Computational study of the molecular mechanism of Lonicera japonica organic acidsagainst influenza
Xuan Luo,Xia Shen#,Ben-Xiang Hu,Hao Wang,Zhen-Yu Zhao,Zi-Hu Guo
1College of pharmacy,Shanxi university of Chinese medicine,Xi'an,712046,Shanxi,China.2College of Life Sciences,Northwest A&FUniversity,Yangling,712100,Shanxi,China.
Introduction
Pandemic caused by a rapidly evolving virus,influenza,exists worldwide and influences human health markedly.Influenza virus causes infections in the human respiratory tract,leading to deaths and economical losses nearly every year[1].For instance,a newly emerged influenza H7N9 in southeast China in March 2013 has caused a total of 37 deaths and has become a major public health concern[2].
Influenza has two major surface glycoproteins[3], hemagglutinin(HA)and neuraminidase(NA),which determine its subtypessuch as H1N1[4]and H5N1[5].In addition,the following proteins are critical for influenza virus:M2 protein,a proton channel to control the pH of the Golgi complex during HA synthesis[6],and PB2,a viral polymerase protein,which provides viral RNA-dependent RNA polymerase activity[7].
As an enzyme present on the surface of the virus,NA has become an ideal target for designing competitive inhibitor analysis assays[8].Studies have shown that oseltamivir and zanamivir,two influenza neuraminidase inhibitors,are NA-targeting first-line therapies for many influenza subtypes[9-11].However,such therapies are limited by the emergence of resistant strains due to the rapid mutations of the influenza virus[12-15].Recent studies revealed that H7N9 strains resistant to NA inhibitors (both oseltamivir and zanamivir) have emerged [16].Therefore,there is an urgent need for new approaches and drugs for pandemic disease control[17].
China has a long history of using herbal medicines as standard treatments against infectious diseases.Lonicera japonica was one of the most commonly used herbs and shown excellent clinical activity.Lonicera japonica has been recommended for the treatment and prevention of influenza [18,19].Numerous in vitro studies have evaluated the anti-viral properties of herbal medicines,such as anti-bacterial,anti-oxidant,and anti-inflammatory activities[20].However,because of disadvantages of experimental analyses of natural herbs at a molecular level(time-consuming,risky,and costly),the molecular mechanism of Lonicera japonica against influenza remains poorly understood.
Computational drug discovery approaches are considered an effective alternative strategy to overcome these limitations.Structure-based drug design(SBDD)is one of the computational approaches,and virtual screening based on molecular docking has become one of the most widely used SBDD method[21].Molecular docking is one solution to explore the possible binding sites of the target in a non-covalent fashion[22,23].It has been most commonly used in the field of drug discovery,such as for determining candidate inhibitors for influenza NA,and is an efficient approach for elucidating the molecular mechanism of anti-influenza action of natural herbs[24].
Lonicera japonica is one of the herbal medicines with anti-influenza virus activities.Previous studies have shown that the anti-influenza activity of Lonicera japonica could be attributed to chlorogenic acid,isochlorogenic acid,and their isomers[25].In this study,we aim to determine the molecular targets of organic acids in L.japonica and to explore the pharmacological effect through a multi-component and multi-target approach.First,the organic acids of Lonicera japonica were virtually screened to elucidate their potential targets.As this part of research,all the vital proteins of influenza virus,such as N1,N2,N3,N4,N5,N6,N8,H1,H2,H7,PB2 and M2,were chosen as potential targets for the screening of organic acids.Second,we constructed compound-target maps to examine the anti-influenza activity and the relationship between herbal medicines and their potential targets.In addition,studies were performed to investigate as to why Lonicera japonica produces universal inhibition of influenza virus mutants and has fewer incidences of resistance.Since N1 and N9 subtypes of influenza virus cause pandemic influenza worldwide,we chose 3 subtypes of N1[26,27]and N9[28,29]as the potential targets in our study.In this study,we explored the potential targets of organic acids in Lonicera japonica and constructed a network to elucidate the anti-influenza virus effects of Lonicera japonica,which will help in the understanding of the biological basis of natural herbs via a multicomponent and multi-target approach.
Methods
Data preparation
We focused on emerged subtypes of NA as the targets,which included 3 subtypes of N1,two mutant substitutions:I223R and H274Y(PDB ID:4B7J and 3CL0)and one non-mutant(PDB ID:3TI3).We also focused on 3 subtypes of N9,two mutant substitutions:R294K and R292K(PDB ID:4MWW and 2QWA)and one non-mutant(PDB ID:4MWV);N2,N3,N4,N5,N6,and N8,6 different subtypes of NA(PDB ID:4GZP,4HZX,2HTW,3SAN,2CML,and 2HTQ);H1,H2,and H7,3 different subtypes of HA(PDB ID:1RUZ,2WRO,and 1TI8);M2 proton channel,and PB2 polymerase protein(PDB ID:2RLF and 4CB6).3D structures of all the proteins were retrieved from RCSB Protein Data Bank(PDB)(http://www.rcsb.org/)[30].Proteins were prepared for molecular docking by removal of metal ions,water molecules,cofactors,additional charges,and hydrogen atoms by using sybyl-x1.1 software.Energy of the target protein was minimized prior to molecular docking.
The structures of organic acids in Lonicera japonica and oseltamivir carboxylate (OTV, the active metabolite of oseltamivir had inhibitory effect on influenza neuraminidase)were derived independently from NCBI PubChem Compound(http://www.ncbi.nlm.nih.gov/pccompound)[31]and TcmSP(Traditional Chinese Medicines for Systems Pharmacology Database and Analysis Platform http://lsp.nwsuaf.edu.cn/tcmsp.php ) [32].Organic acids used in this study include chlorogenic acid(compound A),isochlorogenic acid A,B,and C(compounds F,D,and E),and their isomers 4-caffeoylquinic acid and 5-caffeoylquinic acid(compounds C and B)(table 1).All 3D structures of those compounds were built with ChemBioOffice 2008.Conformations of all ingredients were generated using Confort™.
Target prediction and validation
Target prediction was performed for organic acids in Lonicera japonica by using the sybyl-x1.1.Organic acids of Lonicera japonica were virtually screened.First,3D structure construction and modeling were performed with the sybyl-x1.1[33].Second,energy minimizations were performed using the Tripos force field with a distance-dependent dielectric and the Powell conjugate gradient algorithm by using a convergence criterion of 0.01 kcal/(mol A).PDB Sum was utilized to determine the binding sites with which the ligand can bind the receptor.Finally,each compound was docked to the active sites of different NA,HA,M2,and PB2 proteins by using Surflex-Dock Program.The docking scores and binding energies of docked ligands were estimated to determine potential targets of each compound.In order to validate the reliability of docking results,oseltamivir,the known xxx influenza neuraminidase inhibitor was used as the reference standard.
Disease
The therapeutic efficacy of natural herbs depends on multiple components as opposed to multiple targets.While,the complexity of the herbs becomes a huge obstacle for elucidating its molecular mechanisms.To further clarify the relationship between the ingredients of Lonicera japonica and their relative targets,we constructed the compound-target network by using Cytoscape [34].Compound–target interactions in which a compound and a target protein are connected to each other if the protein is a potential target of the compound, giving rise to a ‘compound–target network’.
Results
Predicted targets of organic acids in Lonicera japonica.
In this study,virtual screening was applied to predict the targets of each organic acid.The 3D structures of N1,N2,N3,N4,N5,N6,N8,N9,HA,M2 and PB2 proteins were downloaded from the PDB database.The structures of organic acids are shown in Table 1.Normally,docking score≥5 was representative of a potential target[35].Based on this,each compound was found to be effective against almost all the tested targets(Table 2).
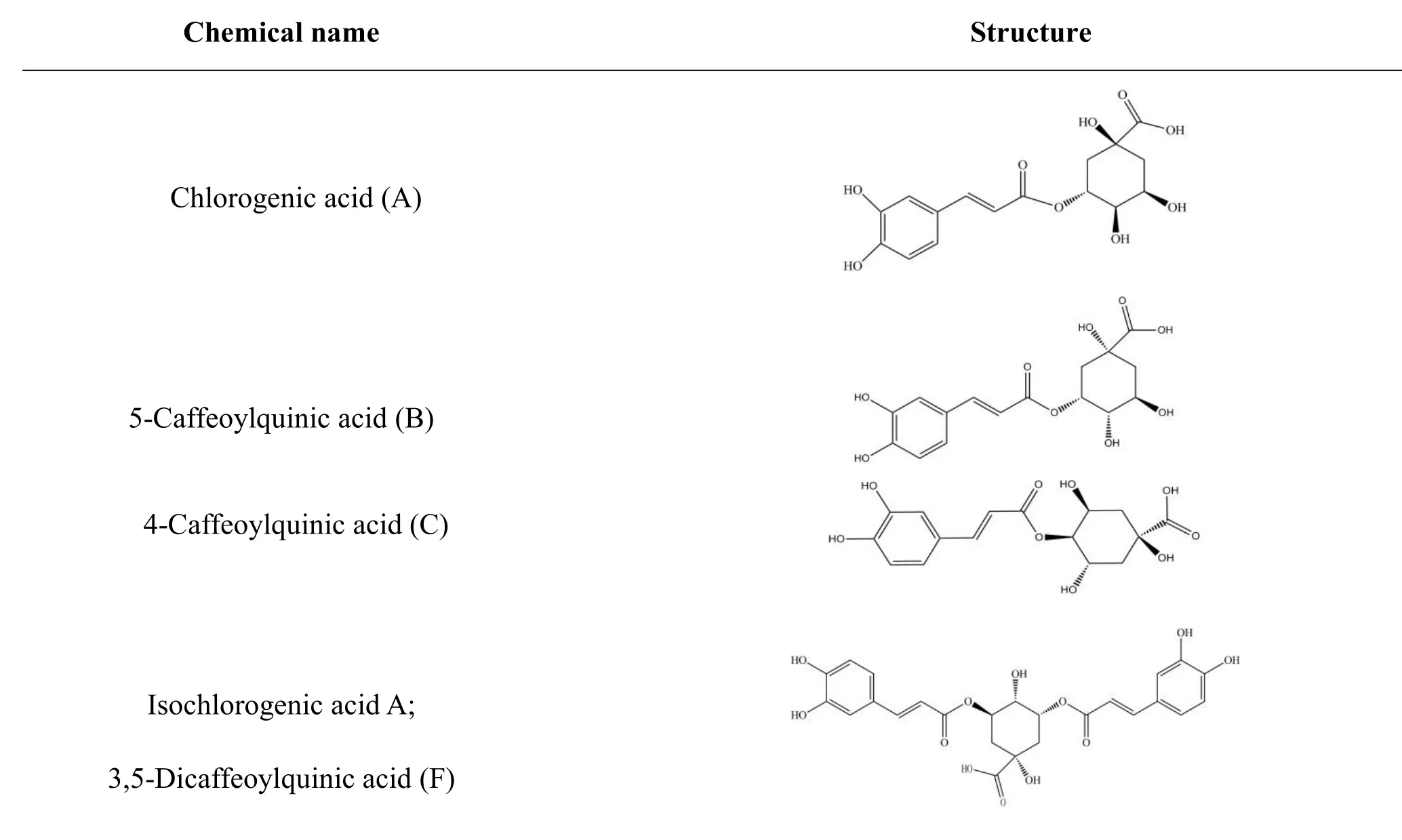
Table 1 Structures of six selected compounds(organic acids)in Lonicera japonica.
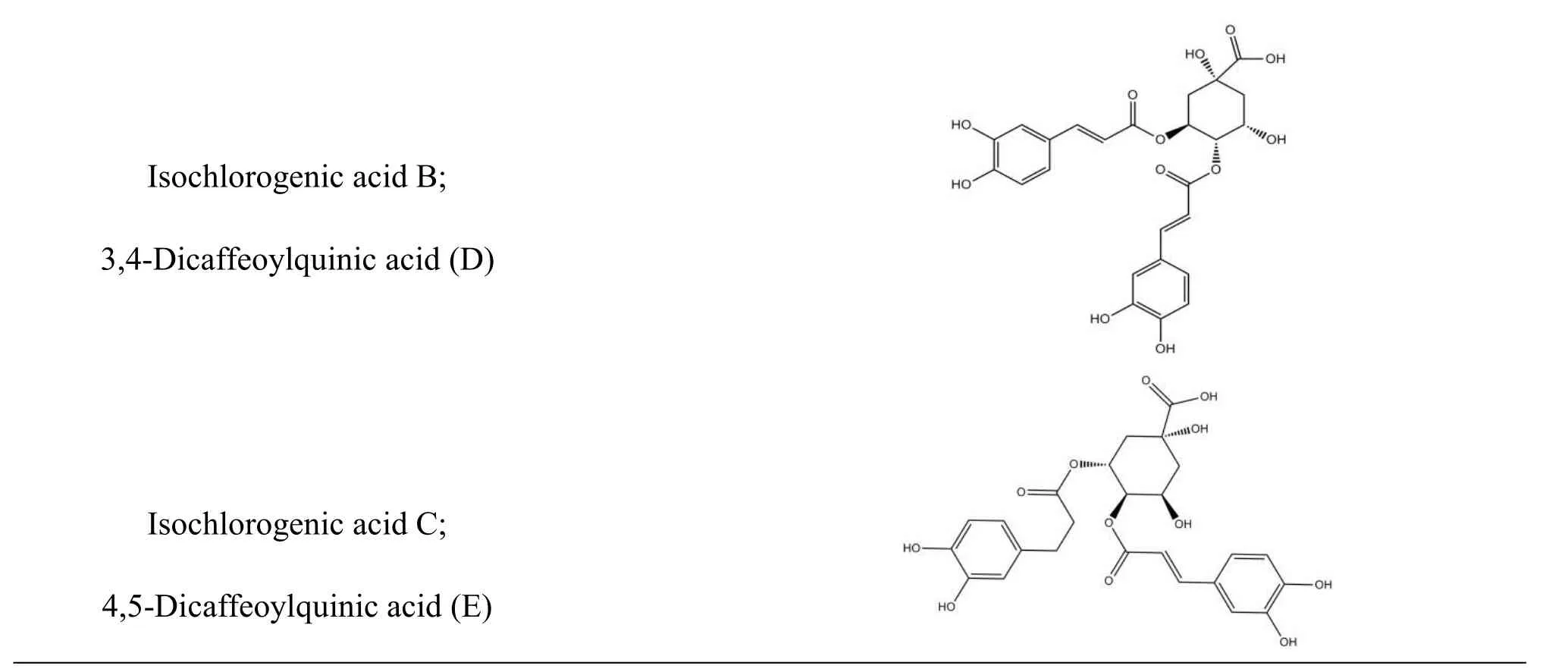
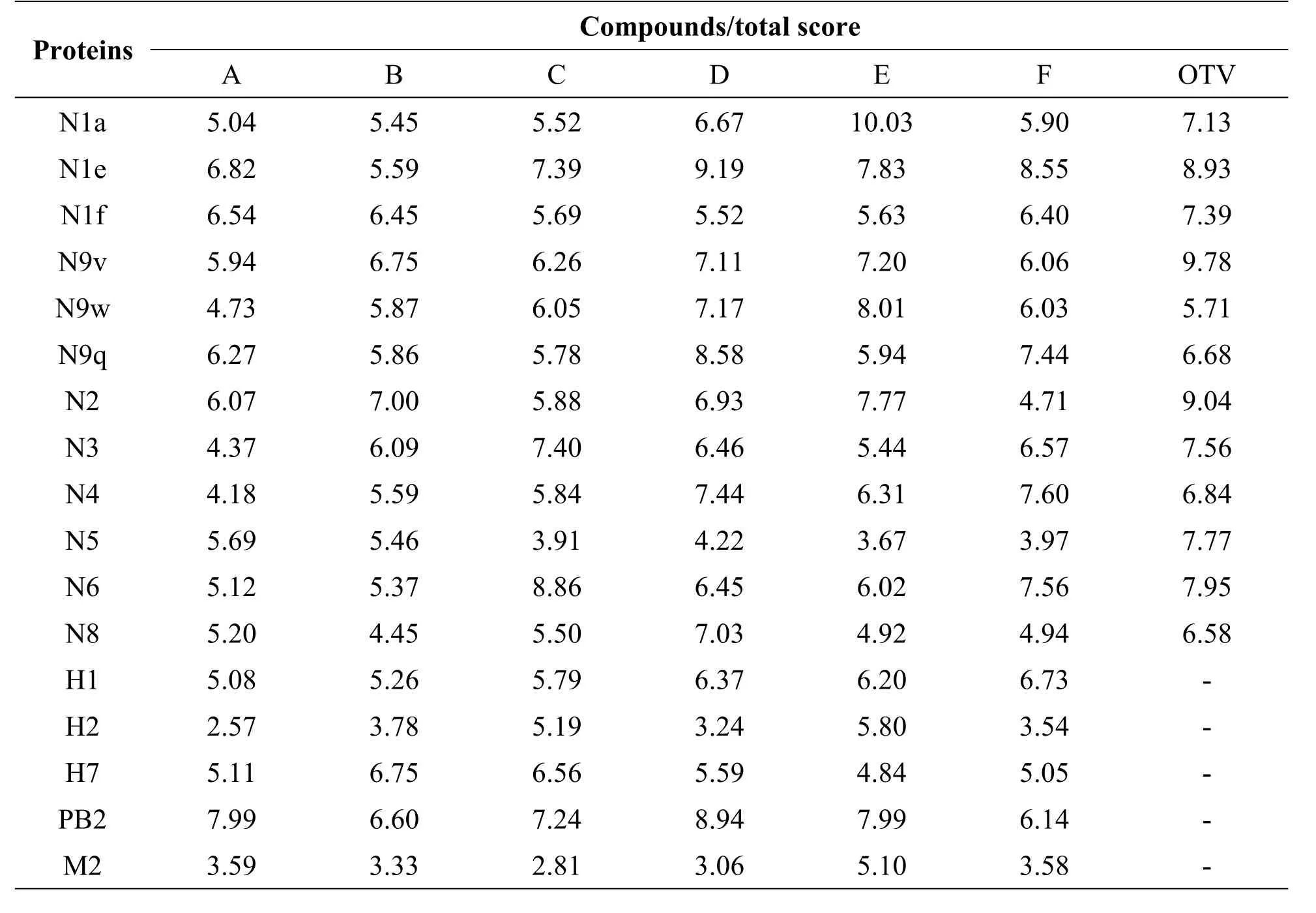
Table 2 Virtual binding scores of chlorogenic acid,isochlorogenic acid and their isomers with proteins of influenza virus;different subtypes of NA and HA,M 2 and PB2 proteins.
In order to determine the optimal potential targets of each compound,oseltamivir carboxylate(OTV),the active metabolite of oseltamivir was chosen as the positive control. As shown in Table 2,the docking scores of Eto N1a(10.03)and N9w(8.01)are much higher than those for the positive control OTV to N1a(7.13)and N9w(5.71).This showed that compound E is a better inhibitor of N1a and N9w than OTV.Moreover,the docking scores of D to N4(7.44),and N8(7.03)are higher than that of OTV to N4 and N8(6.84,6.58).Furthermore,the docking scores of D to PB2(8.94)are above 5.Overall,the respective targets of each compound were discovered.In addition,some of the compounds showed a better inhibition when bound to a certain target than OTV.The results suggested that Lonicera japonica inhibited the influenza virus in a multi-component versus multi-target manner.
Compound–Target network for Lonicera japonica.
In this study,we made progress in multi-component and multi-target analysis of anti-influenza virus activity of Lonicera japonica.The Compound-Target network was constructed for the organic acids of Lonicera japonica and their potential targets(Figure 1).This network comprises 24 nodes and 79 edges.The global view of this network as in Figure 1 showed different numbers of interactions of compounds with their potential targets.For example,most of the targets were hit by more than one compound:N9w was hit by five compounds(B,C,D,E,and F)while N6,PB2,and other subtypes of N1 and N9 were hit by six compounds(A,B,C,D,E,and F).Similarly,each compound targeted more than one protein.N1a,N1e,N1f,N9v,N9w,N9q,N2,N3,N4,N6,N8,H7,H1,and PB2 are the potential targets of compound D as well as compound B.However,M2 is an exception in this network since it has only one compound interaction.Taken together,our study revealed that Lonicera japonica inhibit the influenza virus in a multi-component versus multi-target manner,wherein two or more active ingredients simultaneously target multiple proteins.That is,the inhibitory effect of Lonicera japonica to the influenza virus depends on multiple components as opposed to multiple targets
Organic acids showed effective inhibition of N1 and N9 mutants.
Lonicera japonica has been used clinically for the treatment and prevention of infection caused by influenza A mutants.To validate this,molecular docking was performed to compare their binding modes with that of OTV.The results of docking organic acids to mutants of N1 and N9 are shown in Figure 2.First,the binding scores of compound D,E,and F docking to N1 and N9 showed no statistical differences compared to that of OTV(P>0.05).We noticed that some compounds also exhibited better inhibition of the mutants than OTV.For example,the docking scores of E to N1a(10.03)and N9w(8.01)were much higher than those of OTV to N1a and N9w.The docking scores of D to N1e(9.19),N9w(7.17)and N9q(8.58)were similar.These results provide good explanations as to why Lonicera japonica is effective in the treatment and prevention of infections caused by influenza A mutants.Furthermore,three molecules(D,E,and F)from Lonicera japonica were predicted as effective inhibitors of N1 and N9 as well as their mutants.These compounds could be crucial in dealing with the rapid mutation rate of influenza virus.
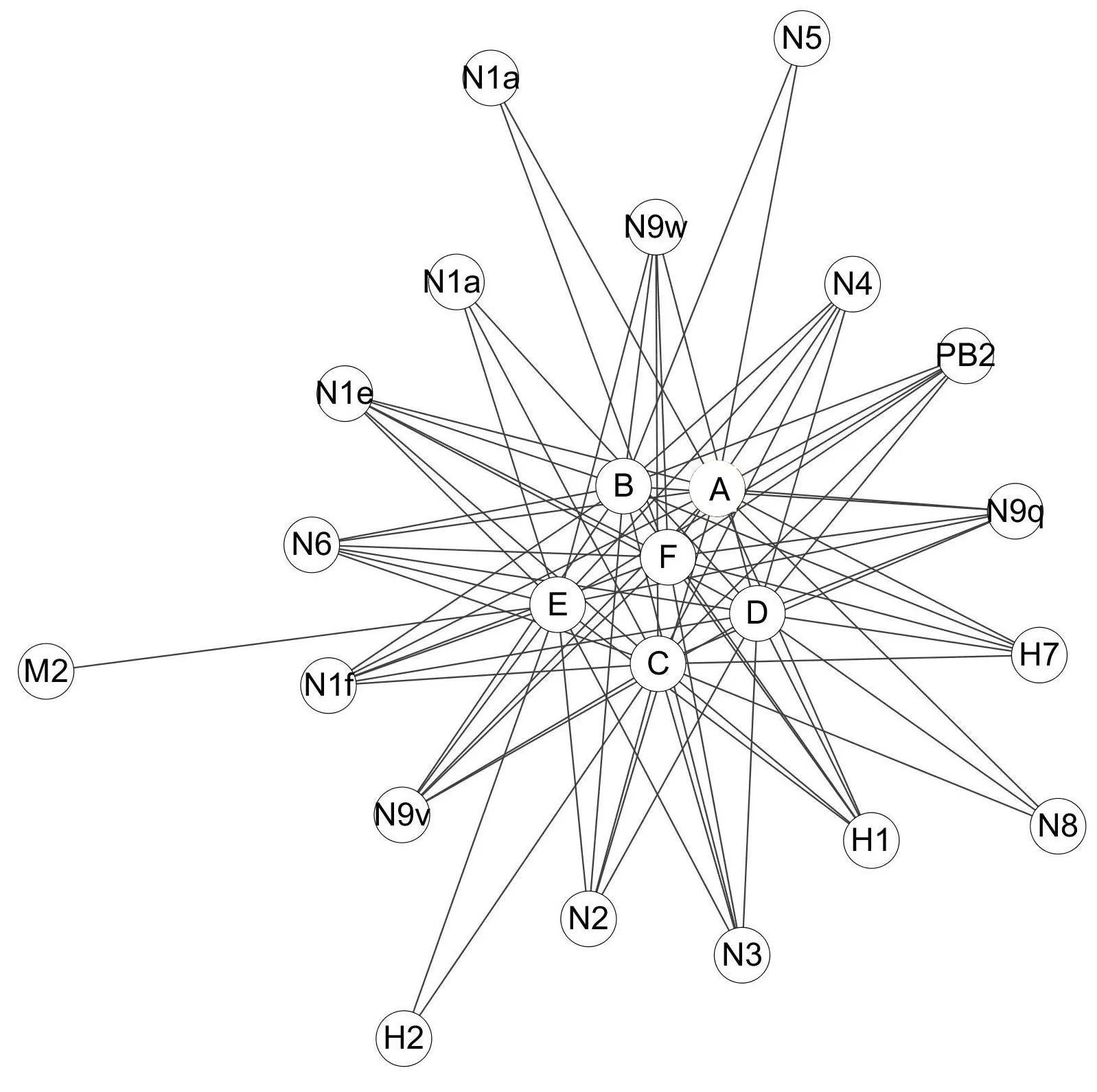
Figure 1 The Compound-Target(C-T)network of Lonicera japonica organic acids and their potential targets.A,B,C,D,E and F represent different organic acids.Proteins that considered as their potential targets are represented by N1,N2,N3,N4,N5,N6,N8,N9,H1,H2,H7,M 2 and PB2.
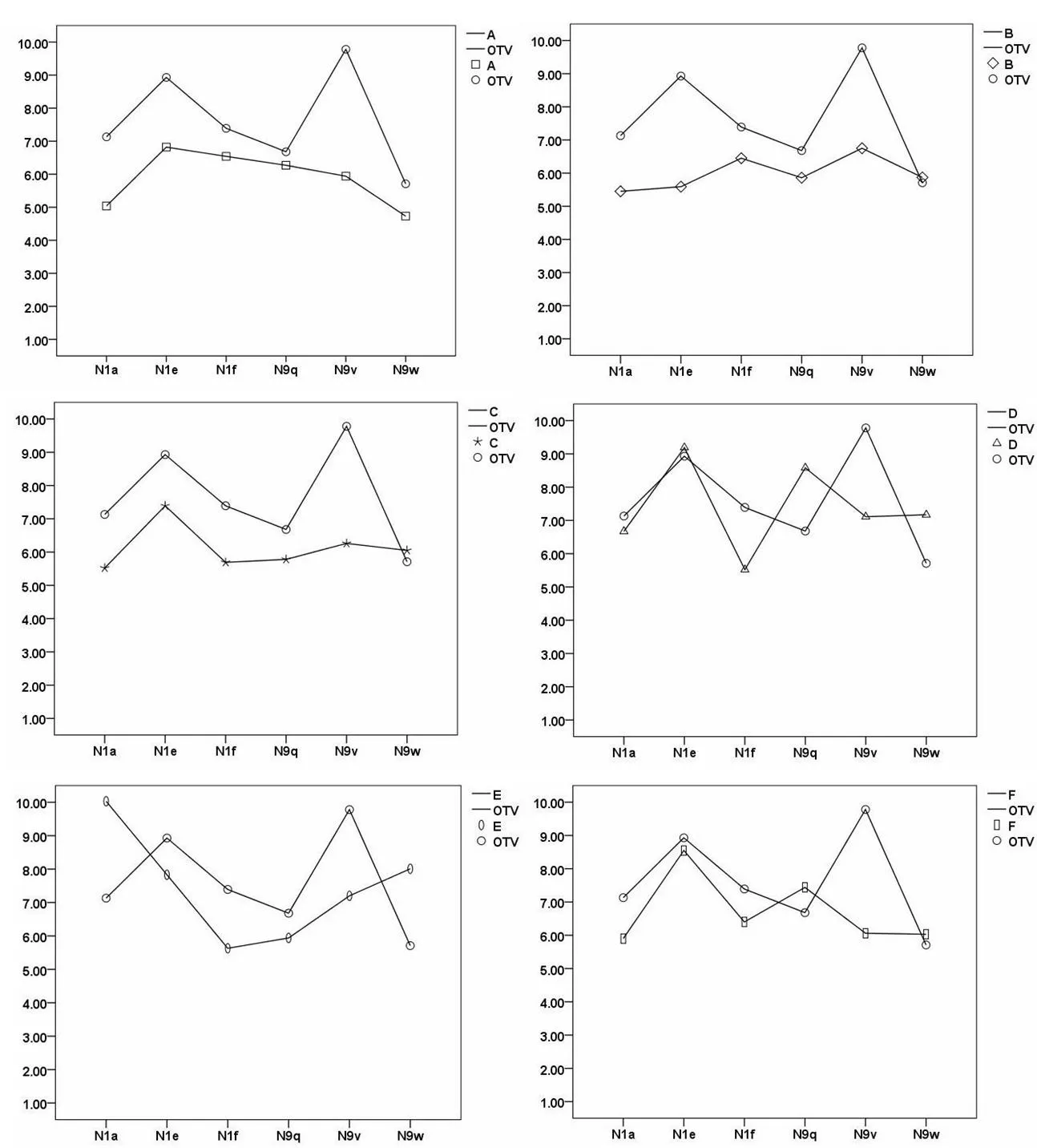
Figure 2 Molecular docking analyses were performed to testify the interactions of organic acids with mutants of N1 and N9 compared with that of oseltamivir carboxylate(displayed as OTV).Y-axis shows the binding score of compounds and x-axis shows the different subtypes of N1 and N9.The letters represent organic acids,chlorogenic acid(compound A),isochlorogenic acid A,B and C(compound F,D and E)and their isomers 4-caffeoylquinic acid and 5-caffeoylquinic acid(compound C and B).The icons£,◇,Ô,△,0, and represent the total scores(docking scores)of compound A,B,C,D,E,Fand OTV respectively.
Discussion
Natural herbs have been used to treat influenza for hundreds of years in Southeast Asia.In the history of herbal medicine against influenza virus,Lonicera japonica has been considered very effective for both the treatment and prevention of influenza[19,20].Numerous studies have been performed to explain and elaborate the underlying molecular mechanisms of Lonicera japonica.However,the complexity of component composition in natural herbs has always been the obstacle to this kind of research.In our work,we developed and employed a computational method to study the molecular mechanism of anti-influenza effect of Lonicera japonica.
Virtual screens were applied to predict the targets of each organic acid.The result showed that each compound could inhibit all tested targets(Table 2).Targets selected herein are vital to the influenza virus life cycle.These results showed why Lonicera japonica could be useful for the treatment of infections caused by different subtypes of influenza virus;the function of Lonicera japonica has been validated in the clinics.However,our results showed that different compounds have different inhibitory activities against different target proteins.Only some of the compounds showed a better inhibition than OTV.Moreover,the inhibitory abilities of 6 organic acids as a whole showed no statistical difference compared to that of OTV.This suggested that inhibition by a single compound against one target might not be enough for prevention of infection caused by all subtypes of influenza virus, which might need multiple components for inhibition.Recently,new subtypes of influenza viruses have been reported that could infect humans with a limited infectious ability[1,5,36].It has been proved that Lonicera japonica could be used to develop an efficient combinatorial intervention through mechanistic redundancy in order to deal with the newly emerged mutants.This highly flexible redundant mechanism that exists in plants will make herbal medicine one of the most promising and effective choices in clinical trials in the future.
The combinational therapy with different herbal medicines against resistant microbes has existed in China for thousands of years[18,19].To uncover the molecular mechanism of combinational effects of Lonicera japonica for the treatment and prevention of influenza virus infection,we developed Compound-Target network to further clarify multiple compounds versus multiple targets effects(Figure 1).Further analysis of the network showed that the organic acids target not only NA and HA,which is considered as the most attractive drug targets of influenza virus[3],but also other proteins(PB2,M2)[6,7],which is proven to be vital in the life cycle of the virus.Notably,the organic acids studied in the present study have been experimentally proven as the active ingredients in Lonicera japonica[25].In addition,it is suggested that the combination of multiple compounds probably are effective at low concentration,which is evidently safer than single-compound drug and causes less potential drug resistance.It appears that the anti-influenza virus activity of Lonicera japonica can be attributed to the synergistic effect contributed by multiple components and less potential drug resistance.
Influenza virus is difficult to prevent from because of its high mutation rate as well as rapid acquisition of drug resistance.The mutants of N1 and N9 subtypes of influenza virus were found to be the obstacles to epidemic prevention and control.Many current therapies do not achieve expected treatment effect because of the emergence of resistant strains caused by rapid mutation of influenza virus[12,15].The newly emerged mutated strains of H7N9 that are resistant to NA inhibitors(both oseltamivir and zanamivir)have been reported[16].In this research,three molecules(D,E,and F)from L japonica were selected and predicted as effective inhibitors of N1 and N9 as well as their mutants.Our study explains the mechanism of anti-influenza activity of Lonicera japonica at a molecular level.Owing to the diverse compounds and mechanisms of Lonicera japonica,it could be a potential candidate for the development of drugs against the newly emerged influenza virus mutants,and this could also hold true for other natural herbs.
For years,Lonicera japonica has been proven to be effective as an anti-influenza therapy.However,the molecular mechanisms of Lonicera japonica remain unclear.To elucidate the underlying mechanisms,computational methods were developed and used for target prediction and validation.Network analysis and examining the inhibitory potential against the mutants elucidated the molecular mechanism of anti-influenza virus activity of Lonicera japonica.Our results suggest that organic acids of Lonicera japonica inhibit influenza by targeting various proteins via a multi-target/multi-component approach. Such versatility is vital for providing treatments to the new emerging virus subtypes and future applications.In addition to Lonicera japonica,other herbal medicines have proven to be effective in the clinical treatment of influenza virus[19].Taken together,herbal medicines,which are still widely used in Asia,could be a potential candidate for the treatment of newly emerged subtypes of influenza virus mutants.
This study is an attempt to investigate the effect of anti-influenza virus of the organic acids from Lonicera japonica.Molecular docking programs use scoring functions to estimate the binding energetics of the predicted ligand-receptor complexes.This shows that the results obtained here through the method based on the computer simulation,which lack of experimental verification.However,this research can provide guideline and the theories basis for the further experimental study.For instance,we can construct herbal medicines pharmacology network by composition screening and target predicting based on molecular docking,and learn the molecular mechanism of herbal medicines from the pharmacology network.
Conclusion
As a botanical drug,Lonicera japonica is usually used to treat influenza in China.This research revealed the mechanism of Lonicera japonica in the inhibition of influenza virus through molecular docking.Our results suggest that Lonicera japonica is useful for the prevention and treatment of influenza and the anti-influenza effect can be attributed to its components.We found that the anti-influenza activity of Lonicera japonica occurs in a multi-component versus multi-target manner,through the actions between components.Moreover,three molecules(D,E,and F)were selected and predicted as potential inhibitors of N1 and N9 as well as their mutants.
Molecular docking was found to be an effective approach to determining the mechanism of antiinfluenza virus action of natural herbs;in this study,the activity of certain herbs on influenza viral proteins was investigated.
Acknowledgements
This research is financially supported by the Natural Science Fund of Science and Technology Department of Shaanxi Province(2014JQ4148)and the National Natural Science Foundation of China(Grant No.81002758)
Conflict of interest
The authors report no declarations of interest.
1.Molinari NA,Ortega-Sanchez IR,Messonnier ML,et al.The annual impact of seasonal influenza in the US:measuring disease burden and costs.Vaccine 2007,25(27):5086-5096.
2.Wiwanitkit V.H7N9 Influenza:The Emerging Infectious Disease.N Am J Med Sci 2013,5(7):395-398.
3.Gamblin SJ,Skehel JJ.Influenza hemagglutinin and neuraminidase membrane glycoproteins.J Biol Chem 2010,285(37):28403-28409.
4.Dawood FS,Jain S,Finelli L,et al.Emergence of a novel swine-origin influenza A(H1N1)virus in humans.N Engl JMed 2009,360(25):2605-2615.
5.Iskander J,Strikas RA,Gensheimer KF,et al.Pandemic influenza planning,United States,1978-2008.Emerg Infect Dis 2013,19(6):879-885.
6.Du QS,Huang RB,Wang SQ,et al.Designing inhibitors of M2 proton channel against H1N1 swine influenza virus.PloSone 2010,5(2):e9388.
7.Armour BL,Barnes SR,Moen SO,et al.Multi-target parallel processing approach for gene-to-structure determination of the influenza polymerase PB2 subunit.J Vis Exp 2013,(76):e4225.
8.Moscona A.Medical management of influenza infection.Annu Rev Med 2008,59:397-413.
9.Michiels B,Van Puyenbroeck K,Verhoeven V,et al.The value of neuraminidase inhibitors for the prevention and treatment of seasonal influenza:a systematic review of systematic reviews.PloSone 2013,8(4):e60348.
10.Smith JR.Oseltamivir in human avian influenza infection.J Antimicrob Chemother 2010,65 Suppl 2:ii25-ii33.
11.Heneghan CJ,Onakpoya I,Thompson M,et al.Zanamivir for influenza in adults and children:systematic review of clinical study reports and summary of regulatory comments.Brit Med J 2014,348:g2547.
12.Butler J,Hooper KA,Petrie S,et al.Estimating the fitness advantage conferred by permissive neuraminidase mutations in recent oseltamivir-resistant A,(H1N1),pdm09 influenza viruses.PLoSPathog 2014,10(4):e1004065.
13.Thorlund K,Awad T,Boivin G,et al.Systematic review of influenza resistance to the neuraminidase inhibitors.BMC Infect Dis 2011,11:134.
14.Hurt AC, Holien JK, Parker M,et al.Zanamivir-resistant influenza viruses with a novel neuraminidase mutation.J Virol 2009,83(20):10366-10373.
15.Yamaya M,Nadine L,Kubo H,et al.The effects of neuraminidase inhibitors on the release of oseltamivir-sensitive and oseltamivir-resistant influenza viruses from primary cultures of human tracheal epithelium.J Med Virol 2015,87(1):25-34.
16.Salathe M,Freifeld CC,Mekaru SR,et al.Influenza A(H7N9)and the importance of digital epidemiology.N Engl J Med 2013,369(5):401-404.
17.Wang YT,Chan CH,Su ZY,et al.Homology modeling,docking,and molecular dynamics reveal HR1039 as a potent inhibitor of 2009 A(H1N1)influenza neuraminidase.Biophys Chem 2010,147(1-2):74-80.
18.Chen W,Lim CE,Kang HJ,et al Chinese herbal medicines for the treatment of type A H1N1 influenza:a systematic review of randomized controlled trials.PloSone 2011,6(12):e28093.
19.Li T,Peng T.Traditional Chinese herbal medicine as a source of molecules with antiviral activity.Antiviral Res 2013,97(1):1-9.
20.Shang X,Pan H,Li M,et al.Lonicera japonica Thunb.:ethnopharmacology,phytochemistry and pharmacology of an important traditional Chinese medicine.JEthnopharmacol 2011,138(1):1-21.
21.Ou-Yang SS,Lu JY,Kong XQ,et al.Computational drug discovery.Acta Pharmacol Sin 2012,33(9):1131-1140.
22.Kontoyianni M,McClellan LM,Sokol GS.Evaluation of docking performance:comparative data on docking algorithms.J Med Chem 2004,47(3):558-565.
23.Cunningham F,McPhillie MJ,Johnson AP,Fishwick CW.An in silico structure-based approach to anti-infective drug discovery.Parasitology 2014,141(1):17-27.
24.Mitrasinovic PM.Advances in the structure-based design of the influenza A neuraminidase inhibitors.Curr Drug Targets 2010,11(3):315-326.
25.Song J,Zhang HM,Guo CJ,et al.Fingerprint of anti-influenza virus constituents from Lonicerae japonicae Flos.Chin Tradit Patent Med 2011,6:033.
26.Van der Vries E,Collins PJ,Vachieri SG,et al.H1N1 2009 pandemic influenza virus:resistance of the I223R neuraminidase mutant explained by kinetic and structural analysis.PLoS Pathog 2012,8(9):e1002914.
27.Collins PJ,Haire LF,Lin YP,et al.Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants.Nature 2008,453(7199):1258-1261.
28.Wu Y,Bi Y,Vavricka CJ,et al.Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses.Cell Res 2013,23(12):1347-1355.
29.Woods CJ,Malaisree M,Long B,et al.Computational assay of H7N9 influenza neuraminidase reveals R292K mutation reduces drug binding affinity.Sci Rep 2013,3:3561.
30.Huang YH,Rose PW,Hsu CN.Citing a Data Repository:A Case Study of the Protein Data Bank.PloSone 2015,10(8):e0136631.
31.Li Q,Cheng T,Wang Y,et al.PubChem as a public resource for drug discovery.Drug Discov Today 2010,15(23-24):1052-1057.
32.Ru J,Li P,Wang J,et al.TCMSP:a database of systems pharmacology for drug discovery from herbal medicines.J Che min form 2014,6:13.
33.Ivanov AS,Skvortsov VS,Archakov AI.Computer modeling of the three-dimensional structure of full-length cytochrome B5.Vopr Med Khim 2000,46(6):615-625.
34.Smoot ME,Ono K,Ruscheinski J,et al.Cytoscape 2.8:new features for data integration and network visualization. Bioinformatics 2011, 27(3):431-432.
35.Jain AN. Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine.JMed Chem 2003,46(4):499-511.
 Traditional Medicine Research2016年3期
Traditional Medicine Research2016年3期
- Traditional Medicine Research的其它文章
- Explore the medication rules of Chinese medicine of Professor Huang Wenzheng for the treatment of chronic kidney disease based on data mining
- Feiji Recipe Reverses Lung Cancer Immune Escapeby Inhibiting the Expression of IDO
- Uncertainty profile for NIR analysis of tanshinone Icontent in tanshinone extract powders
- Molecular docking study on the molecular mechanism of rhaponticin for treatment of chronic myelocytic leukemia
- The present treatment and mechanism progress of arsenic trioxide in hematological malignancies
- Traditional Chinese medicine should not beignored during the development of precision medicine with Chinese characteristics
