Cloning and Expression of Bile Salt Hydrolase Gene from Lactobacillus plantarum M1-UVS29
Yu Chang-qing, and Li Rong
1Department of Organization, Bayi Agricultural University, Daqing 163319, Heilongjiang, China
2Center of Railway Disease Prevention and Control, Harbin 150000, China
Cloning and Expression of Bile Salt Hydrolase Gene from Lactobacillus plantarum M1-UVS29
Yu Chang-qing1, and Li Rong2
1Department of Organization, Bayi Agricultural University, Daqing 163319, Heilongjiang, China
2Center of Railway Disease Prevention and Control, Harbin 150000, China
We cloned and expressed bile salt hydrolase gene of Lactobacillus plantarum M1-UVS29 in Lactococcus lactis NZ9000 successfully. Gene-specific primers for amplification of L. plantarum bsh were designed by using sequence which availabled from GenBank. The production of PCR amplicon was confirmed by sequencing and cloned into pMD18-T vector, and then recombined into expression vector pNZ8148 and yielding vector pNZ8148-BSH. pNZ8148-BSH was transferred into Lactococcus lactis NZ9000. Sequencing indicated that the cloned bsh fragment contained 995 nucleotides, and shared 99.3% sequence homology with bsh gene from L. plantarum MBUL10. Cloned bsh fragment was successfully transduced into NICE expression system and confirmed by PCR and restriction digest. Recombinant BSH protein was analyzed by SDS-PAGE. The molecular weight of BSH protein was approximately 37 ku. Activity of the expressed protein was 0.77 µmol • min-1. The successfully expressed proteins by genetic engineering technology made the function of lactic acid bacteria be abundant and laid the foundation for further researches into cholesterol-lowering lactic acid bacterium food and probiotics.
bile salt hydrolase, gene cloning, expression, Lactococcus lactis NZ9000
Introduction
Although cholesterol has an important physiological function in human body, when presents in elevated levels, it can lead to stroke and other cardiovascular diseases (Maria et al., 2006). Experiments showed that lowering serum cholesterol level helped to reduce vascular disease; therefore, methods to control serum cholesterol level were the focus of many researches. Existing drug treatments were expensive and had many adverse side effects, often with no obvious beneficial effects. Oral administration of probiotics is currently the focus of the therapeutic researches, as probiotics have obvious curative effects, which can significantly reduce serum cholesterol level and play a positive role in controlling high blood lipid level (Pereira and Gibson, 2002). Bile salt hydrolase (BSH) is an intracellular enzyme encoded by bsh gene. It is essential for microbial growth and reproduction. BSH production is mainly restricted to grampositive bacterium, including the probiotic lactic acid bacterium. BSH enzyme hydrolyzes bile salts that are conjugated with taurine and glycine, resulting in bile salts, amino acids and free cholic acids. The conjugated bile salt absorption efficiency is low, which results in high level of the conjugated bile salts being discharged in the fecal matter. This loss of conjugatedbile salts must be compensated by the body, which needs more cholesterol to synthesize bile salts to make up for the loss (Kimoto et al., 2002; Grill et al., 1995). This synthesis gradually reduces serum cholesterol level, mainly through the following two ways: through the synthesis of bile salts from cholic acid to supplement that lost in the feces, which increases cholesterol demand, or through a reduction in the solubility of cholesterol which reduces the absorption of cholesterol in the intestinal cavity. Biochemical and genetic characterization of bile salt hydrolase from the genus Lactobacillus and from Clostridium have been documented (Coleman and Hudson, 1995). However, genetic information for BSH from Bifidobacterium is still lacking. bsh gene from Bifidobacterium longum is only recently isolated and characterized. There are four known bsh genes (LP-bsh1, LP-bsh2, LP-bsh3 and LP-bsh4) in L. plantarum genome (Lambert et al., 2003; Oh et al., 2008). Over-expression and deletion of one or more of these bsh genes suggested that only LP-bsh1 is responsible for the majority of BSH activity in L. plantarum (Lambert et al., 2008). In 2009, BSH genes of Lactobacillus casei is cloned, then the structure of the gene and the sequence of protein that BSH gene translated is analyzed (Zhang et al., 2009). Bile salt tolerance of some microorganisms is possibly, due to the expression of BSH as well as some transporter proteins. These proteins are functionally related to each other and coordinate an efficient response to the stress of the bile salt exposure (Kim and Lee, 2008). Lactobacilli isolated from different sources showed BSH activity could overcome the toxic effect of bile in gut (Mc et al., 2005; Liong and Shah, 2005; Mathara et al., 2008). The gene encoding the conjugated bsh from L. plantarum was cloned and expressed in the heterologous host Lactobacillus casei LK1 with the aid of pSMA23-derived vectors (Sudhamani et al., 2008). Recently, Jones (et al., 2008) reported that BSH activity was an conserved microbial adaptation to the human gut environment, with a high level of redundancy in this ecosystem. Researches focused on the development of a genetic transformation system in Lactococcus lactis to produce food grade metabolites for using in the food and pharmaceutical industries, as well as for vaccine researches now. Here, we described the construction of Lactococcus lactis expression vector pNZ8148-BSH, and the efficient inducible expression of L. plantarum bsh gene for production of food grade BSH.
Materials and Methods
Bacterial strains and plasmids
Bile salt hydrolase gene (bsh) was isolated from L. plantarum M1-UVS29. E. coli DH5α and Lactococcus lactis NZ9000 (NIZO Food Research, Holland) were used for the expression of the target gene. The resulting transformant was grown in GM17 medium containing 10 μg • mL-1chloramphenicol at 30℃. The commercial T-cloning vector pMD18-T (TaKaRa Biomedical Inc., Dalian, China) was used for the direct cloning of PCR products. The expression vector pNZ8148 (NIZO Food Research, Holland) was used for the constitutive expression of bsh genes.
Chemicals and enzymes
Taq DNA polymerase, T4DNA ligase and restriction enzymes were purchased from Roche Applied Science (Mannheim, Germany) and TaKaRa Biomedical Inc., respectively. Plasmid extraction and PCR purification kits were provided by Invitrogen (Carlsbad, California, USA).
PCRamplificationandcloningofbsh
Primer 5.0 was used to design gene-specific primers from L. plantarum bsh sequence which availabled from GenBank (EU477374). Primers were synthesized by Sangon Biotech Co., Ltd (Shanghai, China). Primer sequences were as the followings: up primer: 5' AAT CCA TGG ATG TGT ACT GCC ATA ACT TAT C 3'; down primer: 5' AAT GAG CTC TAT TAG TTA ACT GCA TAG TAT TGT G 3'. Genomic DNA was extracted from L. plantarum M1-UVS29 and statically cultured for 12-24 h at 30℃, using CTABgenomic DNA extraction method (Sambrook et al., 2003). bsh gene was then amplified by PCR. PCR reaction was performed by a Px2 thermal cycler (Thermo-Hybaid, Middlesex, UK) with the following reaction conditions: an initial denaturation step of 5 min at 95℃, followed by 30 cycles of 50 s at 94℃, 50 s at 48℃, 1 min at 72℃, and a final cycle of 10 min at 72℃. The amplified DNA fragment was cloned into pMD-18T cloning vector and designated pMD18-BSH. The integrity of the plasmid was confirmed by PCR and double enzyme reaction, and then the inserted fragment was sequenced by Sangon Biotech Co., Ltd (Shanghai, China). The resulting sequences were analyzed by DNAMAN.
Constructionofrecombinantexpression bacteria
Recombinant plasmid pMD18-BSH was digested by NcoⅠand HindⅢ, releasing a 995 bp fragment that was then ligated downstream of the promoter region of expression vector pNZ8148, resulting in recombinant plasmid pNZ8148-BSH. pNZ8148-BSH was transformed into competent Lactococcus lactis NZ9000 by electroporation. Electroporation was carried out at 2 000 V, 25 µF and 200 Ω. Recombinant expression bacteria were statically-cultured in GM17 medium containing 10 μg • mL-1chloramphenicol at 30℃ for 24-72 h. DNA of recombinant bacteria was extracted and verified by PCR and restriction digestion.
Expressionofbsh in recombinant L. lactis NZ9000 and SDS-PAGE analysis
Recombinant pNZ8148-BSH which verified correctly was statically-cultivated in GM17 (Cm+) medium overnight. This starter culture was inoculated into fresh GM17 (Cm+) medium at a ratio of 1 : 50. This culture was statically-cultivated at 30℃ for approximately 3 h. When OD reached 0.6-0.8, 1 ng • mL-1nisin was added to the culture to induce expression. After the culture was incubated for a further 3 h, centrifuged at 12 000 r • min-1for 10 min. The cell pellet was washed twice with TBS, and then disrupted by sonication (VCX750; Sonics & Materials, Inc., Newtown, CT, USA). After 2×SDS-PAGE buffer was added to cell solution and boiled for 5-10 min, centrifuged at 12 000 r • min-1for 10 min. The resulting protein solution was analyzed by SDS-PAGE on 12% acrylamide gel. Empty vector pNZ8148/NZ9000 was used as a control for protein expression.
Measurementofexpressionproductactivity
Bile salt hydrolase activity was determined by measuring the amount of the amino acids resulting from hydrolysis of amide bond of bile salts by ninhydrin assay (Suresh Kumar et al., 2006). 1) Recombinant bacteria were incubated in GM17-THIO (GM17 with addition of 2% sodium thioglycolate) medium which contained 0.2% taurocholic acid and sodium salt at 30℃ for 24 h. 2) After cultures were centrifuged at 12 000 r • min-1for 10 min, the amount of free acid in the supernatant was analyzed (Dong, 2008). 3) After changing pH into 7 by added 1 mol • L-1NaOH into 5 mL culture fluid, we centrifuged it at 12 000 r • min-1for 10 min. 4) The supernatant was transferred into a 1.5 mL tube and pH changed into 1 with the addition of 10 mol • L-1HCL. After added ethyl acetate into the mixed solution, shocked for 2 min, then staticed and layered. Two mL upper solution which contained ethyl acetate was transferred into a clean tube and dried using nitrogen at 60℃. After dissolved the residue by immediately added 1 mL 0.01 mol • L-1NaOH into it, added 1 mL 1% furfural and 1 mL 9 mol • L-1H2SO4and mixed, and then transferred mixed solution to a 65℃ water bath for 15 min. After decreasing to room temperature, we added 2 mL glacial acetic acid and shocked. Optical density was measured at OD660nm. The amount of free acid was calculated according to the standard curve.
One unit (1 U) of bile salt hydrolase enzyme activity was defined as the amount of the enzyme required to liberate 1 µmol amino acid perminte, measured in µmol • min-1. Enzyme activity was calculated using the following formula:
BSH enzyme activity=[aa (mmol • L-1)]×V/0.1(mL)30 (min). aa was obtained using an absorbance control standard curve. V represented the reactive fluid volume for the final color liquid volume, 0.1 represented the cell culture fluid volume, 30 represented the reaction time. The experiment was generally carried out in a 10 mL volume.
Results
ConstructionandverificationofpMD18-BSH cloningvector
L. plantarum M1-UVS29 bsh gene was PCR-amplified using primers designed from L. plantarum MBUL10 bsh gene sequence available from GenBank. The resulting 995 bp amplicon (Fig. 1) was cloned into E. coli cloning vector pMD18-T and transformed into E. coli DH5α. The integrity of the recombinant vector was verified by PCR (Fig. 2) and restriction digestion with NcoⅠand HindⅢ (Fig. 3).
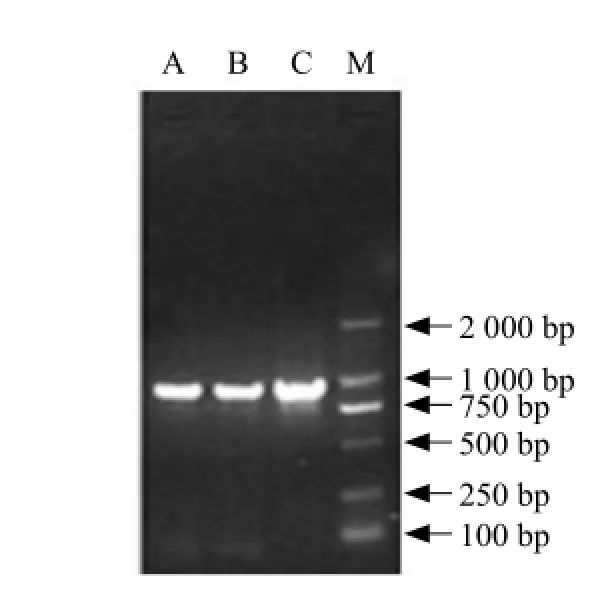
Fig. 1 Amplification of BSH gene by PCR

Fig. 2 Recombinant cloning of pMD18-BSH detected by PCR
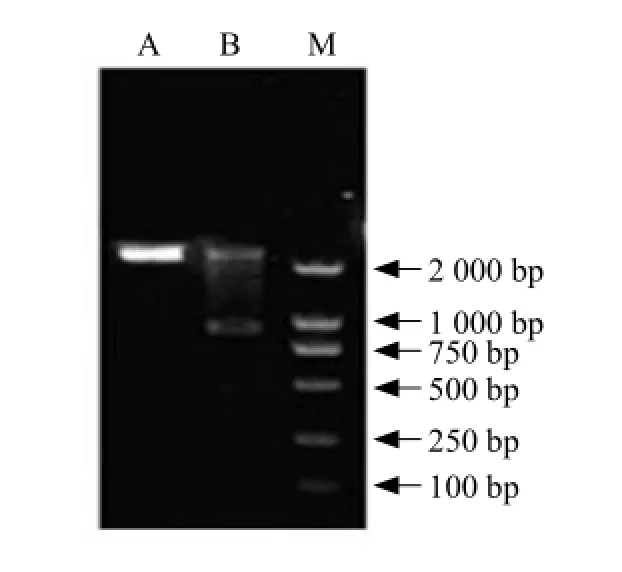
Fig. 3 Recombinant cloning of pMD18-BSH detected by double digested enzymes
Constructionandverificationofrecombinant LactococcusexpressionvectorpNZ8418-BSH
Recombinant vector pMD18-BSH was digested with NcoⅠand HindⅢ and the resulting bsh fragment ligated into Lactococcus lactis expression vector pNZ8418, resulting in vector pNZ8418-BSH. pNZ8418-BSH was then transformed into Lactococcus lactis NZ9000. Plasmid DNA was extracted from resultant colonies and the presence of bsh gene was confirmed using the gene-specific primers described in methods (Fig. 4). A specific 995 bp band was obtained, confirming the presence of bsh gene fragment. Double digestions of pNZ8418-BSH with HindⅢ and NcoⅠwere also used to confirm the integrity of the plasmid (Fig. 5).

Fig. 4 Expression vector detected by PCR
BSHexpressionin L. lactis NZ9000 and SDSPAGE analysis
Following the successful transformation of pNZ8148-BSH into Lactococcus lactis NZ9000, protein expression was induced by the addition of nisin for 3 h. Protein expression was confirmed using SDS-PAGE analysis. A band corresponding to a protein with a molecular mass of 37 ku was visible, indicating the successful expression of BSH. SDS-PAGE analysis showed that L. plantarum M1-UVS29 BSH was highly expressed in Lactococcus lactis NZ9000 (Fig. 6).
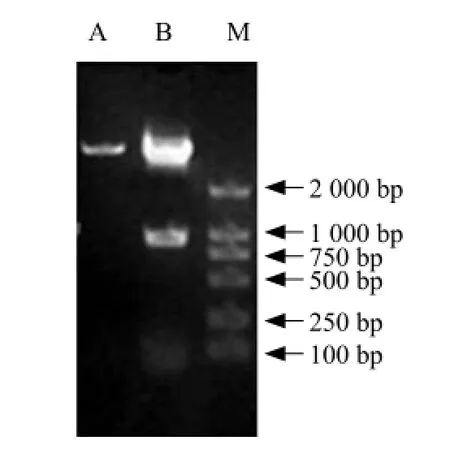
Fig. 5 Expression vector detected by double digested enzymes

Fig. 6 SDS-PAGE profile of recombinant protein
ldentificationofbilesalthydrolaseactivityin recombinant bacteria
Lactic acid bacterium (lactic acid bacteria, LAB) is a general designation of a kind of gram-positive bacterial and fermentable carbohydrate that produces large amounts of lactic acid which is the common bacterial flora within the intestinal of the man and most of the animals (Koletzko et al., 2006). Lactococcus lactis (Lactococcuslactis; L. lactis) is one of the representative strains of the lactic acid bacteria, which exists widely in nature and is recognized as the safety food grade microorganisms (Generally Regarded as Safe, GRAS) (Nieuwenhuise and Van, 2007). Previous researches had found that bile salt hydrolase was capable of hydrolyzing conjugated bile salts, thereby, decreasing serum cholesterol levels and ultimately reduced the incidence of cardiovascular disease. According to the standard curve produced in the current experiment, we calculated that the amount of the free cholic acid was 0.8524 mmol • L-1by hydrolysis of recombinant Lactococcus lactis and the activity of the expressed proteins was 0.77 µmol • min-1.
Discussion
The recombinant cloned vector which contained bsh gene was named pMD18-BSH. The inserted fragment was sequenced and analyzed by DNAMAN. Result showed that the length of inserted fragment was 995 bp and shared 99.3% identity with bsh gene from L. plantarum strain MBUL10. Lactococcus lactis pNZ8418-BSH expression vector was successfully constructed. Lactococcus lactis NZ9000 was a grampositive bacterium with a thicker cell wall, so the preparation of making competent cell was particularly important. Because the higher voltage could lead to a lower survival rate of recombinant bacteria, we transformed pNZ8418-BSH into Lactococcus lactis NZ9000 successfully by lower voltage conditions. We found the following conditions achieved the highest transformation rates: 2 000 V, 25 μF and 200 Ω, with shock time of 4 ms. Compared with the expression of BSH in E. coli that previous researchers had reported, Lactococcus lactis was frequently used for expressing heterologous proteins, because the proteins were directly expressed and without further purification. The most important was that Lactococcus lactis did not produce endotoxin or inclusion bodies. Nisin was used as an induction agent for NZ9000/pNZ8148exogenous gene receptor system. In general, very low level of nisin (0.05 ng • mL-1) could be distinguished by NisK and inducted expression of the target gene. Because nisin acts as a bacteriocin against many other bacterial species, a low induction concentration was particularly useful, so as not to affect the fermentation and growth of other bacteria. This induction effect in proportion to nisin (0.05-50 ng • mL-1) observed in Lactococcus, Enterococcus, and Lactobacillus species. In our experiment, we determined that the optimal concentration for induction of BSH was 1 ng • mL-1. The lower concentration of the nisin might be related to some factors of the recipient bacterium. pNZ8148 was a highly efficient and stable expression vector; but the presence of the chloramphenicol resistance gene did not meet standards for food grade application. In order to avoid the problem of an antibiotic resistance marker, the most effective approach was the use of food-grade selection markers. However, experiments determined that these marker systems did not have a desirable level of protein expression. A possible reason for this decreased expression might lie in the design of PCR primers. When designed the primers, NcoⅠand HindⅢ restriction enzyme sites were introduced into up and down primers. Then, the secretory signal spUSP45 was removed, thereby rendering the plasmid a non-secretory expression vector. Many studies showed that nonsecretory expression vectors were used in Lactococcus lactis, the expressed protein accumulated in the bacterial body, thereby, limiting the release of recombinant protein. This factor ultimately limited the functional capacity of the recombinant lactic acid bacteria.
Conclusions
bsh gene from Lactobacillus plantarum M1-UVS29 was cloned and expressed successfully. Recombinant bacteria had bile salt hydrolase activity by calculation. Therefore, the consumption of food containing BSH-expressing lactic acid bacteria might assist in controlling serum cholesterol levels. Food containing lactic acid bacteria had a special texture, flavor and nutritional characteristics, and had obvious therapeutic effects.
References
地震剖面投影法与气藏剖面投影法道理相同,首先将深度域的井资料转到时间域地震剖面上[4],然后再通过投影确定平面图上投影点与两井点的位置关系,进而确定出气水内外边界线。这种方法的优点同样是简单和直观,而且基于地震资料这种硬数据,对地层的产状更容易把握。缺点是各井点时深转换存在误差,一定程度上对结果产生影响。
Ballongue, Crociani J, Grill J P, Schneider F. 1995. Purification and characterisation of conjugated bile salt hydrolase from Bifidobacterium longumBB536. Appl Environ Microbiol, 61: 2577-2582.
Begley M, Hill C, Jones B V, et al. 2008. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci USA, 105(36): 1358-013585.
Bhathena J, Martoni C, Urbanska A M, et al. 2008. Microencapsulated bile salt hydrolase producing Lactobacillus reuteri for oraltargeted delivery in the gas trointestinal tract. Applied Microbiology and Biotechnology, 81(2): 225-233.
Bontems P, Crone J, Kalach N, et al. 2006. Prospective multicentre study on antibiotic resistance of Helicobacter pylori strains obtained from children living in Europe. Gut, 55: 1711-1716.
Bongers R S, deVos W M, Kleerebezem M, et al. 2008. Functional analysis of four bile salt hydrolase and penicillin acylase family members in Lactobacillus plantarum WCFS1. Appl Environ Microbiol, 74: 4719-4726.
Bongers R S, Kleerebezem M, Lambert J M. 2003. Cre-lox-based system formultiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl Environ Microbiol, 73: 1126-1135.
Branningan J A, Dodson E J, Dodson G G, et al. 2006. Structural and functional analysis of a conjugated bile salt hydrolase from Bifidobacterium longum reveals an evolutionary relationship with penicillin vacylase. Biol Chem, 43: 32516-32525.
Cano R J, Klaenhammer T R, Mc Auliffe O. 2005. Genetic analysis of two bile salt hydrolase activities in Lactobacillus acidophilus NCFM. Appl Environ Microbiol, 71: 4925-4929.
Coleman J P, Hudson L L. 1995. Cloning and characterization of a conjugated bile acid hydrolase gene from Clostridium pefringens. Appl Environ Microbiol, 61: 2514-2520.
Dong G X. 2008. Screening of bile salt hydrolase activity and application of 16Sr DNA molecular methods for identification of lactic acid bacteria. China Dairy Industry, 11: 7-10.
Geis A, Ismaiel E, Sudhamani M, et al. 2008. Characterisation of pSMA23, a 3.5 kbp plasmid of Lactobacillus casei and application for heterologous expression in Lactobacillus. Plasmid, 59(1): 11-19.
Gibson G R, Pereira D I. 2002. Effects of consumption of probiotics andprebiotics on serum lipid levels in humans. Crit Rev Biochem Mol Biol, 37: 259-281.
Guigas C, Mathara J M, Schillinger U, et al. 2008. Functional characteristics of Lactobacillus spp. from traditional Maasai fermented milk products in Kenya. Food Microbiol, 126(12): 57-64.
Hae K, Lee J Y, Oh, et al. 2008. Molecular cloning and characterization of a bile salt hydrolase from Lactobacillus acidophilusPF01. Microbiol, 18: 449-456.
Jeong D W, Lee J H, Lee J M, et al. 2008. Use of the cellulase gene as a selection marker of food-grade integration system in lactic acid bacteria. Food Science and Biotechnology, 17(6): 1221-1227.
Khan B M, Sridevi N, Srivastava S, et al. 2009. Characterization of the smallest dimeric bile salt hydrolase from a thermophile Brevibacillussp. Extremophiles Life under Extreme Conditions, 13(2): 363-370.
Kim G B, Lee B H. 2008. Genetic analysis of a bile salt hydrolase in Bifidobacterium animalis ssp. lactis KL61. Appl Microbiol, 105(3): 778-790.
Kimoto H, Ohmomo S, Okamoto T. 2002. Cholesterol removal from media by lactococci. Dairy Sci, 85: 3182-3188.
Liong M T, Shah N P. 2005. Bile salt deconjugation ability, bile salt hydrolase activity and cholesterol coprecipitation ability of Lactobacilli strains. Int Dairy, 15: 391-398.
Loos M, Steidler L, van H K. 2009. Immunomodulation by genetically engineered lactic acid bacteria. Frontiers in Bilscience, 14(6): 4825-4835.
Luis G. 2009. Lactococcus lactis as a live vector for mucosal delivery of therapeutic proteins. Human Vaccines, 5(4): 264-267.
Maria L M, Michiel K, Sonia P. 2006. Towards understanding molecular modes of probiotic action. Current Opinion in Biotechnology, 17: 204-210.
Nieuwenhuise E, Van B. 2007. Life in commercial probiotics. FEMS Immunol MedMicrobiol, 50(3): 281-283.
Peitang H, Russell D W, Sambrook J. 2003. Molecular Cloning 3. Science Press, Beijing. pp. 1252-1253.
Pereira D I A, Gibson G R. 2002. Cholesterol assimilation by lactic acid bacteria and bifidobacteria isolated from the human. gut. Appl Environ Microbiol, 68: 4689-4693.
Suresh Kumar G, Nayaka H, Dharmesh S M, et al. 2006 Free and bound phenolic antioxidants in amla (Emblica officinalis) and turmeric (Curcuma longa). J Food Comp Anal, 19: 446-452.
Sun Z H, Wu R N, Zhang W Y, et al. 2009. Molecular cloning and characterization of bile salt hydrolase in Lactobacillus casei. Annals of Microbiology, 59(4): 721-726.
S512.1 Document code: A Article ID: 1006-8104(2015)-02-0060-07
16 January 2015
Supported by the National Natural Science Fund Project (31171657); Heilongjiang Province Natural Fund Project (ZD201207); Heilongjiang Province Postdoctoral Special Funds (LBH-Q13133)
Yu Chang-qing (1969-), male, Ph. D, professor, engaged in the research of livestock products processing. E-mail: byndycq@126.com
 Journal of Northeast Agricultural University(English Edition)2015年2期
Journal of Northeast Agricultural University(English Edition)2015年2期
- Journal of Northeast Agricultural University(English Edition)的其它文章
- Factors Hindering Pakistani Farmers' Choices Towards Adoption of Crop Insurance
- Agricultural Production Structure Adjustment Scheme Evaluation and Selection Based on DEA Model for Punjab (Pakistan)
- Where Pakistan Stands Among Top Rice Exporting Countries, an Analysis of Competitiveness
- Design and Experiment of Slave Computer Control System for Applying Variable-rate Liquid Fertilizer
- Rural Power System Load Forecast Based on Principal Component Analysis
- Laboratory Observations Regarding Different Instars of Cyclosainsulana (Costa, 1834) (Araneidae ) During Developmental Stages
