black soils [J]. Frontiers in Microbiology, 2015, 6: 405, doi: 10.3389/fmicb.2015.00405
[18] Pradeep S, Josh M S, Binod P, et al. Achromobacter denitrificans strain SP1 efficiently remediates di (2-ethylhexyl) phthalate [J]. Ecotoxicology and Environmental Safety, 2015, 112: 114-121
[19] 段星春, 易筱筠, 杨晓为, 等. 两株邻苯二甲酸二丁酯降解菌的分离鉴定及降解特性的研究[J]. 农业环境科学学报, 2007, 26(6): 1937-1941
Duan X C, Yi X Y, Yang X W, et al. Isolation and characterization of two di-n-butyl phthalate degrading bacteria [J]. Journal of Agro-Environment Science, 2007, 26(6): 1937-1941 (in Chinese)
[20] Latorre I, Hwang S, Montalvo-Rodriguez R. Isolation and molecular identification of landfill bacteria capable of growing on di-(2-ethylhexyl) phthalate and deteriorating PVC materials [J]. Journal of Environmental Science and Health, Part A: Toxic/Hazardous Substances and Environmental Engineering, 2012, 47(14): 2254-2262
[21] Okamoto Y, Toda C, Ueda K, et al. Transesterification in the microbial degradation of phthalate esters [J]. Journal of Health Science, 2011, 57(3): 293-299
[22] Chatterjee S, Dutta T K. Complete degradation of butyl benzyl phthalate by a defined bacterial consortium: Role of individual isolates in the assimilation pathway [J]. Chemosphere, 2008, 70(5): 933-941
[23] Yang X, Zhang C, He Z, et al. Isolation and characterization of two n-butyl benzyl phthalate degrading bacteria [J]. International Biodeterioration & Biodegradation, 2013, 76: 8-11
[24] Liang R X, Wu X L, Wang X N, et al. Aerobic biodegradation of diethyl phthalate by Acinetobacter sp. JDC-16 isolated from river sludge [J]. Journal of Central South University of Technology, 2010, 17: 959-966
[25] Jackson M, Labeda D, Becker L. Isolation for bacteria and fungi for the hydrolysis of phthalate and terephthalate esters [J]. Journal of Industrial Microbiology, 1996, 16(5): 301-304
[26] Wang J L, Ye Y C, Wu W Z. Comparison of di-n-methyl phthalate biodegradation by free and immobilized microbial cells [J]. Biomedical and Environmental Sciences, 2003, 16(2): 126-132
[27] Rosales E, Cobas M, Tavares T, et al. Removal of di-(2-ethylhexyl) phthalate (DEHP) from water using a LECA-Pseudomonas putida biobarrier [C]. The 12th International Chemical and Biological Engineering Conference (CHEMPOR 2014), Portugal, 2014: 142-144
[28] Sompornpailin D, Siripattanakul-Ratpukdi S, Vangnai A S. Diethyl phthalate degradation by the freeze-dried, entrapped Bacillus subtilis strain 3C3 [J]. International Biodeterioration & Biodegradation, 2014, 91: 138-147
[29] Brar S K, Verma M, Tyagi R, et al. Concurrent degradation of dimethyl phthalate (DMP) during production of Bacillus thuringiensis based biopesticides [J]. Journal of Hazardous Materials, 2009, 171(1): 1016-1023
[30] Surhio M A, Talpur F N, Nizamani S M, et al. Complete degradation of dimethyl phthalate by biochemical cooperation of the Bacillus thuringiensis strain isolated from cotton field soil [J]. RSC Advances, 2014, 4(99): 55960-55966
[31] Wang Y, Yin B, Hong Y, et al. Degradation of dimethyl carboxylic phthalate ester by Burkholderia cepacia DA2 isolated from marine sediment of South China Sea [J]. Ecotoxicology, 2008, 17(8): 845-852
[32] Chen X, Zhang X, Yang Y, et al. Biodegradation of an endocrine-disrupting chemical di-n-butyl phthalate by newly isolated Camelimonas sp. and enzymatic properties of its hydrolase [J]. Biodegradation, 2015, 26(2): 171-182
[33] Wang Y, Fan Y, Gu J D. Aerobic degradation of phthalic acid by Comamonas acidovoran Fy-1 and dimethyl phthalate ester by two reconstituted consortia from sewage sludge at high concentrations [J]. World Journal of Microbiology and Biotechnology, 2003, 19(8): 811-815
[34] Chang B, Yang C, Cheng C, et al. Biodegradation of phthalate esters by two bacteria strains [J]. Chemosphere, 2004, 55(4): 533-538
[35] Patil N K, Kundapur R, Shouche Y S, et al. Degradation of plasticizer di-n-butylphthalate by Delftia sp. TBKNP-05 [J]. Current Microbiology, 2006, 52(5): 369-374
[36] Fang C R, Yao J, Zheng Y G, et al. Dibutyl phthalate degradation by Enterobacter sp. T5 isolated from municipal solid waste in landfill bioreactor [J]. International Biodeterioration & Biodegradation, 2010, 64(6): 442-446
[37] Tanaka T, Yamada K, Iijima T, et al. Complete degradation of the endocrine-disrupting chemical phthalic acid by Flavobacterium sp. [J]. Journal of Health Science, 2006, 52(6): 800-804
[38] Kido Y, Tanaka T, Yamada K, et al. Complete degradation of the endocrine-disrupting chemical dimethyl phthalate ester by Flavobacterium sp. [J]. Journal of Health Science, 2007, 53(6): 740-744
[39] Chatterjee S, Dutta T K. Metabolism of butyl benzyl phthalate by Gordonia sp. strain MTCC 4818 [J]. Biochemical and Biophysical Research Communications, 2003, 309(1): 36-43
[40] Nishioka T, Iwata M, Imaoka T, et al. A mono-2-ethylhexyl phthalate hydrolase from a Gordonia sp. that is able to dissimilate di-2-ethylhexyl phthalate [J]. Applied and Environmental Microbiology, 2006, 72(4): 2394-2399
[41] Chen J A, Li X, Li J, et al. Degradation of environmental endocrine disruptor di-2-ethylhexyl phthalate by a newly discovered bacterium, Microbacterium sp. strain CQ0110Y [J]. Applied Microbiology and Biotechnology, 2007, 74(3): 676-682
[42] Baek J H, Gu M B, Sang B I, et al. Risk reduction of adverse effects due to di-(2-ethylhexyl) phthalate (DEHP) by utilizing microbial degradation [J]. Journal of Toxicology and Environmental Health, Part A: Current Issues, 2009, 72(21-22): 1388-1394
[43] Hu J, Yang Q. Microbial degradation of di-n-butyl phthalate by Micrococcus sp. immobilized with polyvinyl alcohol [J]. Desalination and Water Treatment, 2015, 56(9): 2457-2463
[44] Nakamiya K, Hashimoto S, Ito H, et al. Microbial treatment of bis (2-ethylhexyl) phthalate in polyvinyl chloride with isolated bacteria [J]. Journal of Bioscience and Bioengineering, 2005, 99(2): 115-119
[45] Stingley R L, Brezna B, Khan A A, et al. Novel organization of genes in a phthalate degradation operon of Mycobacterium vanbaalenii PYR-1 [J]. Microbiology, 2004, 150(11): 3749-3761
[46] Wu X L, Wang Y Y, Liang R X, et al. Degradation of di-n-butyl phthalate by newly isolated Ochrobactrum sp. [J]. Bulletin of Environmental Contamination and Toxicology, 2010, 85(3): 235-237
[47] Jin L, Sun X, Zhang X, et al. Co-metabolic biodegradation of DBP by Paenibacillus sp. S-3 and H-2 [J]. Current Microbiology, 2014, 68(6): 708-716
[48] 金雷, 严忠雍, 施慧, 等. 邻苯二甲酸二丁酯DBP降解菌S-3的分离、鉴定及其代谢途径的初步研究[J]. 农业生物技术学报, 2014, 22(1): 101-108
Jin L, Yan Z Y, Shi H, et al.Identification of a dibutyl phthalate (DBP)-degrading strain S-3 and preliminary studies on the metabolic pathway [J]. Journal of Agricultural Biotechnology, 2014, 22(1): 101-108 (in Chinese)
[49] Xu X R, Li H B, Gu J D. Metabolism and biochemical pathway of n-butyl benzyl phthalate by Pseudomonas fluorescens B-1 isolated from a mangrove sediment [J]. Ecotoxicology and Environmental Safety, 2007, 68(3): 379-385
[50] Zeng F, Cui K, Li X, et al. Biodegradation kinetics of phthalate esters by Pseudomonas fluoresences FS1 [J]. Process Biochemistry, 2004, 39(9): 1125-1129
[51] Meng X, Niu G, Yang W, et al. Di (2-ethylhexyl) phthalate biodegradation and denitrification by a Pseudoxanthomonas sp. strain [J]. Bioresource Technology, 2015, 180: 356-359
[52] Sauvageau D, Cooper D G, Nicell J A. Relative rates and mechanisms of biodegradation of diester plasticizers mediated by Rhodococcus rhodochrous [J]. The Canadian Journal of Chemical Engineering, 2009, 87(3): 499-506
[53] He Z, Niu C, Lu Z. Individual or synchronous biodegradation of di-n-butyl phthalate and phenol by Rhodococcus ruber strain DP-2 [J]. Journal of Hazardous Materials, 2014, 273: 104-109
[54] Li J, Gu J-D, Pan L. Transformation of dimethyl phthalate, dimethyl isophthalate and dimethyl terephthalate by Rhodococcus rubber Sa and modeling the processes using the modified Gompertz model [J]. International Biodeterioration & Biodegradation, 2005, 55(3): 223-232
[55] Wang J, Zhang M Y, Chen T, et al. Isolation and identification of a di-(2-ethylhexyl) phthalate-degrading bcterium and its role in the bioremediation of a contaminated soil [J]. Pedosphere, 2015, 25(2): 202-211
[56] 曹相生, 孟雪征, 任书魁, 等. 一株DEHP降解菌的筛选和分子鉴定[J]. 环境科学与技术, 2011, 34(12): 217-220
Cao X S, Meng X Z, Ren S K, et al. Isolation and phylogenitic analysis of a bis (2-ethylhexyl) phthalate degrading bacterial strain [J].Environmental Science & Technology, 2011, 34(12): 217-220 (in Chinese)
[57] Li C, Tian X, Chen Z, et al. Biodegradation of an endocrine-disrupting chemical di-n-butyl phthalate by Serratia marcescens C9 isolated from activated sludge [J]. African Journal of Microbiology Research, 2012, 6(11): 2686-2693
[58] Zeng P, Moy B Y P, Song Y H, et al. Biodegradation of dimethyl phthalate by Sphingomonas sp. isolated from phthalic-acid-degrading aerobic granules [J]. Applied Microbiology and Biotechnology, 2008, 80(5): 899-905
[59] Vega D, Bastide J. Dimethylphthalate hydrolysis by specific microbial esterase [J]. Chemosphere, 2003, 51(8): 663-668
[60] Zhang X Y, Fan X, Qiu Y J, et al. Newly identified thermostable esterase from Sulfobacillus acidophilus: Properties and performance in phthalate ester degradation [J]. Applied and Environmental Microbiology, 2014, 80(22): 6870-6878
[61] Pranaw K, Singh S, Dutta D, et al. Biodegradation of dimethyl phthalate by an entomopathogenic nematode symbiont Xenorhabdus indica strain KB-3 [J]. International Biodeterioration & Biodegradation, 2014, 89: 23-28
[62] Prasad B, Suresh S. Biodegradation of dimethyl phthalate ester using free cells, entrapped cells of Variovorax sp. BS1 and cell free enzyme extracts: A comparative study [J]. International Biodeterioration & Biodegradation, 2015, 97: 179-187
[63] Zeng F, Cui K, Fu J, et al. Biodegradability of di (2-ethylhexyl) phthalate by Pseudomonas fluorescens FS1 [J]. Water, Air, & Soil Pollution, 2002, 140(1-4): 297-305
[64] Quan C, Liu Q, Tian W, et al. Biodegradation of an endocrine-disrupting chemical, di-2-ethylhexyl phthalate, by Bacillus subtilis No. 66 [J]. Applied Microbiology and Biotechnology, 2005, 66(6): 702-710
[65] Amir S, Hafidi M, Merlina G, et al. Fate of phthalic acid esters during composting of both lagooning and activated sludges [J]. Process Biochemistry, 2005, 40(6): 2183-2190
[66] Cartwright C D, Owen S A, Thompson I P, et al. Biodegradation of diethyl phthalate in soil by a novel pathway [J]. FEMS Microbiology Letters, 2000, 186(1): 27-34
[67] Eaton R W, Ribbons D W. Metabolism of dibutylphthalate and phthalate by Micrococcus sp. strain 12B [J]. Journal of Bacteriology, 1982, 151(1): 48-57
[68] Chauret C, Mayfield C I, Inniss W E. Biotransformation of di-n-butyl phthalate by a psychrotrophic Pseudomonas fluoresceins (BGW) isolated from subsurface environment [J]. Canadian Journal of Microbiology, 1995, 41(1): 54-63
[69] Wang J, Liu P, Qian Y. Microbial degradation of di-n-butyl phthalate [J]. Chemosphere, 1995, 31(9): 4051-4056
[70] Chang B, Wang T, Yuan S. Biodegradation of four phthalate esters in sludge [J]. Chemosphere, 2007, 69(7): 1116-1123
[71] Wang J, Chen L, Shi H, et al. Microbial degradation of phthalic acid esters under anaerobic digestion of sludge [J]. Chemosphere, 2000, 41(8): 1245-1248
[72] Di Gennaro P, Collina E, Franzetti A, et al. Bioremediation of diethylhexyl phthalate contaminated soil: A feasibility study in slurry-and solid-phase reactors [J]. Environmental Science & Technology, 2005, 39(1): 325-330
◆
Phthalic Acid Esters-degrading Bacteria: Biodiversity, Degradation Mechanisms and Environmental Applications
Han Yonghe, He Ruiwen, Li Chao, Xiang Ping, Luo Jun, Cui Xinyi*
State Key Laboratory of Pollution Control and Resource Reuse, School of the Environment, Nanjing University, Nanjing 210046, China
Phthalic acid esters (PAEs) are typical persistent organic pollutants (POPs) that can disrupt endocrine systems in humans. Long-term exposure to PAEs can induce toxic effects to living organisms in various environmental matrixes, including water body, sediment and soil. PAEs are persistent and frequently detected with high concentrations in environment, which consequently limit their effective removal from contaminated sites. With high tolerance for contaminants and outstanding ability to biodegrade contaminants, microbes show great potentials to bio-remediate PAE-contaminated sites. Compared to physical and chemical approaches, microbial remediation technology has several advantages, including easy manipulation, large-scale application in remediation and high flexibility. The aims of this review are to summarize most of the reported bacterial species, metabolic mechanisms mediated by PAE-degrading bacteria, and the applications of these bacteria in remediation of PAEs-polluted water and soil. This review can provide some new information for further studies on the bio-remediation of PAEs.
endocrine disrupter; persistent organic pollutants; phthalic acid esters; microbes; biodegradation; health risk
10.7524/AJE.1673-5897.20151107002
江苏省自然科学基金青年科学基金项目(BK20130558);国家自然科学基金青年科学基金项目(21307055)
韩永和(1986-),男,博士研究生,研究方向为污染物的环境微生物行为,E-mail:hanyonghe0423@163.com
*通讯作者(Corresponding author), E-mail: lizzycui@nju.edu.cn
2015-11-07 录用日期:2015-12-22
1673-5897(2016)2-037-13
X171.5
A
简介:崔昕毅(1983-),女,环境工程博士,副教授,主要研究有机污染物的环境行为、生态风险评价及修复和痕量有机物的环境分析监测,发表SCI论文20余篇。
韩永和, 何睿文, 李超, 等. 邻苯二甲酸酯降解细菌的多样性、降解机理及环境应用[J]. 生态毒理学报,2016, 11(2): 37-49
Han Y H, He R W, Li C, et al. Phthalic acid esters-degrading bacteria: Biodiversity, degradation mechanisms and environmental applications [J]. Asian Journal of Ecotoxicology, 2016, 11(2): 37-49 (in Chinese)

