Treatment with analgesics after mouse sciatic nerve injury does not alter expression of wound healingassociated genes
Matt C. Danzi, Dario Motti, Donna L. Avison, John L. Bixby,, Vance P. Lemmon,
1 The Miami Project to Cure Paralysis, University of Miami, Lois Pope LIFE Center, Miami, FL, USA
2 Department of Surgery, Leonard M. Miller School of Medicine, University of Miami, Miami, FL, USA
3 Department of Neurological Surgery, University of Miami, Miami, FL, USA
4 Department of Molecular and Cellular Pharmacology, University of Miami, Miami, FL, USA
RESEARCH ARTICLE
Treatment with analgesics after mouse sciatic nerve injury does not alter expression of wound healingassociated genes
Matt C. Danzi1, Dario Motti1, Donna L. Avison2, John L. Bixby1,3,4, Vance P. Lemmon1,3,*
1 The Miami Project to Cure Paralysis, University of Miami, Lois Pope LIFE Center, Miami, FL, USA
2 Department of Surgery, Leonard M. Miller School of Medicine, University of Miami, Miami, FL, USA
3 Department of Neurological Surgery, University of Miami, Miami, FL, USA
4 Department of Molecular and Cellular Pharmacology, University of Miami, Miami, FL, USA
Animal models of sciatic nerve injury are commonly used to study neuropathic pain as well as axon regeneration. Administration of post-surgical analgesics is an important consideration for animal welfare, but the actions of the analgesic must not interfere with the scientific goals of the experiment. In this study, we show that treatment with either buprenorphine or acetaminophen following a bilateral sciatic nerve crush surgery does not alter the expression in dorsal root ganglion (DRG) sensory neurons of a panel of genes associated with wound healing. These findings indicate that the post-operative use of buprenorphine or acetaminophen at doses commonly suggested by Institutional Animal Care and Use Committees does not change the intrinsic gene expression response of DRG neurons to a sciatic nerve crush injury, for many wound healing-associated genes. Therefore, administration of post-operative analgesics may not confound the results of transcriptomic studies employing this injury model.
acetaminophen; analgesics; axon; buprenorphine; dorsal root ganglia; gene expression; peripheral nerve injuries; regeneration; sciatic nerve; wound healing
http://www.nrronline.org/
Accepted: 2015-08-17
Introduction
Sciatic nerve injuries are the most commonly used model to study peripheral axon regeneration (Griffin et al., 2010; Wood et al., 2011; Geuna, 2015) and are also often used in animal models of neuropathic pain (Sorkin and Yaksh, 2009). Since efforts to understand axon regeneration do not require the animals to experience neuropathic pain, relief from this post-operative pain is an important aspect of animal welfare. To be useful in a research project, however, any analgesic used must not interfere with the scientific goals of the experiment. In the case of sciatic nerve injury as a model for axon regeneration, it is necessary that the analgesics not alter the injury response of the neurons that relates to axon regrowth, such as changes in gene expression (Huebner and Strittmatter, 2009).
Buprenorphine is a synthetic opioid with mixed agonist-antagonist activity against mu-, kappa-, and delta-opioid receptors (Lutfy and Cowan, 2004). It has been demonstrated to be effective in reducing neuropathic pain (Kouya et al., 2002; Christoph et al., 2005; Hans, 2007; Pergolizzi et al., 2010) and is commonly used as a post-operative analgesic in murine models of nerve injury (Yu et al., 2001; Kouya et al., 2002; Christoph et al., 2005). However, due to recent reports on the potential ability of opioids (synthetic or endogenous) to modulate the processes of inflammation and wound healing (see Stein and Küchler, 2013 for review), such drugs may not be appropriate for use as analgesics in studies of inflammatory or wound healing processes. Notably, both inflammation and scarring are likely to affect the process of axon regeneration (Gaudet et al., 2011; Chew et al., 2012; Lang et al., 2014).
Acetaminophen is an analgesic with numerous actions similar to nonsteroidal anti-inflammatory drugs (NSAIDs). Acetaminophen produces its analgesic effect by inhibiting the cyclooxygenase enzymes PTGS1 and PTGS2, thereby decreasing prostaglandin synthesis (Graham et al., 2013).
In this study, we examined whether treatment with buprenorphine or acetaminophen following a bilateral sciatic nerve crush surgery altered the response of DRG neurons to the injury as measured by the mRNA expression of a panel of genes associated with wound healing.
Materials and Methods
Sciatic nerve crush and animal handling
Nine female C57BL/6 mice aged 6 to 8 weeks were used for sciatic nerve crush surgeries. All procedures were performed in accordance with the protocols approved by the Institutional Animal Care and Use Committee at the University of Miami. Each animal was anesthetized with 80 mg/kg of Ketamine and 5 mg/kg of Xylazine. The sciatic nerve was exposed at the level of the thigh and then crushed with a pair of #5 forceps (Dumont; Switzerland) for 10 seconds before closing the incision. This procedure was repeated on the other leg to obtain a bilateral sciatic nerve crush. After the surgery, the animals were given 1 mLof Lactated Ringers subcutaneously. Gentamicin was given to the animals at a 0.2 mg/100 g dose once per day for 7 days. Animals were housed individually after surgery and animal recovery gel (DietGel Recovery from ClearH2O; USA) was placed in each cage for nutritional support and hydration. Nine animals were chosen so that there would be three animals per treatment group. We chose to use three animals per sample group because that is the standard used for microarray projects, and yields suitable power for that application.
Analgesic treatments
The mice that underwent bilateral sciatic nerve crush surgery were divided into three groups of three mice each for treatments. Three mice were given no post-operative analgesia. Three mice were injected with 0.1 mg/kg of buprenorphine subcutaneously once on the day of surgery, twice on the day following the surgery, and twice on the second day after the surgery. The final three mice were given 1 mg/mL of acetaminophen (Children’s Tylenol) in their water supply for seven days after surgery. The animals were observed daily by a veterinarian blinded to the treatments who monitored the animals’ behavior, especially that related to neuropathic pain.
Laser Capture Microdissection of DRG neurons and RNA isolation
On the seventh day after surgery, animals were euthanized with CO2inhalation. Animals were then perfused with 15-20 mL of a Zinc fixative solution (BD Pharmingen; USA). DRGs from lumbar levels 3-5 were then dissected bilaterally, embedded in Tissue Freezing Medium (Triangle Biomedical Sciences, Inc., USA), and flash-frozen on a layer of 2-methylbutane floating in liquid nitrogen. The frozen tissue was sectioned in a cryostat at 10 µm and sections were mounted on RNase-free PEN-membrane slides (Leica, USA). The slides were stained by submersion in the following solutions the specified number of times for the designated length of time: 3 times of 30 seconds each in 70% ethanol, 30 seconds in 0.5% Toluidine blue (Sigma, USA) dissolved in 70% ethanol, 2 times for 30 seconds each in 70% ethanol, 2 times for 30 seconds each in 90% ethanol, and finally 30 seconds in 100% ethanol. All recognizable DRG neuronal bodies in all sections of the ganglia were microdissected using a Leica LMD 6000 microscope. The cell bodies were collected in 50 µL of Extraction Buffer from the Arcturus PicoPure RNA Isolation Kit (Life Technologies, USA). RNA extraction was performed following the manufacturer’s instructions. The whole procedure was performed in RNase-free conditions and all solutions were made with diethyl pyrocarbonate (DEPC, Sigma) treated water in order to maintain RNA integrity. RNA quantity and quality was checked via Bioanalyzer 2100 (Agilent, USA).
qPCR panels
For each animal, 40 ng of RNA was synthesized into cDNA and pre-amplified for genes in the Qiagen Mouse Wound Healing PCR Array using the Qiagen RT2 PreAMP cDNA Synthesis Kit and the RT2 PreAMP cDNA Synthesis Primer Mix for Mouse Wound Healing PCR Array (Qiagen, USA). The cDNA was used on a RT2 Profiler PCR Array for Mouse Wound Healing genes (Qiagen, USA). Reactions were performed using 2X Power SYBR Green PCR Master Mix (Applied Biosystems, USA) on a 7300 Real-Time PCR System (Applied Biosystems).
Data analysis
Relative expression for each gene in each sample was calculated using the 2-ΔCqmethod (Schmittgen and Livak, 2008) with beta-actin as the reference gene. Tests for statistical significance of changes in expression level were conducted using a oneway ANOVA in SPSS 22 (IBM, USA) with a post-hoc Dunnett’s correction on the relative gene expression data. Fold change of expression was calculated for the buprenorphine- and acetaminophen-treated groups by averaging the relative expression of each gene across its replicates and dividing by the average relative expression for the untreated group. Of the 5 housekeeping genes present in the qPCR panel, beta-actin was used as the reference gene for the data analysis because it exhibited the least variance among samples. The analysis was repeated using each of the four other housekeeping genes as the reference gene, but this did not change the result of the significance testing for any genes. Volcano plot was generated in Matlab 2014b (Mathworks; USA). The data analysis and reporting in this work endeavors to conform to the Minimum Information for publication of Quantitative real-time PCR Experiments (MIQE) guidelines (Bustin et al., 2009) as well as the Minimal Information about a Spinal Cord Injury Experiment reporting guidelines (Lemmon et al., 2014).
Results
Gene expression of wound healing-associated genes does not change with buprenorphine or acetaminophen treatment after the crush injury
Nine mice underwent bilateral sciatic nerve crush surgeries and were divided into three treatment groups of three mice each. The first group received no post-operative analgesia, the second group was treated with buprenorphine, and the third group was treated with acetaminophen (see Materials and Methods for a description of the doses and regimens of treatment). Animals were sacrificed 7 days after surgery and RNA was extracted from the DRG cell bodies at lumbar levels 3-5 from each animal after collection with laser capture microdissection. This time point post-injury was chosen because the study was designed as a preliminary experiment for an RNA-Seq project. The goal was to perform RNA-Seq on DRGs while they were in the elongation phase of axon regeneration, and we hypothesized that the DRG neurons would be in the elongation phase at 7 days post-injury. These measurements were performed to see if the analgesics would have an effect on the RNA-Seq data. Commercially available qPCR panels for mouse wound healing genes were used to assay the expression levels of 84 genes thought to be involved in wound healing, along with 5 housekeeping genes (Table 1). A one-way ANOVA with a post-hoc Dunnett’s correction revealed that none of the genes had any statistically significant change in expression among the treatment groups and provided the p-values for the individual comparisons (Table 2). Two genes (Fga and Mmp7) were expressed at such low levels in all three treatment groups that they received a Cq value of ‘undetermined’ too often to be usable for significance testing. These two genes were not plotted in Figure 1 because it was not possible to assign them a P-value. Although several genes were scored as having Log2 (Fold Change) >1 or < -1, which could be an indicator of biological significance, none of the genes come close to achieving a statistically significant change in expression relative to the untreated control (denoted by horizontal dashed line in Figure 1).
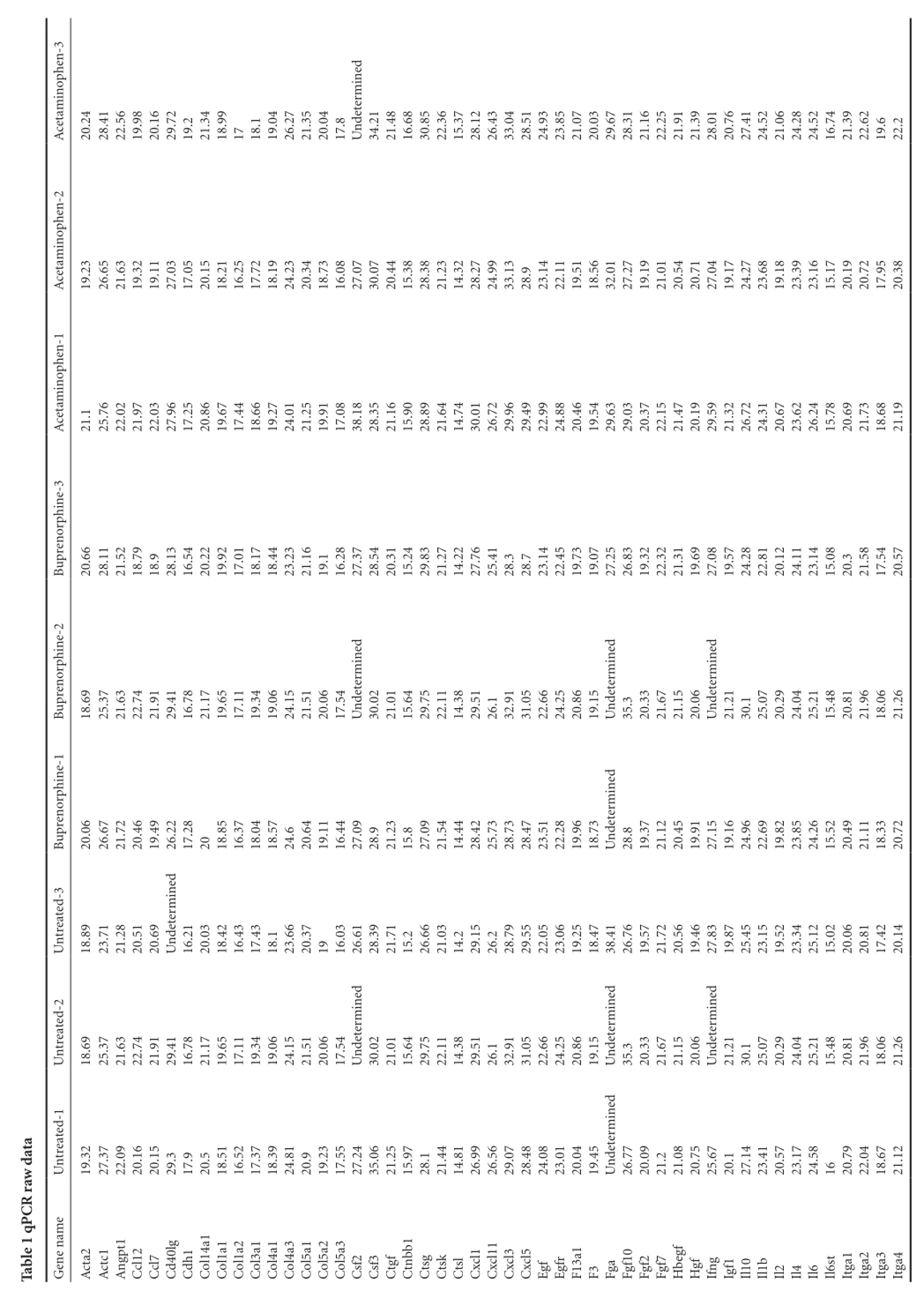
ata rawdCRTable 1 qPhen-3 opininedAcetam20.24 28.41 22.56 19.98 20.16 29.72 19.221.34 18.99 1718.119.04 26.27 21.35 20.04 17.8determUn34.21 21.48 16.68 30.85 22.36 15.37 28.12 26.43 33.04 28.51 24.93 23.85 21.07 20.03 29.67 28.31 21.16 22.25 21.91 21.39 28.01 20.76 27.41 24.52 21.06 24.28 24.52 16.74 21.39 22.62 19.622.2ophen-2 inAcetam19.23 26.65 21.63 19.32 19.11 27.03 17.05 20.15 18.21 16.25 17.72 18.19 24.23 20.34 18.73 16.08 27.07 30.07 20.44 15.38 28.38 21.23 14.32 28.27 24.99 33.13 28.923.14 22.11 19.51 18.56 32.01 27.27 19.19 21.01 20.54 20.71 27.04 19.17 24.27 23.68 19.18 23.39 23.16 15.17 20.19 20.72 17.95 20.38 ophen-1 inAcetam21.125.76 22.02 21.97 22.03 27.96 17.25 20.86 19.67 17.44 18.66 19.27 24.01 21.25 19.91 17.08 38.18 28.35 21.16 15.90 28.89 21.64 14.74 30.01 26.72 29.96 29.49 22.99 24.88 20.46 19.54 29.63 29.03 20.37 22.15 21.47 20.19 29.59 21.32 26.72 24.31 20.67 23.62 26.24 15.78 20.69 21.73 18.68 21.19 e-3 orphinprenBu20.66 28.11 21.52 18.79 18.928.13 16.54 20.22 19.92 17.01 18.17 18.44 23.23 21.16 19.116.28 27.37 28.54 20.31 15.24 29.83 21.27 14.22 27.76 25.41 28.328.723.14 22.45 19.73 19.07 27.25 26.83 19.32 22.32 21.31 19.69 27.08 19.57 24.28 22.81 20.12 24.11 23.14 15.08 20.321.58 17.54 20.57 e-2 inorphedinedinedinprenBu18. 6925. 3721. 6322. 7421. 9129. 4116. 7821. 1719. 6517. 1119. 3419. 0624. 1521. 5120. 0617. 54determUn30. 0221. 0115. 6429. 7522. 1114. 3829. 5126. 1 32. 9131. 0522. 6624. 2520. 8619. 15determUn35. 3 20. 3321. 6721. 1520. 06determUn21. 2130. 1 25. 0720. 2924. 0425. 2115. 4820. 8121. 9618. 0621. 26e-1 inorphedinprenBu20.06 26.67 21.72 20.46 19.49 26.22 17.28 2018.85 16.37 18.04 18.57 24.620.64 19.11 16.44 27.09 28.921.23 15.827.09 21.54 14.44 28.42 25.73 28.73 28.47 23.51 22.28 19.96 18.73 determUn28.819.37 21.12 20.45 19.91 27.15 19.16 24.96 22.69 19.82 23.85 24.26 15.52 20.49 21.11 18.33 20.72 treated-3 edinUn18.89 23.71 21.28 20.51 20.69 determUn16.21 20.03 18.42 16.43 17.43 18.123.66 20.37 1916.03 26.61 28.39 21.71 15.226.66 21.03 14.229.15 26.228.79 29.55 22.05 23.06 19.25 18.47 38.41 26.76 19.57 21.72 20.56 19.46 27.83 19.87 25.45 23.15 19.52 23.34 25.12 15.02 20.06 20.81 17.42 20.14 treated-2 edinedinedinUn18.69 25.37 21.63 22.74 21.91 29.41 16.78 21.17 19.65 17.11 19.34 19.06 24.15 21.51 20.06 17.54 determUn30.02 21.01 15.64 29.75 22.11 14.38 29.51 26.132.91 31.05 22.66 24.25 20.86 19.15 determUn35.320.33 21.67 21.15 20.06 determUn21.21 30.125.07 20.29 24.04 25.21 15.48 20.81 21.96 18.06 21.26 treated-1 edinUn19.32 27.37 22.09 20.16 20.15 29.317.920.518.51 16.52 17.37 18.39 24.81 20.919.23 17.55 27.24 35.06 21.25 15.97 28.121.44 14.81 26.99 26.56 29.07 28.48 24.08 23.01 20.04 19.45 determUn26.77 20.09 21.221.08 20.75 25.67 20.127.14 23.41 20.57 23.17 24.58 1620.79 22.04 18.67 21.12 GenenameActa2 Actc1 gpt1AnCcl12 Ccl740lgCdh1CdCol14a1 Col1a1Col1a2Col3a1Col4a1Col4a3Col5a1Col5a2Col5a3Csf2Csf3CtgfCtnbb1CtsgCtskCtslCxcl1 Cxcl11Cxcl3 Cxcl5 Egf EgfrF13a1 F3Fga Fgf10 Fgf2Fgf7Hbegf Hgf IfngIgf1Il10Il1bIl2 Il4 Il6 Il6st Itga1 Itga2 Itga3 Itga4
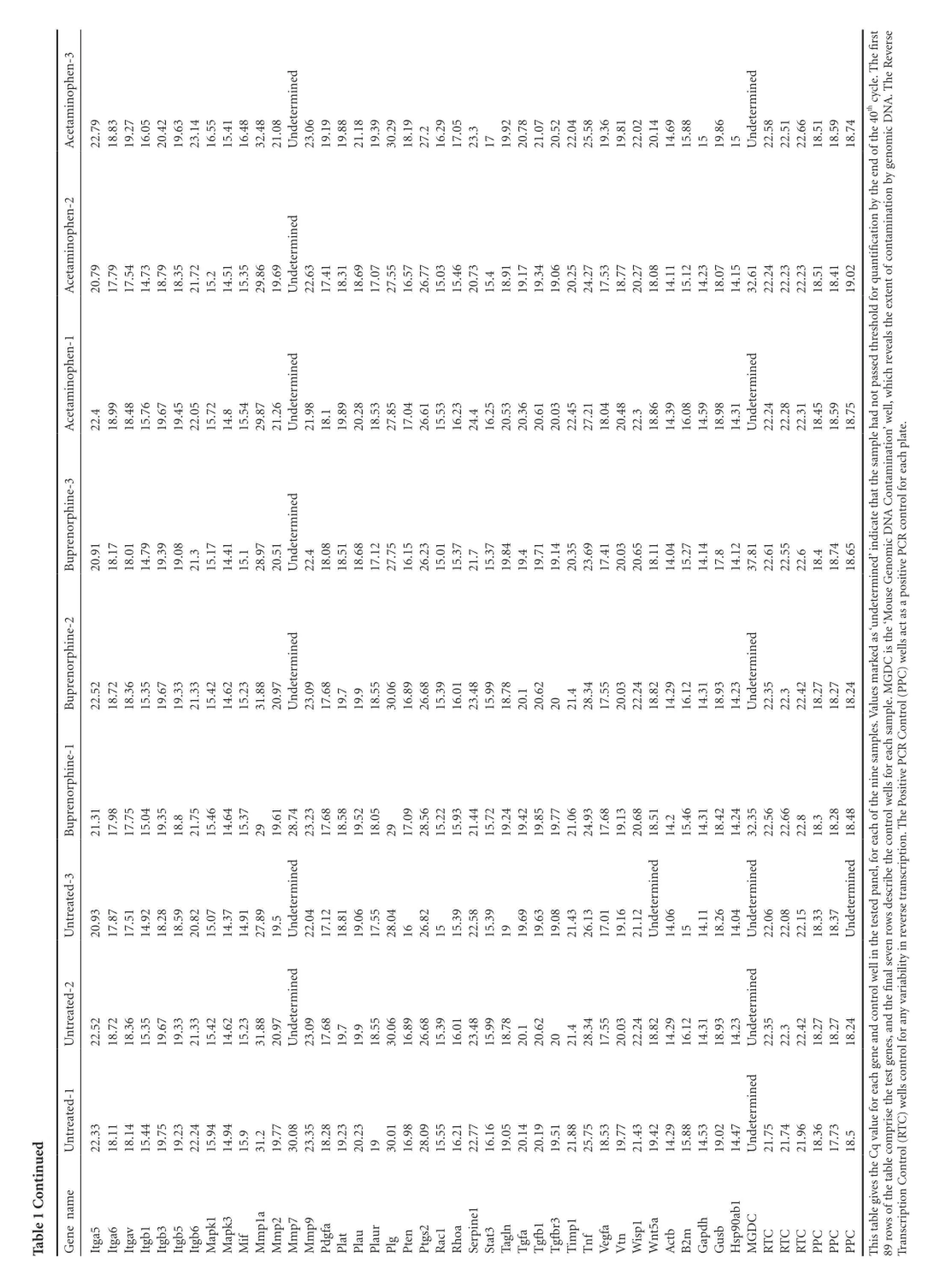
ued ntinTable 1 Cohen-3 ofthe40thcycle. Thefi rst everseopheRininedined.TAcetam22.79 18.83 19.27 16.05 20.42 19.63 23.14 16.55 15.41 16.48 32.48 21.08determNAUn23.06 19.19 19.88 21.18 19.39 30.29 18.19 27.216.29 17.05 23.31719.92 20.78 21.07 20.52 22.04 25.58 19.36 19.81 22.02 20.14 14.69 15.88 1519.86 15Undeterm22.58 22.51 22.66 18.51 18.59 18.74icDomophen-2 edbytheendinintaminationbygenAcetam20.79 17.79 17.54 14.73 18.79 18.35 21.72 15.214.51 15.35 29.86 19.69determUn22.63 17.41 18.31 18.69 17.07 27.55 16.57 26.77 15.03 15.46 20.73 15.418.91 19.17 19.34 19.06 20.25 24.27 17.53 18.77 20.27 18.08 14.11 15.12 14.23 18.07 14.15 32.61 22.24 22.23 22.23 18.51 18.41 19.02 forquantifi cationtofconophen-1 ededinininAcetamotpassedthresholdhichrevealstheexten22.418.99 18.48 15.76 19.67 19.45 22.05 15.72 14.815.54 29.87 21.26determUn21.98 18.119.89 20.28 18.53 27.85 17.04 26.61 15.53 16.23 24.416.25 20.53 20.36 20.61 20.03 22.45 27.21 18.04 20.48 22.318.86 14.39 16.08 14.59 18.98 14.31determUn22.24 22.28 22.31 18.45 18.59 18.75 ple hadnell,wation’wine-3 orphinedinContamNAprencontrolforeachplate. CR20.91 18.17 Bu18.01 14.79 19.39 19.08 21.315.17 14.41 15.128.97 20.51determUn22.418.08 18.51 18.68 17.12 27.75 16.15 26.23 15.01 15.37 21.715.37 19.84 19.419.71 19.14 20.35 23.69 17.41 20.03 20.65 18.11 14.04 15.27 14.14 17.814.12 37.81 22.61 22.55 22.618.418.74 18.65 ed’iomndicatethatthesamicDinenseGoue-2determinorphedinedinCisthe‘Mprenarkedas ‘un) wells actasapositivePPCBu22.5218.7218.3615.3519.6719.3321.3315.4214.6215.2331.8820.97determUn23.0917.6819.7 19.9 18.5530.0616.8926.6815.3916.0123.4815.9918.7820.1 20.622021.4 28.3417.5520.0322.2418.8214.2916.1214.3118.9314.23determUn22.3522.3 22.4218.2718.2718.24esmGDple.Me-1ples. ValuControl (PinCRorphesaminprenellsforeachsamePositivePBu21.31 17.98 17.75 15.04 19.35 18.821.75 15.46 14.64 15.37 2919.61 28.74 23.23 17.68 18.58 19.52 18.05 2917.09 28.56 15.22 15.93 21.44 15.72 19.24 19.42 19.85 19.77 21.06 24.93 17.68 19.13 20.68 18.51 14.215.46 14.31 18.42 14.24 32.35 22.56 22.66 22.818.318.28 18.48 trolwtreated-3edinedinedinedinel,foreachofthenescribetheconscription.ThUn20.93 17.87 17.51 14.92 18.28 18.59 20.82 15.07 14.37 14.91 27.89 19.5determUn22.04 17.12 18.81 19.06 17.55 28.04 1626.82 1515.39 22.58 15.39 1919.69 19.63 19.08 21.43 26.13 17.01 19.16 21.12determUn14.06 1514.11 18.26 14.04determUn22.06 22.08 22.15 18.33 18.37determUnthe testedpanrowsdreverse trantreated-2ededell ininincontrolwefi nalseventhUn22.52 18.72 18.36 15.35 19.67 19.33 21.33 15.42 14.62 15.23 31.88 20.97determUn23.09 17.68 19.719.918.55 30.06 16.89 26.68 15.39 16.01 23.48 15.99 18.78 20.120.62 2021.428.34 17.55 20.03 22.24 18.82 14.29 16.12 14.31 18.93 14.23determUn22.35 22.322.42 18.27 18.27 18.24 es,andtrolforanyvariabilityintreated-1edinellsconC)wUn22.33 18.11 18.14 15.44 19.75 19.23 22.24 15.94 14.94 15.931.219.77 30.08 23.35 18.28 19.23 20.23 1930.01 16.98 28.09 15.55 16.21 22.77 16.16 19.05 20.14 20.19 19.51 21.88 25.75 18.53 19.77 21.43 19.42 14.29 15.88 14.53 19.02 14.47determUn21.75 21.74 21.96 18.36 17.73 18.5qvalueforeach geneandprisethetestgene amn Control(RTGenene1legivestheCItga5 Itga6 Itgav Itgb1 Itgb3 Itgb5 Itgb6 Mapk1 Mapk3 h softhetablecomMifp1a 1r31 Mmp2p7p9MmMmMmPdgfa PlatPlaurPlauPlg PtenPtgs2 Rac1RhoaSerpinStat3 Tagln TgfaTgfbTgfbTimpf DCscriptioTnVegfa Vtn Wisp1t5a WnActbB2m GapdGusbHsp90ab1MGCCCCCC RTRTRTPPPPPPistabTh89rowTran
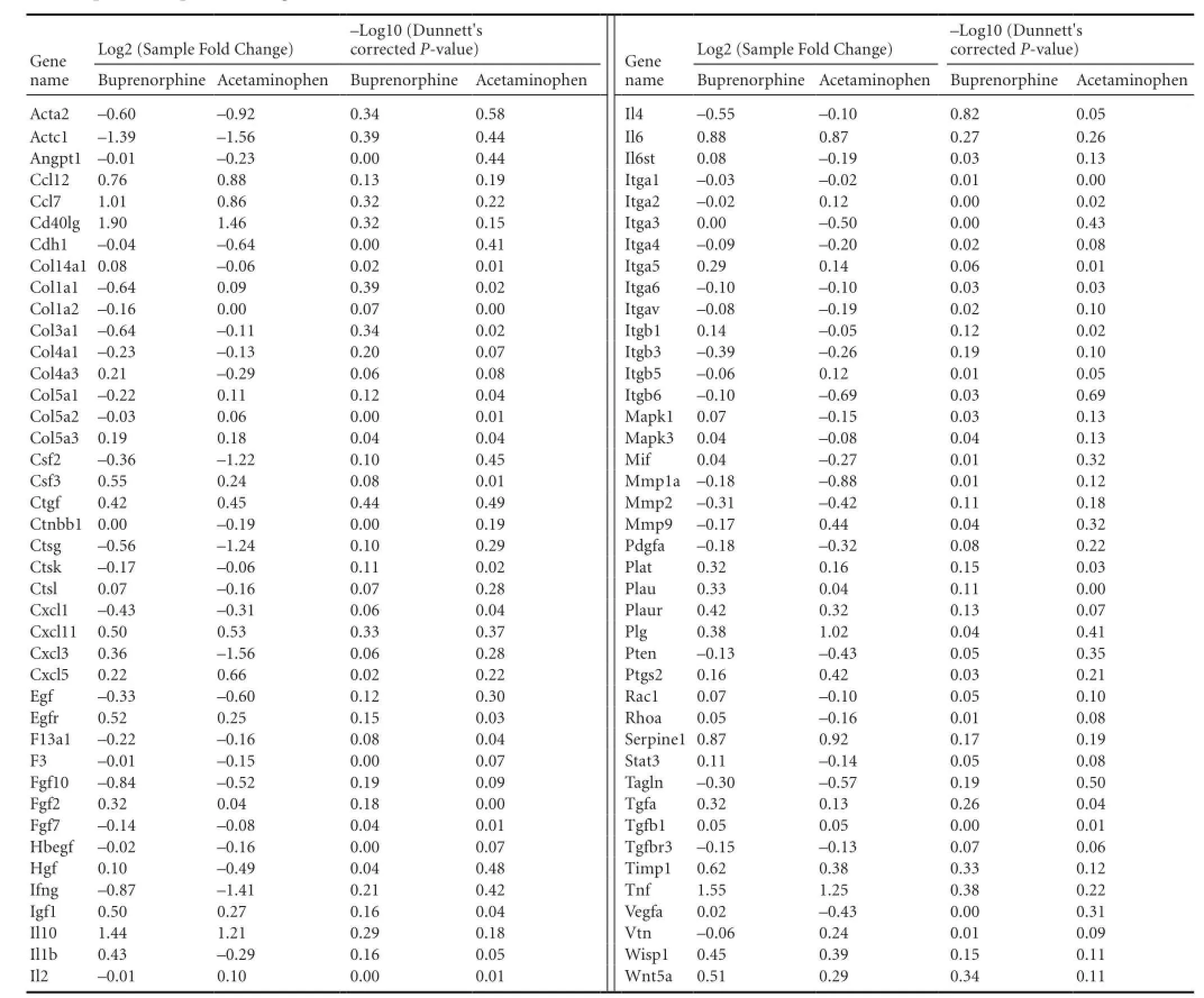
Table 2 qPCR data plotted in Figure 1
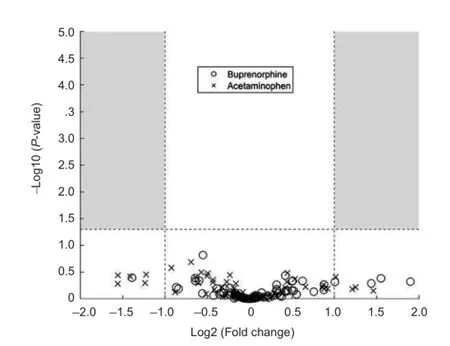
Figure 1 qPCR shows no significant changes in gene expression of wound healing-associated genes due to treatment with buprenorphine or acetaminophen after a sciatic nerve injury.
Discussion
Despite evidence that opioids may play a role in modulating inflammatory and wound healing responses, we did not observe any significant changes in the expression of wound healing associated genes by DRG neurons upon treatment with buprenorphine. Similarly, we did not observe any significant changes in the expression of the tested genes upon treatment with acetaminophen. These results suggest that these analgesics at the doses used do not interfere with intrinsic transcriptomic responses of the damaged neurons to the injury. Therefore, it appears that treatment with either of these analgesics can be used post-operatively without affecting the outcome of transcriptomic studies of DRG neurons performed in this model system.
One limitation of this study is that we did not test whether the administered dose of analgesic alleviated post-operative pain in the mice. The doses of the analgesics were chosen based on suggestions from our university’s Institutional Animal Care and Use Committee. Our primary goal was to test whether satisfying this institutional requirement would interfere with the results of our study in the form of altered gene expression data. While we succeeded in demonstrating that the expression of the tested genes was not altered, the study is limited by the fact that we did not test the extent to which the animals achieved a state of analgesia, nor did we identify a dose of analgesic treatment at which animals had an altered transcriptional response to the injury. Further, because we did not quantify the volume of acetaminophen-treated water drunk by each mouse, the actual dosage of acetaminophen administered is not calculable. Rather, we included this treatment at the recommendation of our Institutional Animal Care and Use Committee, which considers it to be a standard and viable treatment option.
A second limitation of this study is that the qPCR panel used was for general wound healing genes, and not genes specific to axon growth, neuropathic pain or neuroinflammatory processes. This panel was chosen because it contained more of the genes found to be differentially expressed between DRGs and Cerebellar Granule Neurons identified in a previous study (Lerch et al., 2012) than any of the other qPCR panels inspected. Although RNA-Seq would have been more informative than the qPCR panels, that method was outside of the scope of this project.
A final limitation of this study is that there was no positive control for the effect of the analgesic treatments. This leaves open numerous explanations for the negative gene expression results. One possibility is that the expression of these genes does not change in response to the injury, so the analgesics could not be seen to affect such changes. This explanation is unlikely, since we found previously that 15 of the 84 wound healing associated genes measured in this study were significantly upregulated in DRG neurons 7 days after sciatic nerve crush without post-surgical analgesic treatment (unpublished RNA-Seq observations). Other possibilities are that a higher dosage of analgesics might be necessary to evoke a measurable gene expression response, or that the effect of analgesics on gene expression was transient, and no longer significant by 7 days post-injury. While these open questions remain, it is nevertheless possible to conclude that these commonly used (and IACUC-recommended) doses of buprenorphine and acetaminophen do not significantly alter expression of this suite of wound healing associated genes in DRG neurons 7 days after a nerve crush injury.
Author contributions: MCD, JLB, and VPL conceived and designed the experiments. DLA designed analgesics regime. MCD, DM, and DLA conducted the experiments. MCD wrote the paper and other authors edited the paper.
Conflicts of interest: None declared.
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: Minimum Information for publication of quantitative real-time PCR experiments. Clin Chem 55:611-622.
Chew DJ, Fawcett JW, Andrews MR (2012) The challenges of long-distance axon regeneration in the injured CNS. Elsevier B.V. Available at: http://dx.doi.org/10.1016/B978-0-444-59544-7.00013-5.
Christoph T, Kögel B, Schiene K, Méen M, De Vry J, Friderichs E (2005) Broad analgesic profile of buprenorphine in rodent models of acute and chronic pain. Eur J Pharmacol 507:87-98.
Gaudet AD, Popovich PG, Ramer MS (2011) Wallerian degeneration: gaining perspective on inflammatory events after peripheral nerve injury. J Neuroinflammation 8:110.
Geuna S (2015) The sciatic nerve injury model in pre-clinical research. J Neurosci Methods 243:39-46.
Graham GG, Davies MJ, Day RO, Mohamudally A, Scott KF (2013) The modern pharmacology of paracetamol: therapeutic actions, mechanism of action, metabolism, toxicity and recent pharmacological findings. Inflammopharmacology 21:201-232.
Griffin JW, Pan B, Polley MA, Hoffman PN, Farah MH (2010) Measuring nerve regeneration in the mouse. Exp Neurol 223:60-71.
Hans G (2007) Buprenorphine--a review of its role in neuropathic pain. J Opioid Manag 3:195-206.
Huebner EA, Strittmatter SM (2009) Axon regeneration in the peripheral and central nervous systems. Results Probl Cell Differ 48:339-351.
Kouya PF, Hao JX, Xu XJ (2002) Buprenorphine alleviates neuropathic pain-like behaviors in rats after spinal cord and peripheral nerve injury. Eur J Pharmacol 450:49-53.
Lang BT, Wang J, Filous AR, Au NP, Ma CH, Shen Y (2014) Pleiotropic molecules in axon regeneration and neuroinflammation. Exp Neurol 258:17-23.
Lemmon VP, Ferguson AR, Popovich PG, Xu XM, Snow DM, Igarashi M, Beattie CE, Bixby JL (2014) Minimum information about a spinal cord injury experiment: a proposed reporting standard for spinal cord injury experiments. J Neurotrauma 1361:1354-1361.
Lerch JK, Kuo F, Motti D, Morris R, Bixby JL, Lemmon VP (2012) Isoform diversity and regulation in peripheral and central neurons revealed through RNA-Seq. PLoS One 7:e30417.
Lutfy K, Cowan A (2004) Buprenorphine: a unique drug with complex pharmacology. Curr Neuropharmacol 2:395-402.
Pergolizzi J, Aloisi AM, Dahan A, Filitz J, Langford R, Likar R, Mercadante S, Morlion B, Raffa RB, Sabatowski R, Sacerdote P, Torres LM, Weinbroum AA (2010) Current knowledge of buprenorphine and its unique pharmacological profile. Pain Pract 10:428-450.
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101-1108.
Sorkin LS, Yaksh TL (2009) Behavioral models of pain states evoked by physical injury to the peripheral nerve. Neurotherapeutics 6:609-619.
Stein C, Küchler S (2013) Targeting inflammation and wound healing by opioids. Trends Pharmacol Sci 34:303-312.
Wood MD, Kemp SW, Weber C, Borschel GH, Gordon T (2011) Outcome measures of peripheral nerve regeneration. Ann Anat 193:321-333.
Yu P, Matloub HS, Sanger JR, Narini P (2001) Gait analysis in rats with peripheral nerve injury. Muscle Nerve 24:231-239.
10.4103/1673-5374.169637
How to cite this article: Danzi MC, Motti D, Avison DJ, Bixby JL, Lemmon VP (2016) Treatment with analgesics after mouse sciatic nerve injury does not alter expression of wound healing-associated genes. Neural Regen Res 11(1)∶144-149.
Funding: This work was supported by National Institutes of Health HD057632, the Buoniconti Fund and the Walter G. Ross Distinguished Chair in Developmental Neuroscience (to VPL).
*Correspondence to: Vance P. Lemmon, Ph.D., vlemmon@med.miami.edu.
orcid: 0000-0003-3550-7576 (Vance P. Lemmon) 0000-0003-1568-5965 (Matt C. Danzi)
- 中国神经再生研究(英文版)的其它文章
- Vascular endothelial growth factor: an attractive target in the treatment of hypoxic/ischemic brain injury
- Angiogenesis in tissue-engineered nerves evaluated objectively using MICROFIL perfusion and micro-CT scanning
- Dexamethasone prevents vascular damage in earlystage non-freezing cold injury of the sciatic nerve
- Cerebrolysin improves sciatic nerve dysfunction in a mouse model of diabetic peripheral neuropathy
- A novel bioactive nerve conduit for the repair of peripheral nerve injury
- Time representation of mitochondrial morphology and function after acute spinal cord injury

