Isolation and Pathogenicity Analyses on Yersinia enterocolitica from Pelteobagrus vachelli
Zhao Jing, and Wang Li
College of Life Science and Technology, Southwest University for Nationalities, Chengdu 610041, China
Isolation and Pathogenicity Analyses on Yersinia enterocolitica from Pelteobagrus vachelli
Zhao Jing, and Wang Li*
College of Life Science and Technology, Southwest University for Nationalities, Chengdu 610041, China
Yersinia enterocolitica is an important zoonotic pathogen that can induce disease outbreaks in a wide host range. Strain YER6022 was isolated from Pelteobagrus vachelli and identified using bacterial morphology and 16S rDNA sequence analysis. Five virulence factors were detected, then artificial infection experiment and histopathological method were carried out. These results showed that strain YER6022 was one of Y. enterocolitica family members. In addition, ail, ystb, virF, yadA and HPIint were dectected. In artificial infection experiment, with 80% mortality and 100% morbidity, injected Pelteobagrus vachellis showed red swollen of the anus, abdomen swelling and fim bleeding. There existed serious hyperaemia and edema in kidney, spleen, intestine and liver at the light microscope. Ultrastructural observation indicated that mitochondria of the liver, kidney, spleen and intestine swelled and mitochondrial cristae broke. The data had further shed light on its pathogenicity in Pelteobagrus vachelli. It would benefit for further studies on pathogenesis of Pelteobagrus vachelli infected with Y. enterocolitica.
Yersinia enterocolitica, isolation, pathogenicity, Pelteobagrus vachelli
Introduction
Darkbarbel catfish (Pelteobagrus vachelli) are vital economic aquaculture species in China, which poss abundant nutrition and economy. To date, a lot of bacterial diseases have been outbroken in Pelteobagrus vachelli, such as Edwardsiella ictaluri, which causes large economic losses in aquaculture breeding (Li et al., 2014). However, there is no information about Pelteobagrus vachelli infected with Yersinia enterocolitica.
Y. enterocolitica is one of the three vital pathogenic gram-negative bacteria of Yersinia strains, which can cause various clinical symptoms and pathologies, such as enteric infection and septicemia, which has been isolated from a wide variety of the sources included vertebrates and invertebrates in most countries of the world, particularly in Sweden, European Union (EU), and United States (Rosner et al., 2013; Thoerner et al., 2003; Mcnally et al., 2004). The outbreak of Y. enterocolitia has caused high mortality and great economic loss in aquaculture of China in 2013, while Y. enterocolitia has been isolated from marine fish and water environment before (Dhayanithi et al., 2010).
Y. enterocolitia is classically divided into different serotypes and six biotypes. Apart from biotype 1A, all of them are considered pathogenic to human and other animals. The potential pathogenicity of Y. enterocolitia is mainly attributed to the virulence factors locatedon chromosome and plasmid (Marceau, 2005; Marta et al., 2005). The virulence plasmid and chromosome play a crucial role in helping bacterium colonize the intestinal tract and resisting host immune mechanisms to evade infection (Pepe et al., 1995; Okwori et al., 2009; Nesbakken et al., 2007). For instance, yadA located in virulence plasmid pYV is involved in host defence and serum resistance to protect the bacteria from the immune mechanism, but its mutant will give rise to avirulence (Danica et al., 2007; Mikula et al., 2012). ail has been studied by means of the assay in vitro, having a commom to another pathogenic bacterium (Pederson et al., 1995; Uliczka et al., 2011). These plamids and chromosomes could provide adhesive properties to escape host innate immune mechanisms.
Molecular epidemiology, biotypes, serotypes and remarkable virulence factors of Y. enterocolitia have been studied on human, pig, cattle and so on, but the reports about fish were really rare (Mcnally et al., 2004; Dhayanithi et al., 2010; Marta et al., 2005). In this study, the isolate YER6022 was confirmed through molecular technology. The pathogenicity of Y. enterocolitia in Pelteobagrus vachellis was discussed using artificial infection experiment and histopathological method. It was intended to provide more information for further researches on pathogenesis and prophylactic measures in Pelteobagrus vachelli infected with Y. enterocolitia.
Materials and Methods
Bacterial isolation and culture condition
YER6022 was isolated aseptically from the liver and heart of naturally infected Pelteobagrus vachelli and cultured by nutrient agar at 28℃ for 24 h. Then, the single colony was selected randomly and incubated by Cepulodin Irgasan Novobiocin agar (CIN), which was a dedicated agar to cultured Y. enterocolitia, at 28℃ for 24 h. The pure isolates were stored at–20℃ in luria bertani (LB) with 20% glycerol for further studies.
Amplification and sequence analyses on 16S rDNA
A pour colony was enriched in LB with constant shaking at 180 r • min-1at 28℃, 18 h. After centrifugation, bacterial DNA were extracted by Bacterial DNA Kit (OMEGA), according to the manufacturer's instructions. Then it was amplified with universal primer of bacteria using PCR (5'-AGAGTTTGATCCTGGC TTAG-3'; 5'-ACGGCTACCTTGTTACGACTT-3'). The products of PCR were detected with 1.0% agarose and analyzed by Shanghai Sangon Biotech Companies. DNAMAN were applied to sequence analysis. Compared with the data from GenBank, isolated YER6022, and some strains of Y. enterocolitia and the strains of Vibrio, Edwardsiella, and Enterobacter were respectively selected to construct phylogenetic tree using ClustalX and MEGA5.0.
Detection of virulence factors
The primers of virulence genes were designed using software Primer 5.0 referred to the data published by GenBank. These primers are shown in Table 1, and synthesized by Shanghai Sangon Biotech Companies. Bacterial DNA was extracted by the method described above, then virulence factors (ail, yadA, ystB, virF, and HIPint) were amplified using PCR. The reaction mixture was 20 μL containing 10 μL of Mix, 5 μL of DNA template, 0.8 μL of each primer and ultrapure water. Optimized condition of PCR: initial denaturation at 94℃ for 4 min, then 30 cycles were followed by denaturation at 94℃ for 40 s, annealing for 45 s, elongation at 72℃ for 40 s, finally extension at 72℃ for 5 min. All PCR products were separated in 1.2% agarose gel stained with GreenView and identified using 2 000 bp DNA Ladder.
Artificialinfectionexperiment
Twenty health pelteobagrus vachelli weighting about 40 g were divided into two groups randomly and equally, the infected group and the control group. Feeding after 10 days, the infected group were injectedintraperitoneally 0.2 mL with 2×108cfu • mL-1bacteria diluted by 0.9% normal saline, while the control group were injected at a same dosage of 0.9% normal saline. These fishes were fed in a fish tank with fresh water and adequate air condition, then the growth and symptoms of the fishes were recorded during the experimental periods 15 days.
Histopathological observation
With serious symptoms, these diseased Pelteobagrus vachellis were sacrificed, and then the heart, liver, intestine, kidney, gill, spleen, lymph, skin, eyes, and brain were respectively removed and fixed in 10% neutral buffered formalin solution. Then the specimens were embedded in paraffin and cut with 5 µm thickness, stained with hematoxylin and eosin (HE). The pictures were taken in light microscope Leica (RM2235).
Ultrastructural observation
The procedure for ultrastructural observation referred to the description of Yue et al (2010). These slices were observed and taken in transmission electron microscope (TEM).

Table 1 Primer of virulence factors
Results
ldentificationofY. enterocolitia
Strain YER6022 was cultured and showed that the colony appearance was small and existing single in CIN agar. The sequence of partial 16S rDNA gene was 1 408 bp length, which shared 99% similarity with other Y. enterocolitica family members. The sequence was submitted and acquired the accession number that was JX434637. Furthermore, phylogenetic tree indicated that the isolated strain was clustered with Y. enterocolitica isolated from the other animal (Fig. 1). Therefore, it could be inferred that strain YER6022 was Y. enterocolitia.
Analysis of virulence factors
Five significant virulence genes were detected by method previously described. The results showed that ail, yadA, ystB, virF, and HIPint were positive, which presented 351 bp band to ail, 800 bp to yadA, 200 bp to ystB, 561 bp to virF, and 714 bp to HPIint, which were similar to expected fragment (Fig. 2).
Artificialinfectionexperiment
After infection, the control group were growing well without any clinical symptoms up to the end of experiment. The studied group showed 80% morbidity and 50% mortality with red and swollen of anus, abdomen swelling, fim bleeding. With necro-scopy, intestine showed severe edema, kidney and liver revealed mild swelling and necrotic. The symptoms were similar with these infected Y. enterocolitica. naturally.
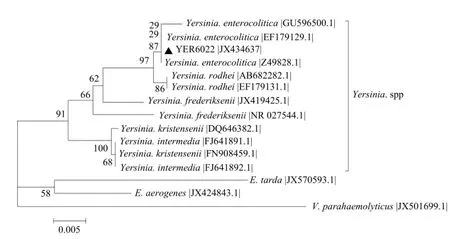
Fig. 1 Phylogenetic tree of 16S rDNA of strain YER6022 constructed by neighbor-joining

Fig. 2 Results of amplification of virulence factors
Histopathological observation
These slices were captured in light microscope. There exsited obvious pathological lesions characterized by cell swelling and hyperaemia in kidney, intestine, liver and spleen. The kidney demonstrated glomerular atrophy and swollen and necrosis in renal tubular epithelial cells. A large number of the red blood cells distributed in the spleen. The detailed changes in kidney, intestine, liver, heart, and spleen were marked with arrowhead in Fig. 3. But no severe lesions were evident in slices of the brain, muscle, eyes, or other examined organs of the suffering Pelteobagrus vachellis.
Ultrastructural observation
As shown in Fig. 4, in the liver, the mitochondria swelled first, then the cristae of the mitochondria disordered, the quantity of lysosome increased (a); in the spleen, the lysosome increased, the nucleus of lymphocyte exhibited pyknosis and split (b); in the kidney, the epithelial cells were edema, then the nucleus of the lymphocyte exhibited pyknosis (c); in the intestine, the epithelial cells swelled, the microvilli underwent degeneration, the karyotheca fractured and the chromatin decreased, and the mitochondria of the enterocyte swelled (d).

Fig. 3 Histopathological analysis of tissues paraffin slices of Pelteobagrus vachelli infected with Y. enterocolitia stained with HE in ×400

Fig. 4 Ultrastructural observation in Pelteobagrus vachelli
Discussion
Y. enterocolitica has become an important zoonotic pathogen around the world; however, there was few reports on aquaculture. In this study, strain YER6022 isolated from naturally infected Pelteobagrus vachelli was successfully identified with the morphology and molecular level. 16S rDNA sequence analysis was a quite simple and more economic method for identification, this molecular technology had been widely applied to the identification of other bacteria, such as Vibrio harveyi, Aeromonas hydrophila (Jiang et al., 2013; Nagar et al., 2013). However, several valuable methods had been used to identify pathogenic Y. enterocolitica strains, such as Fourier Transform Infrared Spectroscopy (FT-IR), Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) (Stamm et al., 2013; Balakrishna et al., 2010). These methods accumulated some desirable data for identification, thus the best method could be chosen depended on the experiment need.
It was known that the pathogenicity of Y. entero-colitia was closely related with its virulence factors. In present study, ail, ystb, virF, yadA and HPIint were positive. ail gene has been considered a vital virulence maker for pathogenic Y. enterocolitia, because its mutation may change the virulence (Danica et al., 2007; Pierson et al., 1993). However, ail was also abtained from nonpathogenic Y. enterocolitia (Lepka et al., 2009; Sivonen et al., 2011; Batzilla et al., 2011). yadA endoced on virulence plasmid could mediate the change of virulence plasmid to promote its adhension (Gierczynski, 2000). It had been found that the incidences of the arthritis causing by pathogenic strain was less low than that of yadA mutant in rats (Platt-Samoraj et al., 2006). Both of ail and yadA were able to mediate the adhesion mechanism, but their virulence plasmids were easily lost in vitro (Huang et al., 2010). Although the mechanisms of these virulence factors were not established, the virulence factors played a vital role in pathogenicity of Y. enterocolitica. These results indicated that this strain YER6022 was potential for pathogenicity.
After being infected with 0.2 mL bacteria, there were 80% morbidity and 50% mortality occured. To some content, the artificial infection experiment and histopathological observation explained its virulence. The pathogenic Y. enterocolitia colonized intestinal lumen, and then it would invaded epithelial cell giving rise to clinical symptoms. The cytokines of the rats infected with pathogenic Y. enterocolitia increased in early period and then quickly decreased (Wang et al., 2013). In the present study, the severe lesions mainly happened at the intestine and some important immune organs. However, there were some differences in the results shown by the models of mice and rabbits established. Several rats died at different phases after injection with pathogenic strain, when the survival ones developed arthritis and irritated claws (Gripenberg-Lerche et al., 1994). The histopathological examination displayed a large number of the blood cells existed obviously in liver and spleen, and the intestinal tract was representative with hyoeraemia, which was consistent with previously described (Marta et al., 2005). These differences between them explained that they might be caused by different strains and different species.
In conclusion, strain YER6022 was pathogenic and could cause death in Pelteobagrus vachelli. To our knowledge, it is the first time to describe Y. enterocolitia infection in Pelteobagrus vachelli. It will contribute to the identification and pathogenicity of Y. enterocolitia infection in fish.
References
Balakrishna K, Murali H S, Batra H V. 2010. Cloning, expression and characterization of attachment-invasion locus protein (Ail) of Yersinia enterocolitica and its utilization in rapid detection by immunoassays. Lett Appl Microbiol, 50: 131-137.
Batzilla J, Heesemann J, Rakin A. 2011. The pathogenic potential of Yersinia enterocolitica 1A. Int J Med Microbiol, 301: 556-561.
Danica G O, Barbara S, Kofitsyo S, et al. 2007. Outbreak of Yersinia enterocolitica Serogroup 0:9 infection and processed pork, Norway. Emerg Infect Dis, 13: 754-756.
Dhayanithi N B, Kumar T T, Kathiresan K. 2010. Effect of neem extract against the bacteria isolated from marine fish. J Environ biol, 31: 409-412.
Gierczynski R. 2000. Evaluation of the usefulness of selected virulence markers for identification of virulent Yersinia enterocolitica strains. II. Genotypic markers associated with the pYV plasmid. Med Dosw Mikrobiol, 52(1): 35-49.
Gripenberg-Lerche C, Skurnik M, Zhang L. 1994. Role of YadA in arthritogenicity of Yersinia enterocolitica serotype O : 8 experimental studies with rats. Infect Immun, 62(12): 5568-5575.
Jiang Q, Shi L, Ke C, et al. 2013. Identification and characterization of Vibrio harveyi associated with diseased abalone Haliotis diversicolor. Dis Aquat Organ, 103(2): 133-139.
Huang Y, Wang X, Cui Z G, et al. 2010. Possible use of ail and foxA polymorphisms for detecting pathogenic Yersinia enterocolitica. BMC Microbiol, 10: 211-217.
Lepka D, Tobias K, Skiebe E, et al. 2009. Adding to Yersinia enterocolitica gene pool diversity: two cryptic plasmids from a biotype 1A isolate. J Biomed Biotechnol, 10: 2-10.
Li M, Yu N, Qin J G, et al. 2014. Effects of ammonia stress, dietary linseed oil and Edwardsiella ictaluri challenge on juvenile darkbarbel catfish Pelteobagrus vachelli. Fish Shellfish Immunol, 38(1):158-165.
Marceau M. 2005. Transcriptional regulation in Yersinia: an update. Cur Issues Mol Biol, 7(2): 151-177.
Marta B S, Venho R, Skurnik M. 2005. Role of YadA, Ail, and Lipopolysaccharide in serum resistance of Yersinia enterocolitica serotype O : 3. Infect Immun, 73(4): 2232-2244.
McNally A, Cheasty T, Fearnley C, et al. 2004. Comparison of the biotypes of Yersinia enterocolitia isolated from pigs, cattle and sheep at slaugher and from humans with yersiniosis in Great Britain during 1999-2000. Lett Appl Microbiol, 39: 103-108.
Mikula K M, Kolodziejczyk R, Goldman A. 2012. Yersinia enterocolitica tools-characterization of structure and function of adhesions. Front Cell Infect Microbiol, 2: 1-14.
Nagar V, Shashidhar R, Bandekar I R. 2013. Characterization of Aeromonas strains isolated from Indian food using rpoD gene sequencing and whole cell protein analysis. World J Microbiol Biotechnol, 29(4): 745-752.
Nesbakken T, Iversen T, Lium B. 2007. Pig herds free from human pathogenic Yersinia enterocolitica. Emerg Infect Dis, 13(12): 1860-1864.
Okwori A E, Martinez P O, Fredriksson-Ahomaa M, et al. 2009. Pathogenic Yersinia enterocolitica 2/O : 9 and Yersinia pseudotuberculosis 1/O : 1 strains isolated from human and non-human sources in Plateau State of Nigeria. Food Microbiol, 26(8): 872-875.
Pederson K J, Pierson D E. 1995. Ail expression in Yersinia enterocolitica is affected by oxygen tension. Infect Immun, 63(10): 4199-4201.
Pepe J C, Wachtel M R, Wagar E, et al. 1995. Pathogenesis of defined invasion mutants of Yersinia enterocolitica in a BALB/c mouse model of infection. Infect Immun, 63(12): 4837-4848.
Pierson D E, Falkow S. 1993. The ail gene of Yersinia enterocolitica has a role in the ability of the organism to survive serum killing. Infect Immun, 61(5): 1846-1852.
Platt-Samoraj A, Ugorski M, Szweda W, et al. 2006. Analysis of the presence of ail, ystA and ystB genes in Yersinia enterocolitica strains isolated from aborting sows and aborted fetuses. J Vet Med B Infect Dis Vet Public Health, 53(7): 341-346.
Rosner B M, Wichael D, Hohle M, et al. 2013. Clinical aspects and selfreported symptoms of sequelae of Yersinia enterocolitica infections in a population-base study. Germany 2009-2010. BMC Infect Dis, 13: 236.
Sihvonen L M, Hallanvuo S, Haukka K, et al. 2011. The ail gene is present in some Yersinia enterocolitica biotype 1A strains. Foodborne Pathog Dis, 8(3): 455-457.
Stamm I, Hailer M, Depner B, et al. 2013. Yersinia enterocolitica in diagnostic fecal samples of European dogs and cats: identification by FT-IR and MALDI-TOF MS. J Clin Microbiol, 51(3): 887-893.
Thoerner P, Bin Kingombe C I, Bogli-Stuber K, et al. 2003. PCR detection of virulence genes in Yersinia enterocolitica and Yersinia pseudotuberculosis and investigation of virulence gene distribution. Appl Environ Microbiol, 69(3): 1810-1816.
Uliczka F, Pisano F, Schaake J, et al. 2011. Unique cell adhesion and invasion properties of Yersinia enterocolitica O : 3, the most frequent cause of human. Plos Pathog, 7(7): 1-24.
Wang X, Gu W, Qiu H, et al. 2013. Comparison of the cytokine immune response to pathogenic Yersinia enterocolitica bioserotype 1B/O : 8 and 2/O : 9 in susceptible BALB/C and resistant C57BL/6 mice. Mol Immunol, 55(3/4): 365-371.
Yue X, Liu B Z, Xiang J, et al. 2010. Identification and characterization of the pathogenic effect of a Vibrio parahaemolyticus-related bacterium isolated from clam Meretrix meretrix with mass mortality. J Invertebr Pathol, 103(2): 109-115.
Q176 Document code: A Article ID: 1006-8104(2015)-02-0029-07
1 October 2014
Supported by the Science & Technology Department of Sichuan Province (2013FZ0014); the Construction Project of the Postgraduate Academic Degree in Southwest University for Nationalities (2013XWD-S071007)
Zhao Jing (1989-), female, Master, engaged in the research of zoopathology and molecular biology. E-mail: 782144909@qq.com
. Wang Li, Ph. D, associate professor, engaged in the research of zoopathology and molecular biology. E-mail: qinxin916@ aliyun.com
 Journal of Northeast Agricultural University(English Edition)2015年2期
Journal of Northeast Agricultural University(English Edition)2015年2期
- Journal of Northeast Agricultural University(English Edition)的其它文章
- Effects of Rice Yield and Quality Across Accumulated Temperature Zone Planting in Cold Area
- Separation and Purification of Total Phloroglucinols in Dryopteris crassirhizoma with DM-130 Macroporous Adsorption Resin
- Characterization and Expression of Outer Membrane Protein A I Gene of Aeromonas veronii
- Construction and Expression of Methionine-rich and Lysine-rich Fusion Gene in Bacillus natto
- Effects of Three Different Diluents on Quality of Boar Semen Stored at 17℃
- Effects of Sub-chronic Aluminum Exposure on Renal Structure in Rats
