Simultaneous heterotrophic nitrification and aerobic denitrification at high initial phenol concentration by isolated bacterium Diaphorobacter sp.PD-7☆
Qilong Ge,Xiuping Yue*,Guoying Wang
School of Environmental Science and Engineering,Taiyuan University of Technology,Taiyuan 030024,China
Keywords:Diaphorobacter sp.Phenol biodegradation Kinetics Heterotrophic nitrification Aerobic denitrification Enzyme activity
A B S T R A C T A strain capable of phenol degradation,heterotrophic nitrification and aerobic denitrification was isolated from activated sludge of coking-plant wastewater ponds under aerobic condition.Based on its morphology,physiology,biochemical analysis and phylogenetic characteristics,the isolate was identified as Diaphorobacter sp.PD-7.Biodegradation tests of phenol showed that the maximum phenol degradation occurred at the late phase of exponential growth stages,with 1400 mg·L−1 phenol completely degraded within 85 h.Diaphorobacter sp.PD-7 accumulated a vast quantity of phenol hydroxylase in this physiological phase,ensuring that the cells quickly utilize phenol as a sole carbon and energy source.The kinetic behavior of Diaphorobacter sp.PD-7 in batch cultures was investigated over a wide range of initial phenol concentrations(0–1400 mg·L−1)by using the Haldane model,which adequately describes the dynamic behavior of phenol biodegradation by strain Diaphorobacter sp.PD-7.At initial phenol concentration of 1400 mg·L−1,batch experiments(0.25 L fl ask)of nitrogen removal under aerobic condition gave almost entirely removal of 120.69 mg·L−1 ammonium nitrogen within 75 h,while nitrate nitrogen removal reached 91%within 65 h.Moreover,hydroxylamine oxidase,periplasmic nitrate reductase and nitrite reductase were successfully expressed in the isolate.
1.Introduction
Phenol is a characteristic contaminant in many industrial effluents and waste wasters[1].It is recommended that human exposure to phenol does not exceed 20 mg on average per day.Besides,phenol is toxic to fish and lethal at concentrations of 5–25 ppm[2].Therefore,treatment of phenol effluents is critical to maintaining human and wildlife environments.Several methods have been reported for removal of phenol from waste waters,including solvent extraction[3],ozonization[4],and biological degradation[5].Biological treatment is considered advantageous over the other methods because of its environmental friendliness,lower cost,economic and practical viability as it mineralizes phenol and forms less hazardous byproducts[6,7].
Since nitrate commonly occurs in many phenolic waste waters,simultaneous degradation of phenolics and removal of nitrogen offer an attractive option.Bacteria capable of combining heterotrophic nitrification and aerobic denitrification have been investigated as potential microorganisms in soils and wastewater treatment systems[8,9].Because of their high growth rate,these microorganisms have many advantages for the removal of nitrogen:(1)less acclimation problems;(2)procedural simplicity,with simultaneous nitrification and denitrification;and(3)less buffer quantity needed,be cause alkalinity generated during denitrification can partly compensate for its consumption in nitrification[10].Some groups of heterotrophic nitrification and aerobic denitrification bacteria,such as Alcaligenes faecalis,Bacillus sp.,and Providencia rettgeri,have been isolated[11–13].There are many differences between bacteria with the ability for nitrification and denitrification[14].Generalization of the biochemical mechanisms is still difficult due to the limitation of the number of tested species[15].Therefore,further investigation on a broader range of species is necessary.
The objectives of this study are to isolate a newly stable and metabolically versatile strain for simultaneous heterotrophic nitrification and aerobic denitrification with high initial phenol as a sole source of carbon,to investigate the intrinsic kinetics of cell growth and phenol degradation of the isolate,and to determine the character of enzyme activities for phenol biodegradation,heterotrophic nitrification and aerobic denitrification.This study is of particular importance to efficient phenolic wastewater treatment where simultaneous removal of nitrogen and phenol is desired.
2.Materials and Methods
2.1.Sample preparation,growth medium and culture condition
Activated sludge was collected from coking-plant wastewater ponds in Taiyuan City,Shanxi Province,China in April 2012 and used to isolate the strain for simultaneous phenol degradation and heterotrophic nitrification–aerobic denitrification.
The strain was grown and enriched in LB medium(L−1)containing 10 g trypt one,10 g soudium chloride,and 5 g yeast extract leaching powder with initial pH 7.2–7.4.The composition of basal inorganic medium(B)for biodegradaion study of phenol and aerobic nitrification was as follow s(L−1):0.5 g NH4Cl,0.5 g KH2PO4,0.2 g MgSO4·7H2O,0.1 g CaCl2,0.5 g K2HPO4,and 1 ml of trace mineral solution.The trace mineral solution contained(L−1)2.86 g H3BO3,0.22 g Zn SO4·7H2O,0.08 g Cu SO4·5H2O,2.03 g Mn SO4·4H2O,and 1.26 g Na2MoO4·2H2O(pH value of 7.2–7.4).Giltay and nitrite(GN)medium[16]at pH 7.2–7.4 contained(L−1)1.0 g KNO3and certain concentration of phenol,to which 5.0 ml of 1%alcoholic solution of bromothymol blue,1.0 g KH2PO4,1.0 g MgSO4·7H2O,0.2 g CaCl2,and 0.05 g FeCl3were added.The medium(C)used for denitrification and nitrate reduction study contained(L−1)1 g NaNO3rather than 0.5 g NH4Cl,other compositions were the same with medium(B).Phenol,as a sole source of carbon,was added to the medium as needed,and then autoclaved at 121°C for 30 min.The degradation was conducted in 250 ml Erlenmeyer flasks(n=3)sealed with autoclaved gauzes at 30°C in a rotary shaker with a speed of 180 r·min−1.Solid media were prepared from growth media with an addition of 2%agar.All chemicals were of analytical grade.
A 5 ml mixed bacterium liquid was added to autoclaved 100 ml inorganic medium(B),adjusted pH value to 7.2–7.4 with NaOH(aq)and HCl(aq)as neutral conditions to favor the growth of bacteria,and then incubated on a rotary shaker(180 r·min−1)at 30 °C in an Erlenmeyer lf ask.After cultivation for 3 days,the culture was periodically sampled and tested for the presence of nitrite and residual phenol.When nitrite was detected and phenol was degraded,5 ml of the enrichment suspension was transferred to 100 ml of fresh inorganic medium and incubated for another 3 days.Ammonium removal was also measured to screen the bacterial cultures.The most promising culture(with the best ammonium and phenol removal)was spread onto agar LB medium plates using the dilution plate method and incubated at 30°C for 72 h.Isolated colonies were individually recultured on agar plates to obtain pure strains.The capability of aerobic denitrification of isolated strains was tested on the solid GN media.The strain PD-7 show ed the highest aerobic denitrification activity,as indicated by the color change of GN medium from green to bright blue[16].This isolate was identified and further tested for phenol degradation and heterotrophic nitrification–aerobic denitrification.
2.2.Enrichment and isolation of bacteria
2.3.DNA extraction,PCR amplification and 16S rDNA gene sequence analysis
DNA was extracted from a bacterial suspension(1 ml,OD600~1.2)by using the EZ Spin Column Bacterial Genomic DNA Isolation Kit(Sangon,Shanghai).Micrographs of strain PD-7 were taken with a scanning electron microscope(Quanta200,Holland).The physiological characteristics of PD-7 were examined according to the methods in[17].16S r DNA gene was amplified by PCR using the universal primers F27(5′-AGAGTT TGATCMTGGCTCAG-3′)and R1492(5′-TAC GGYTACCTTGTTACGACTT-3′).16S rDNA sequence was compared with that of other strains by BLAST(http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi).Related sequences were obtained from the GenBank database using the BLAST search program.
2.4.Phenol biodegradation
After the strain was activated twice,the cells in the phases OD600=1.217 were harvested as inocula.5 ml of this subculture was transfused into 100 ml mineral medium(B)containing varying phenol concentrations over the range from 0 to 200 mg·L−1at an interval of 20 mg·L−1,200–600 mg·L−1at an interval of 100 mg·L−1,and 600–1400 mg·L−1at an interval of 200 mg·L−1.Samples were periodically taken for determining biomass and phenol concentrations.
2.5.Measurements of heterotrophic nitrification and aerobic denitrification with shake flask experiments
Strain PD-7 was separately cultivated in basal media B and C at 30°C with a shaking speed of 180 r·min−1.Centrifugation was conducted(4°C,15 min)after 72 h of cultivation and then the pellets were washed with sterilized water.Centrifugation and w ashing were repeated three times to purify the bacterial suspension.To start experiments,different basal media(B and C,100 ml each)and bacterial suspension(5 ml)were placed into conical flasks(250 ml)and cultivated at 30°C and 180 r·min−1.During incubation,the cultures were sampled periodically to determine cell optical density(OD600)and then centrifuged(4°C,15 min)to obtain supernatants for determination of phenol concentration,ammonium,hydroxylamine,nitrite and nitrate.
2.6.Enzyme activity
Enzyme activities about phenol degradation were spectrophotometrically determined in cell-free extracts using quartz cuvettes of 1 cm path length.Cells grown in different exponential stages in basal medium(B)were harvested and centrifuged at 7500 r·min−1for 10 min.After being w ashed twice with 0.1 mol·L−1phosphate sodium buffer(pH 7.2)and resuspended in the same buffer,the cell pellet was disrupted by ultrasonication for 5 min,and then cell debris were removed by centrifugation of the homogenized cell suspension at 12000 r·min−1and 4 °C for 20 min.The cleared supernatant was immediately used for assays of enzyme and total protein.The phenol hydroxylase activity depended on the presence of NADPH and was assayed spectrophotometrically according to NADPH absorbance at 340 nm[18].Catechol 1,2-dioxygenase and 2,3-dioxygenase activities were spectrophotometrically determined following the formation of cis,cis-muconic acid and 2-hydroxymuconic semialdehyde at 260 and 375 nm.The former was monitored in the presence of 1 mmol·L−1EDTA and 33 mmol·L−1Tris–HCl at pH 7.2 and 10 μl enzyme extract with the total volume of 2 ml.The reaction was started by adding 0.1 mmol·L−1catechol.The latter,2,3-dioxygenase,was evaluated by measuring the formation of 2-hydroxymuconic semialdehyde at 375 nm.The reaction mixture contained 0.1 mmol·L−1catechol,50 mmol·L−1Tris–HCl at pH 7.2 and 10 μl enzyme extract,w ithin a total volume of 2 ml[19].
Three reaction mixtures I,II and III were prepared for detecting the hydroxylamine oxidase(HAO),nitrate reductase(NAR)and nitrite reductase(NIR),respectively.The disappearance of hydroxylamine from reaction mixture I in the presence of cell-free extract was taken as a measure of HAO activity.Reaction mixture I in 10 ml contained enzyme extract,0.11 mmol·L−1cytochrome c and 10 mmol·L−1Tris–HCl buffer(pH 7.2),and the reaction was initiated by the addition of hydroxylamine.The production of nitrite from nitrate in the presence of cell-free extract in reaction mixture II was taken as a measure of NAR activity.Reaction mixture II in 20 ml contained enzyme extract,0.2 mmol·L−1NADH and 10 mmol·L−1potassium phosphate buffer(pH 7.2),and the reaction was started by the addition of NaNO3.The reduction of nitrite from reaction mixture III in the presence of cell free extract was taken as a measure of NIR activity.Reaction mixture III in 20 ml contained enzyme extract,0.2 mmol·L−1NADH and 10 mmol·L−1potassium phosphate buffer(pH 7.2),and the reaction was started by the addition of NaNO2.Total protein concentration in the crude extract was quantified by the method of Bradford to determine the specific activity of enzymes[20].One unit of enzyme activity(U)is defined as the amount of enzyme which catalyzes the trans formation of 1 μmol of substrate per minute.The specific activity(U·mg−1)is defined as the amount of enzyme units divided by the concentration of protein in mg.
2.7.Analysis methods
During incubation,the optical density of the culture was measured at OD600using a spectrophotometer(UV-5500,China).The cell dry w eight was determined[21].NH4+-N was determined by photometric determination with Nessler's reagent;NO2−-N was measured by N-(1-naphtha-lene)-diaminoethane photometry method;NO3−-N was analyzed by phenol disulfonic acid method according to[22].The cell free supernatants were used to determine the phenol concentration by high performance liquid chromatography with a C18 column(250 mm×4.6 mm).Elution was performed with 400/300(volume ratio)methanol/water at a flow rate of 1.0 ml·min−1,and detection was realized with the UV detector at 280 nm.
3.Results and Discussion
3.1.Identification and characterization of strain PD-7
A heterotrophic nitrification and aerobic denitrification strain PD-7 at high initial phenol concentration was isolated from coking-plant wastewater pond.Fig.1(a)illustrates the colony of strain PD-7 on GN plate.The colony is rounded,raised and shiny,with a diameter of 1–2 mm.Fig.1(b)shows that strain PD-7 is rod-shaped and motile with a diameter 0.7–0.9 μm.The cells are Gram-negative and demonstrate unipolar flagellum during their growth phase.Other physiological and biochemical characteristics are summarized in Table 1.The partial 16S r DNA sequence(1491 bp)of strain PD-7 is determined and deposited in the GenBank database(GenBank accession No:KF022039).A phylogenetic tree is constructed based on 16S r DNA sequences as shown in Fig.2.Comparison of its sequence with those in the GenBank database show s 99%similarity to Diaphorobacter nitroreducens.Therefore,PD-7 is proposed to be a Diaphorobacter species.

Fig.1.The colony of strain PD-7 on GNplate(a)and scanning electron micrograph of a cell with a single polar fl agellum of strain PD-7(b).
3.2.Phenol degradation
It has been observed that phenol of high concentration inhibits cell growth and its ow n degradation[23,24].The hydrogen bond,formed by phenolic hydroxyl hydrogen and lone pair electrons of amino or imine group on the microprotein,changes the secondary structure of microproteins,making the interaction between charged particlesmore intensely.Protein particles gather and precipitate easily.At high concentration of phenol,more hydrogen bonds form,so the inhibition of cell growth is more severely.

Table 1Morphological,physiological,and biochemical characteristics of strain Diaphorobacter sp.PD-7(+:positive;−:negative)
As shown in Fig.3,a total of 1200 mg·L−1phenol was degraded in 67 h.At higher phenol concentration,the lag phase of cell growth lasts longer and the specific degradation rate decreases,demonstrating that high phenol concentration inhibits cell growth and phenol degradation.Moreover,as the initial phenol concentration increases,cells obtain more carbon source and the final biomass increases slightly.How ever,the biomass yield is not in accordance with the stepwise increase of phenol concentration,which increased 41.86 mg·L−1with the concentration from 800 to 1000 mg·L−1and merely 22.64 mg·L−1from 1000 to 1200 mg·L−1.Cell growth is not in proportion to phenol consumption,so that no relation exists between cell growth and phenol biodegradation,especially at high phenol concentrations[25].The higher the phenol concentration,the stronger the substrate inhibition exhibits.Thus the consumption of phenol is not entirely used to synthesize new cells and most is used to counteract strong substrate inhibition at the utmost phenol concentration.In the biodegradation tests,the highest initial phenol concentration that could be degraded completely by strain Diaphorobacter sp.PD-7 was 1400 mg·L−1.1500 mg·L−1phenol was also examined,which could not be completely degraded.
3.3.Intrinsic kinetics of phenol degradation
It is presumed that the growth rate of Diaphorobacter sp.PD-7 and substrate degradation rate are only limited by phenol concentration at fixed initial pH,temperature and shaking rate.Dissolved oxygen value in a shaking flask is determined beyond 6.0 mg·L−1by an oxygen electrode(HACH LDO10103,USA).We also assume that the aeration provided by shaking the flasks is sufficient and keeps the oxygen concentration constant.
Batch cultures of Diaphorobacter sp.PD-7 were conducted in the mineral medium(B)containing initial phenol concentrations ranging from 0 to 1400 mg·L−1with inoculum.For each batch culture with a certain initial substrate concentration,the specific cell growth rate is calculated by:
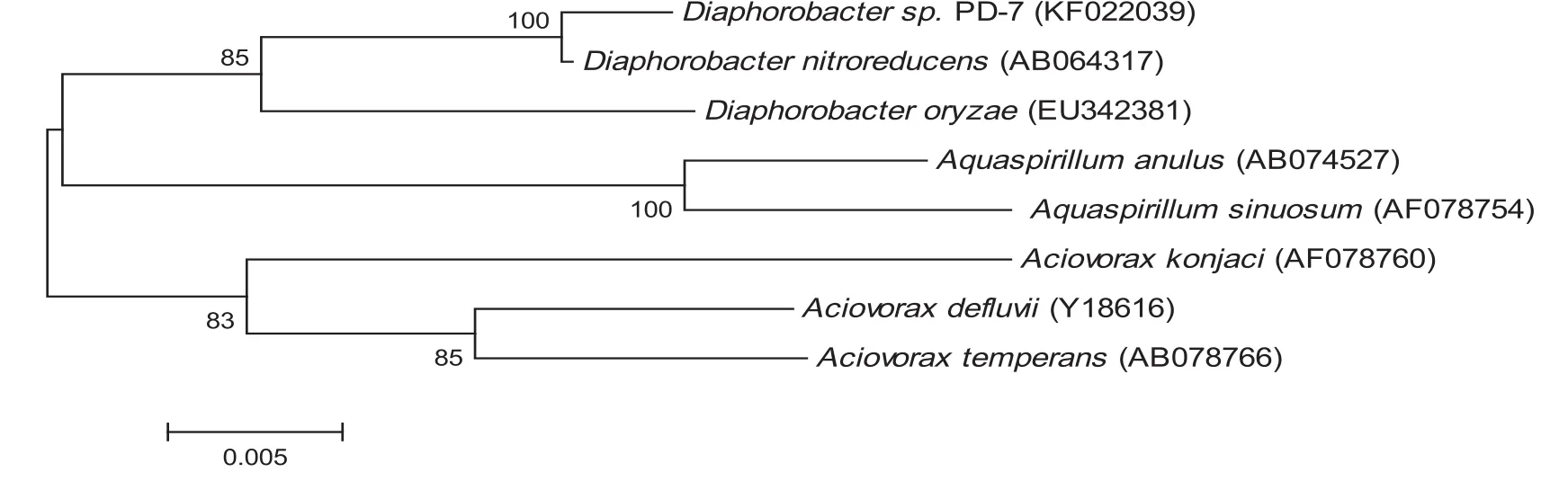
Fig.2.The neighbor-joining tree constructed and bootstrapped by software Mega4.0 to represent the relationship between strain PD-7 and representative species of genus Diaphorobacter nitroreducens and related genera,with Bootstrap values noted on the branch.

where μXis the specific growth rate(h−1),γXis the cell growth rate,and CXis the cell concentration(mg·L−1).
Because of the inhibition of high phenol concentration on cell growth,Haldane's equation is selected for assessing the dynamic behavior of Diaphorobacter sp.PD-7 grown on phenol:

where CSis the phenol concentration(mg·L−1),μX,maxis the model constant,KSis the saturation coefficient,and Kiis the inhibition coefficient.The values of parameters μX,max=0.323 h−1,KS=9.65 mg·L−1,and Ki=152.40 mg·L−1are derived using a nonlinear least-squares regression analysis of MATLAB based on the experimental data in the phenol biodegradation.The value of the squared 2-norm of the residual at these parameters is very small(1.23× 10−3),indicating that the regression curve agrees well with the experimental data.

Fig.3.Cell growth and phenol specific degradation rate at different initial phenol concentrations.
As shown in Fig.4,the maximum specific growth rate occurred at very low phenol concentration,38.35 mg·L−1.Generally,a low specific growth rate indicates very intense substrate inhibition to strain Diaphorobacter sp.PD-7.With the decrease of initial phenol concentrations in the mineral medium(B),the specific growth rates gradually increased from 0.031 to 0.213 h−1until a sharp drop lacking of carbon source in the mineral medium[26].Lower phenol concentration in the medium leads to weaker substrate inhibition.How ever,in spite of a lower specific growth rate,bacteria keep a high phenol-degrading potential.More energy is required to overcome the effect of substrate inhibition at high phenol concentrations.Thus both specific growth rate and biomassyield(g·g−1)are low at the initial phase of biodegradation,and with the consumption of phenol they increase with declining inhibition of phenol.The production and accumulation of various intermediates might also account for the decrease of cell mass yield[27].That is why there is no essential relation between cell growth and phenol degradation,though phenol is mainly consumed for the energy for cell growth and for assimilation into biomass and maintenance[28].
Haldane's equation is widely used to describe the cell growth kinetics on phenol and predict degradation rate of phenol by assuming a constant cell mass yield.

To analyze the utilization of substrate in cells,the consumption of substrate for growth and maintenance and for possible product formation has to be considered[29].The substrate consumption rate of phenol biodegradation is



Fig.4.Kinetic prediction and experimentally determined specific growth and degradation rates at initial phenol concentrations of 0–1400 mg·L−1.
where γP=αγX+βCXis the formation rate of product.Because YX/S,m,YP/S,α,and β are all constants,Eq.(4)could be reduced to where A and B are kinetic constants and regressed using MATLAB based on the experimental data,A=0.83,B=0.0297 h−1,with R2of 0.991,which indicates that degradation kinetics agrees well with the experimental data.
3.4.Ammonium oxidation and nitrate removal efficiency
Fig.5(a)illustrates cell growth and removal of ammonium by Diaphorobacter sp.PD-7 in a 250 ml flask at initial phenol concentration of 1400 mg·L−1.Cell growth reached a stationary phase in 75 h as the cell concentration increased from 67.34 mg·L−1to 512.89 mg·L−1.Ammonium(initial 120.69 mg·L−1-N)was almost entirely transformed in 75 h and the nitrification ratio of-N was about 1.61 mg·L−1·h−1.It was reported that Bacillus sp.LY removed-N at a rate of 0.43 mg·L−1·h−1with initial ammonium of 41.1 mg·L−1,C/N ratio of 15,and sodium glucose as carbon resource[30].TheN removal rate of Pseudomonas alcaligenes AS-1 was 1.15 mg·L−1·h−1with ammonium acetate as carbon and nitrogen resources[31].It appears that Diaphorobacter sp.PD-7 presents a much higher-N removal rate than Bacillus sp.LY and P.alcaligenes AS-1.Nitrification products-N and-N were detected during the experiments.The amount of-N increased very slow ly,w hile the amount of-N changed little in the initial bacterial growth stage(0–25 h).The amount of-N and-N increased quickly in the bacterial rapid growth stage(25–75 h),but decreased dramatically in the stationary growth stage(after 75 h of cultivation).Less than 5%-N and less than 13%-N accumulated.Experimental results indicate that strain PD-7 utilizes NH4+-N or converts it to other nitrogen species,particularly during the growing phase.

Fig.5.Grow th and biodegradation curves in the basal medium(B)(a)and medium(C)(b)with initial 1400 mg·L−1 phenol.
Ignoring the accumulated impact of nitrite nitrogen and nitrate nitrogen,the first order kinetic equation is selected to describe the degradation of ammonia nitrogen.


where C is the ammonia nitrogen concentration(mg·L−1),C0is the initial ammonia nitrogen concentration(mg·L−1),k is the first-order kinetic coefficient,and t is the time.The fitted equation is ln C=4.98−0.047 t(R2=0.978),and simulated kinetic curve agrees well with the experimental data.
To confirm the denitrification by Diaphorobacter sp.PD-7,nitrified intermediate nitrate(NaNO3)was used as the nitrogen source in the basal medium(C).Fig.5(b)shows the growth and nitrate reduction of stain PD-7 in a 250 ml flask with initial phenol concentration of 1400 mg·L−1under an aerobic condition.A significant decrease of nitrate by Diaphorobacter sp.PD-7 can be observed.Approximately 60%of-N was removed in 45 h.By 65 h of cultivation,91%of NO3−-N was removed.The nitrate removal rate was 1.16 mg·L−1·h−1.The capability of aerobic denitrification by Rhodococcus sp.CPZ24 was demonstrated[32],with a nitrate removal rate of 0.93 mg·L−1·h−1after incubation for 36 h.Grow th in the basic medium(C),strain PD-7 removed-N during the rapid growth phase and the removal of-N reached the peak at 80 h.Although low concentrations of-N(less than 37%)relative to-N were detected during the entire culturing period,-N concentrations increased rapidly as-N did in the 65 h of incubation.However,-N concentrations quickly decreased betw een 65 h and 85 h of incubation.Its change trends matched well with-N concentration changes and bacterial population changes.Both concentrations of-N and-N were probably under equilibrium as regulated between heterotrophic nitrification and aerobic denitrification.It is known that many heterotrophic nitrification bacteria are capable of aerobic denitrification[15].
It can be written as

w here q is the specific denitrification rate(d−1),qmaxis the maximum specific denitrification rate(d−1),KDis the half saturation constant(mg·L−1),D is the concentration of nitrate nitrogen(mg·L−1),and CXis the cell concentration(mg·L−1).Using Eq.(9)to fit the experimental data,the model parameter values are as follow s:qmax=0.43 d−1and KD=103.4 mg·L−1.
3.5.Enzyme activity
The present study demonstrates that Diaphorobacter sp.PD-7 has the ability for efficient phenol-degrading with simultaneous heterotrophic nitrification and aerobic denitrification.The efficiency of a certain catabolic pathway often depends on the properties of the key enzyme.In the metabolism of phenol,phenol hydroxylase hydroxylates phenol to catechol,then the ring of catechol is cleaved by the catalysis of catechol 1,2-dioxygenase or 2,3-dioxygenase,following ortho or meta fission.In the assay of enzyme activity,the activity of catechol 1,2-dioxygenase was not found in cell-free extracts,which proves that no cis,cis-muconic acid is produced in the ring fission products of catechol.Thus,phenol is assimilated by meta fission of catechol.
Fig.6 show s that cells excrete phenol hydroxygenase and catechol 2,3-dioxygenase heavily at the middle phase,reaching the maximum at the late phase of exponential stages.It is an important factor leading to higher phenol-degrading potential at the late phase of exponential stages,especially the change of phenol hydroxylase activity.In the two enzymes,phenol hydroxylase,hydroxylating phenol to catechol,is the key enzyme for phenol biodegradation[33].The biodegradation rate chiefly relies on the reaction that phenol is hydroxylated in phenol metabolism.Accordingly,phenol hydroxylase activities in different exponential phases play an important role in the phenol biodegradation.How ever,it could not be excluded that higher cell concentration in the late phase also slightly raises the phenol-degrading rate.

Fig.6.The cell growth and specific enzyme activities of Diaphorobacter sp.PD-7 grown in different physiological phases in basal medium(B)with initial phenol of 1400 mg·L−1.
To identify possible pathways of combined nitrification and aerobic denitrification of different enzymes,HAO(hydroxylamine oxido reductase),NAP(periplasmic nitratereductase)and NIR(nitritereductase)in cell periplasm were performed.Table 2 shows that they are successfully expressed in the isolate.The specific activities of enzyme HAO,NAP and NIR are 0.019,0.0076 and 0.013 U·(mg protein)−1,respectively.
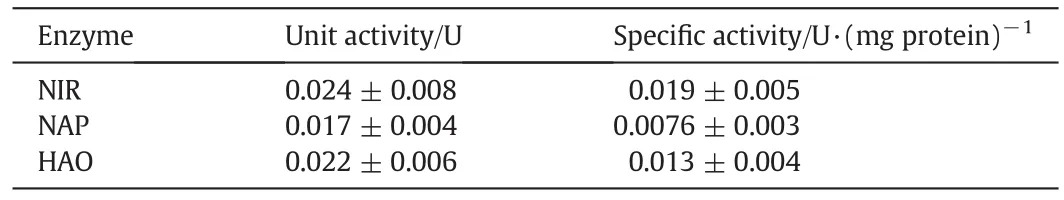
Table 2Unit and specific activities of three enzymes
Enzyme activity study has shown that ammonia,nitrite,and nitrate could all be used by the isolate under aerobic condition and HAO,NAP and NIP are successfully expressed in the cell-free extracts.Similar results have been obtained with Pseudomonas sp.and Bacillus sp.[31,34].These bacteria exhibit a full nitrification and denitrification pathway.In the metabolism of nitrogen,NH4+or NH3is fi rst catalyzed to NH2OH under aerobic condition.Then NH2OH is transformed to NO2−-N by catalytic oxidation of NIR and NO2−-N is further oxidated to NO3−-N with the catalysis of NAR.NO3−-N eventually turns to N2O or N2.However,neither nitrite nor nitrate is utilized as electron accepters by Agrobacterium sp.LAD9,Alcaligenes faecalis and Acinetobacter calcoaceticus[35],without the detection of activity of nitrite or nitrate reductase[36].As a result,denitrification in these bacteria is via hydroxylamine rather than nitrite or nitrate.Namely NH2OH is directly converted into N2O or N2.These results suggest that there may be two main pathways in the combined nitrification and denitrification process.Their difference is in the aerobic denitrification route.One is through nitrite or nitrate,and the other is by hydroxylamine.
4.Conclusions
Strain Diaphorobacter sp.PD-7 isolated from activated sludge cokingplant wastewater pond presents high phenol-degrading with simultaneous heterotrophic nitrification and aerobic denitrification potential.When the subculture at late phase of exponential stages was used as inoculum,1400 mg·L−1phenol was degraded within 85 h.After the cell growth reached the middle phase,cells began to heavily excrete phenol hydroxylase and catechol 2,3-dioxygenase until the late phase of exponential stages.For Diaphorobacter sp.PD-7,phenol was assimilated by meta- fission of catechol.Haldane'sequation well describes the growth kinetics of Diaphorobacter sp.PD-7 at 30 °C and pH 7.2:μX=0.323CS/(9.65+CS+CS2/152.40).The phenol degradation process is associated to cell growth kinetics:μS=0.83μX+0.0297 h−1.Diaphorobacter sp.PD-7 could remove ammonium and nitrate at a rate of 1.61 and 1.16 mg·L−1·h−1,respectively,in batch flasks.Besides,the present investigation demonstratesthat phenol,ammonia,nitrite and nitratecan all be used by the isolate under aerobic condition and phenol hydroxylase,2,3-dioxygenase,hydroxylamine reductase,nitrate reductase and nitrite reductase are expressed in cell-free extract.
 Chinese Journal of Chemical Engineering2015年5期
Chinese Journal of Chemical Engineering2015年5期
- Chinese Journal of Chemical Engineering的其它文章
- Biogas by two-stage microbial anaerobic and semi-continuous digestion of Chinese cabbage waste☆
- Crystal structure and thermal decomposition kinetics of 1-(pyridinium-1-yl)propane-(1-methylpiperidinium)bi[bis(trifluoromethanesulfonyl)imide],[PyC3Pi][NTf2]2☆
- Synthesis of magnetically modi fi ed palygorskite composite for immobilization of Candida sp.99–125 lipase via adsorption☆
- Nitrogen removal characteristics of heterotrophic nitrification-aerobic denitrification by Alcaligenes faecalis C16☆
- Key factors governing alkaline pretreatment of waste activated sludge☆
- Experimental study by online measurement of the precipitation of nickel hydroxide:Effects of operating conditions☆
