Genetic Variation Analysis of Watermelon Genomes with Different Ploidy
, , , , *
1. Tropical Crops Genetic Resources Institute, Chinese Academy of Tropical Agricultural Sciences/Key Laboratory of Crop Gene Resources and Germplasm Enhancement in Southern China, Ministry of Agriculture, Danzhou 571737, China; 2.College of Horticulture and Landscape Architecture, Hainan University, Haikou 570228, China
1 Introduction
Polyploidization, a form of plant’s adaptation to environment, has a significant impact on species diversity[1]. About 70% of angiosperms in nature experience polyploidization in evolutionary history[2]. Due to increase in the number of chromosomes and gene interaction, there are changes in the chromosome structure and gene expression patterns, finally showing difference in morphological feature[3]. Polyploid plants have organs, significantly increased biomass and strong ability to adapt to the environment[4-5], often showing drought-resistant, cold-resistant and disease-resistant features, in line with the needs of agricultural production. Polyploid material has a very important value in the study of crop genetic improvement and new variety breeding, so it is of great importance to analyze the genetic variation of polyploid material[6]. As for the differences in plant diploid and tetraploid genome sequences, different researchers draw varying conclusions. Jiao Fengetal.[7]use RAPD markers to analyze the differences in mulberry diploid and tetraploid genomes, but do not find polymorphic bands. Wang Zhuoweietal.[8]use AFLP markers to compare the differences in mulberry diploid and tetraploid genomes, and results show that there are some changes in the genome sequence of them. Nie Lijuanetal.[9]use AFLP markers to detect changes in watermelon diploid and tetraploid genomes. Compared with RAPD and AFLP markers, SSR and InDel markers have advantages of simple operation, codominant inheritance, amplification stability and good reproducibility, so the result is more accurate and reliable. In recent years, as the watermelon genome sequencing and resequencing studies are carried out, it has provided favorable conditions for the development and application of watermelon SSR and InDel markers[10]. Up to now, the use of SSR and InDel markers to analyze the differences in watermelon genomes with different ploidy is rarely reported. In this study, with the watermelon of different ploidy as test material, we use SSR and InDel molecular marker technology to study the genetic variation of watermelon genomes with different ploidy, which is conducive to explaining the formation mechanism of polyploid superior agronomic traits, thereby providing a scientific basis for watermelon ploidy breeding, new variety improvement and germplasm innovation.
2 Materials and methods
2.1TestmaterialsThe material was the elite inbred line of diploid watermelon FR-32-1B (2N) (round fruit, red flesh, medium size), bred by Tropical Crops Genetic Resources Institute, Chinese Academy of Tropical Agricultural Sciences after years of breeding. FR-32-1B (2N) was induced by oryzalin, and the homologous tetraploid material FR-32-1B (4N) was identified by root tip chromosome counting and flow cytometry ploidy detection. With FR-32-1B (4N) as female parent, FR-32-1B (2N) as male parent, the cross was made up to obtain the triploid material FR-32-1B (3N). The materials were sown in nutrition bowl.
2.2Methods
2.2.1Genomic DNA extraction. 10 plants were selected for each ploidy, and equal amount of young leaves were taken and mixed for genomic DNA extraction. The CTAB method of Sharpetal.[11]was used to extract genomic DNA of diploid material FR-32-1B (2N), triploid material FR-32-1B (3N) and tetraploid material FR-32-1B (4N).
2.2.2Analysis of polymorphic loci. Based on the developed watermelon SSR, InDel markers and established genetic linkage map[12], 120 pairs of SSR markers and 63 pairs of InDel markers evenly distributed on 11 watermelon chromosomes were selected. 25 pairs were on chromosome 1; 13 pairs were on chromosome 2; 16 pairs were on chromosome 3; 11 pairs were on chromosome 4; 22 pairs were on chromosome 5; 12 pairs were on chromosome 6; 14 pairs were on chromosome 7; 7 pairs were on chromosome 8; 27 pairs were on chromosome 9; 24 pairs were on chromosome 10; 12 pairs were on chromosome 11. The genomic DNA of FR-32-1B (2N), FR-32-1B (3N) and FR-32-1B (4N) was regarded as template to amplify and screen polymorphic SSR and InDel markers. PCR reaction system (10 μL) included 8 mmol L-1Tris-HCl (pH 7.6), 50 mmol L-1KCl, 1.0 mmol L-1MgCl2, 0.20 mmol L-1dNTPs, 50 ng primer, 0.60 UTaqDNA polymerase, and 50 ng template DNA. The amplification program: 94℃ initial denaturation for 5 min; 94℃ denaturation for 35 s; 55℃ annealing for 35 s; 72℃ extension for 1 min, 35 cycles; 72℃ final extension for 8 min. PCR product was stored at 10℃. 4 μL of amplification product was mixed with 2 μL of loading buffer solution for 4 h of 200 V electrophoresis by 12% non-denaturing polyacrylamide gel (acrylamide: bis-acrylamide=29: 1), and the band was stained and analyzed.
2.2.3Gene prediction and functional annotation in polymorphic interval. Using watermelon line 97103 genome database (http://www.icugi.org/cgi-bin/gb2/gbrowse/watermelon_v1/), the polymorphic interval was identified; using NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi), the functional annotation was conducted on the genes in the polymorphic interval.
3 Results and analysis
3.1DNAextractionanddetectionUsing modified CTAB method, the genomic DNA of watermelon with different ploidy was extracted, and the gel electrophoresis results showed that DNA bands were clear, having good integrity and high purity, without trailing and degrading effect. The ratio of A260 to A280 was 1.90, indicating that the extracted DNA was not contaminated by proteins, polysaccharides and RNA, fully meeting the requirements of the experiment.

M:DL1000 DNA Marker, 1:FR-32-1B (2N), 2:FR-32-1B (3N), 3:FR-32-1B (4N)
Fig.1GenomicDNAdetectionofwatermelonwithdifferentploidy
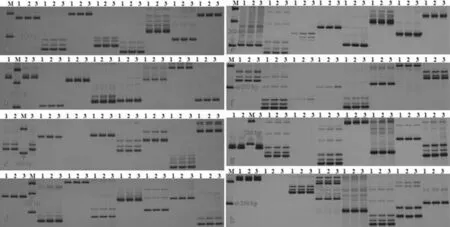

M:DL2000 DNA Marker,1:FR-32-1B (2N), 2:FR-32-1B (3N), 3:FR-32-1B (4N)
Fig.2GenomicSSRanalysisofwatermelonwithdifferentploidy
Table1PolymorphicSSRprimersequencefordifferentploidywatermelongenomes

NameForwardprimersequence(5-3)Reverseprimersequence(5-3)BVWS02494TGCCGTTTGGATCACATAGACAATACGCACAAAAAGCGAABVWS00209TGCTTCAAAATCTATTCACAATTTGCTTCTTGGTTTCGGGTTTCTTTACA

M:DL2000 DNA Marker,1:FR-32-1B (2N), 2:FR-32-1B (3N), 3:FR-32-1B (4N)
Fig.3GenomicInDelanalysisofwatermelonwithdifferentploidy
3.2GenomicSSRanalysisofwatermelonwithdifferentploidyWith genomic DNA of watermelon with different ploidy as template, 120 pairs of SSR markers uniformly distributed on 11 watermelon chromosomes were selected for amplification, and 113 pairs of primers could amplify clear bands. The amplified band pattern of 111 pairs of primers was consistent for different ploidy watermelon materials, and the polymorphic fragments were not found, indicating that the 111 pairs of primers had the same binding site among the test materials, and the target region had the same number of repeating units. In diploid and tetraploid, 2 pairs of SSR markers (BVWS00209 and BVWS02494) showed significant polymorphism, accounting for 1.67% of all primers (Fig. 2, Table 1). These polymorphic loci may be related to chromosome breakage and repair, mitosis and meiotic mutation, transposon deletion and insertion, and gene translocation.
3.3GenomicInDelanalysisofwatermelonwithdifferentploidyWith genomic DNA of different ploidy watermelon as template, 63 pairs of InDel markers evenly distributed on 11 watermelon chromosomes were selected to amplify, and the amplification results showed that there was no difference in amplified fragment size and number between different ploidy watermelon materials (Fig. 3). The results showed that there was no change in genomic insertion and deletion site before and after watermelon diploid doubling.
3.4Genefunctionalannotationinthepolymorphicinterval
The polymorphic SSR marker BVWS02494 was corresponding to the interval of watermelon chromosome 9 (34047313-34047470), and the polymorphic SSR marker BVWS00209 was corresponding to the interval of watermelon chromosome 9 (34063455-34063581). In the interval of 34047313-34063581, one gene was predicted, with gene code of Cla005553. This gene encoded a zinc finger protein, and it was likely to regulate gene expression at the transcriptional and translational level by the specific binding with target molecule DNA, RNA, DNA-RNA sequence, thereby participating in cell differentiation, embryonic development and other life processes.

Fig.4Genepredictioninthepolymorphicgenomicregion
4 Discussions
Polyploid breeding is an important area of plant genetics and breeding. Since the 1930s, China’s polyploid application has been wide and there have been great social and economic benefits[13]. Polyploidization results in partial or complete duplication of genome. However, it is not the passive simple fusion of two genomes, and it involves a wide range of molecular and physiological adjustments. Numerous studies show that compared with diploid, there are varying degrees of variation in the expression pattern and level of polyploid genomes[14]. In the process of polyploidization of different species, there are differences in the genomic change level and manner. Compared with the diploid, there is slight variation in the polyploid genomes of cotton[15]and gorse[16], and the polyploid genomes of pomelo[17]undergo great loss after polyploidization. The genomes of "Zaohong 3" loquat also undergo significant correction after polyploidization[6]. The cause of this result may be related to the difference in mutagens, dosages and materials used by different researchers in induction of tetraploid. In this study, SSR and InDel markers were used to conduct genetic variation analysis on different ploidy watermelon genomes, and the results showed that there were also some differences in DNA level between watermelon tetraploid and diploid, indicating that in the induction of watermelon chromosome doubling, it caused changes in the nucleotide sequence of genomic DNA. The small changes in genomic sequence before and after watermelon chromosome doubling might be related to inadequate application of oryzalin in the induction of watermelon tetraploid. Plant polyploids often exhibit some morphological or physiological traits significantly different from the parental diploid. Then how are the traits of polyploid different from those of the parental diploid formed? Recent studies have found that it may be closely related to another regulation way (epigenetic regulation)[18]. Epigenetic regulation is the genetic regulation on gene expression without changing the gene sequence[19-20], including DNA methylation, histone modification and non-coding RNA regulation. And DNA methylation is a major epigenetic modification form[21]. Related studies show that the DNA methylation level of P.fortuneitetraploid is significantly higher than that of diploid, and DNA methylation may be one of the main reasons for the lack of doubling in morphological characters of tetraploid[22]. This conclusion has been verified in pear[23],Eragrostiscurvula[24]andPaspalumnotatum[25]. The next research step is to explore the mechanism of difference in traits between watermelon polyploid and diploid from the perspective of epigenetics.
[1] ADAMS KL, WENDEL JF. Polyploidy and genome evolution in plants[J]. Current Opinion in Plant Biology, 2005, 8(2): 135-141.
[2] WENDEL JF. Genome evolution in polyploids[J]. Plant Molecular Biology, 2000, 42: 225-249.
[3] ZHANG XQ, CHEN JF, LEI C,etal.AFLP analysis of genetic differences among cucumber materials with different pioidies[J]. Acta Botanica Boreali-Occidentalia Sinica,2006,26(11):2265-2269. (in Chinese).
[4] HILU KW. Polyploidy and the evolution of domesticated plants[J]. American Journal of Botany, 1993, 80(12): 1494-1499.
[5] LIU B, WENDEL JF. Non-mendelian phenomena in allopolyploid genome evolution[J]. Current Genomics, 2002, 3(6): 489-505.
[6] BIAN Y, WANG WX, HAN GH,etal.AFLP analysis of Eriobotrya japonica genome in different ploidies[J]. Journal of Shanxi Agricultural University,2010,30(3):209-211. (in Chinese).
[7] JIAO F, LIU CF, ZHANG YZ,etal. DNA amplified polymorphism among mulberry mutants[J]. Acta Sericologica Sinica,2001,27(3):165-169. (in Chinese).
[8] WANG ZW, YU MD, LU C.The AFLP analysis of genetic diversity between diploid and artificially induced homologous tetraploid of mulberry[J]. Chinese Bulletin of Botany,2002,19(2):194-200. (in Chinese).
[9] NIE LJ, WANG ZC, WANG YF,etal. Analysis on DNA methylation diversity of diploid and tetraploid of watermelon( Citrullus Lanatus )[J]. Acta Agriculturae Nucleatae Sinica,2009,23(1):80-84. (in Chinese).
[10] GUO S, ZHANG J, SUN H,etal. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions[J]. Nature Genetics, 2013, 45(1): 51-58.
[11] SHARP PG, KREIS M, SHEWRY PR,etal. Location of β-amylase sequences in wheat and its relatives[J]. Theoretical and Applied Genetics, 1988, 75 (2): 286-290.
[12] REN Y, MCGREGOR C, ZHANG Y,etal. An integrated genetic map based on four mapping populations and quantitative trait loci associated with economically important traits in watermelon (Citrullus lanatus)[J]. BMC Plant Biology, 2014, 14(1): 33.
[13] YANG L, LI H, XIANG ZX. Analysis on DNA methylation diversity of diploid and autotetraploid of stevia rebaudiana bertoni[J]. Acta Agriculturae Nucleatae Sinica,2013,27(8):1125-1130. (in Chinese).
[14] ADAMS KL. Evolution of duplicate gene expression in polyploid and hybrid plants[J]. Journal of Heredity, 2007, 98(2): 136-141.
[15] LIU B, BRUBAKER CL, MERGEAI G,etal. Polyploid formation in cotton is not accompanied by rapid genomic changes[J]. Genome, 2001, 44(3): 321-330.
[16] BAUMEL A, AINOUCHE M, KALENDAR R,etal. Retrotransposons and genomic stability in populations of the young allopolyploid species Spartina anglica C.E. Hubbard (Poaceae).[J]. Molecular Biology and Evolution, 2002, 19(8): 1218-1227.
[17]HE B, WANG WX, XIANG SQ,etal. Genome analysis of nature and artificial polyploids by AFLP in Citrus grandis, cv. Shatianyou[J]. Southwest China Journal of Agricultural Sciences,2009,22(3):746-749. (in Chinese).
[18] CHENG ZJ, QIN RZ, ZHANG X,etal. Molecular mechanism for phenotypic mutation arisen from polyploidization in plant[J]. Acta Agronomica Sinica,2005,31(7):940-943. (in Chinese).
[19]MARTELOTTO LG, ORTIZ JPA, STEIN J,etal. A comprehensive analysis of gene expression alterations in a newly synthesized Paspalum notatum autotetraploid[J]. Plant Science, 2005, 169(1): 211-220.
[20] MARTELOTTO LG, ORTIZ JPA, STEIN J,etal. Genome rearrangements derived from autopolyploidization in Paspalum sp[J]. Plant Science, 2007, 172(5): 970-977.
[21] FINNEGAN EJ, PEACOCK WJ, DENNIS ES. DNA methylation, a key regulator of plant development and other processes[J]. Current Opinion in Genetics & Development, 2000, 10(2): 217-223.
[22] ZHAI XQ, ZHANG XS, FAN GQ,etal. Analysis of diploid and its autotetraploid Paulownia tomentosa with AFLP and MSAP[J]. Journal of Central South Forestry University,2014,34(1):89-93. (in Chinese).
[23] HU BQ, WANG CG, FANG CQ,etal. Analyzing the levels and patterns of DNA methylation in Yali pear autopolyploid[J]. Acta Scientiarum Naturalium University Nankaiensis,2011,44(2):32-37. (in Chinese).
[24] OCHOGAVIA AC, CERVIGNI G, SELVA JP,etal. Variation in cytosine methylation patterns during ploidy level conversions in Eragrostis curvula[J]. Plant Molecular Biology, 2009, 70(1-2): 17-29.
[25] RODRIGUEZ MP, CERVIGNI GDL, QUARIN CL,etal. Frequencies and variation in cytosine methylation patterns in diploid and tetraploid cytotypes of Paspalum notatum[J]. Biologia Plantarum, 2012, 56(2): 276-282.
 Asian Agricultural Research2016年10期
Asian Agricultural Research2016年10期
- Asian Agricultural Research的其它文章
- Static and Dynamic Analysis on the Environmental Efficiency of 267 Cities in China during 2004-2012
- Analysis of Patterns and Benefits of Cultivated Land Transfer in Rural Areas in the Loess Plateau
——A Case Study of Yuanzhou District of Ningxia - A Study on the Adaptability of Yunnan Tea Cultivars in Southern Fujian
- An Empirical Study on Rural Economic Growth in Hubei Province Based on New C-D Production Function
- Evaluation and Comparative Study of Industrial Competitiveness in the Beijing-Tianjin-Hebei Area
- Demands of New Professional Farmers for Agricultural Leading Industry in Anhui Province
