Characterization of ploidy levels in Chrysanthemum L.by flow cytometry
Yue-ping Ma•Jiang-xue Wei•Zhi-yang Yu•Bing Qin•Si-lan Dai
Characterization of ploidy levels in Chrysanthemum L.by flow cytometry
Yue-ping Ma1•Jiang-xue Wei1•Zhi-yang Yu1•Bing Qin1•Si-lan Dai2
Analyzing the ploidy levels ofplants isimportant foridentifying species,selecting parentallines,identifying the relationships between species,and determining evolutionary patterns.The genus Chrysanthemum is widely distributed throughoutthe world and exhibitsdifferentploidy levels.We used flow cytometry to analyze the ploidy levels of nine speciesof Chrysanthemum L.collected from differentregions and geographical locations in China.Three diploids from Henan and Wuhan provinces corresponded to Chrysanthemum lavandulifolium and two species of C.nankingense, while three tetraploids from various regions corresponded to C.indicum and two species of C.chanetii.Two hexaploids corresponding to C.vestitum were collected atFuniu mountain(Henan province),and C.zawadskii was collected at Huangshan mountain(Anhuiprovince).Wefound thatOTTO extraction bufferwassuitable forextracting nucleifrom most species,apartfrom C.zawadskii.Flow cytometry proved to bea simple,rapid,and highly accurate method for identifying ploidy levels in Chrysanthemum species.
Chrysanthemum L.·Flow cytometry·OTTO extraction buffer·Ploidy identification
Introduction
Chromosomes are the carriers of genetic material.Changes in the number of chromosomes affect the morphology, anatomy,physiology,and biochemistry ofan organism and lead to many changes in genetic characteristics.Therefore, elucidating the ploidy of a species lays foundation for understanding its genetic background,which is helpfulfor germplasm conservation and other applications(Bennett and Leitch 2005).In addition,knowing the ploidy levelofa species is essential if we are to better understand its population structure and evolution;this knowledge can also help answer some taxonomic questions.
Chrysanthemum(Chrysanthemum morifolium Ramat.) is one of the most well-known and popular ornamentals throughoutthe world.Although species of Chrysanthemum have gradually become the world’s most abundant cultivated plants,the origin of this species is still debated and there is no universal identification system for chrysanthemum taxonomy.Hybridization and allopolyploid formation are important for the development of modern chrysanthemums(Dai et al.2005).Many species of this genus are directly or indirectly involved in the evolution of modern chrysanthemums.Chromosomes of species in this genus often have different degrees of differentiation,both between and within species.A basic levelofknowledge about the genetic resources ofchrysanthemum is fundamentalfor the implementation of successfulbreeding programs and tohelp understand the varieties of modern chrysanthemum. The chromosome numbers of many species of wild Chrysanthemum have been determined by cytological approaches.There is a wide range of ploidy variation among Chrysanthemum germplasm,including diploids,tetraploids,hexaploids,octoploids,enneaploids,and even pentaploids(Tahara 1915;Dowrick 1952,1953;Nakata 1987;Nakata et al.1992;Zhou and Wang 1997;Fukai etal.1998;Kondo etal.2003).Correlations existbetween ploidy level and the mode of reproduction.Cytological irregularities lead to erratic flowering and reproductive behavior.Therefore,identification of each accession in a polyploidy species is necessary for efficient breeding schemes.This knowledge is also of practical importance for chrysanthemum breeders to help them utilize new introductions in their breeding programs.

Table 1 Main components of four nuclei extraction solutions
Ploidy identification in chrysanthemum was traditionally performed by chromosome counting on microscope slides. However,this technique is tedious,difficultto perform,and time consuming.Although stomatalsize and density,as well as pollen size,have been used to determine ploidy in some species(Hamill et al.1992;Dolezˇel et al.2004),these methods are often irreproducible and unreliable.Flow cytometry is currently used routinely forploidy analysis and it is regarded as the mostaccurate tool for ploidy determination(Dolezˇel et al.1998).The main advantages of flow cytometry include its simplicity,speed,accuracy,convenience,and ability to screen large numbers of samples per day.In addition,this method is non-destructive and only requires a small amount of tissue(Dolezˇel 1997).Flow cytometry is a very successful and convenient method for analyzing ploidy level and/or estimating genome size in many plantspecies(Obidiegwu etal.2009,2010;Ono and Hosaka 2010;Schlaepfer etal.2010;Kiszczak et al.2011; Kumar et al.2011;Burson et al.2012;Scaldaferro et al. 2012;Angeles etal.2013;Moon etal.2013).
The goalof this study was to determine the ploidy levels of some wild species of Chrysanthemum collected from different regions and geographical locations using flow cytometry.The results provide clues about the origin and evolution of Chrysanthemum species.Moreover,this study may help generate new ideas for overcoming the current problems of chrysanthemum cultivar classification and may lead to more precise,efficientuse ofthe genetic diversity of this genus for the purpose of systematic hybridization.
Materials and methods
The plantmaterialconsisted of ten Chrysanthemum species collected from different locations(Table 1).All plants were cultivated in 30 cm pots in a greenhouse under natural light at25°C(16 h/8 h of light/dark).
Sample preparation
Young leaves were collected from individual plants and samples were prepared for flow cytometry according to the procedure of Dolezˇel et al.(1994),with some modifications.For each sample,approximately 100 mg of healthy leaf tissue was excised and placed into a 35×10 mm plastic Petridish on ice.The tissue was chopped into thin strips(0.25–0.5 mm wide)in 1 ml of ice cold extraction buffer(Table 1).The suspension was collected with a pipette and filtered through 30μm nylon mesh into a 1.5 ml microfuge tube to remove cell fragments and large debris.The nuclei in the microfuge tube were purified by centrifugation at 150 g at 4°C for 5 min,resuspended in 1 mL of PBS,and centrifuged at150 g at4°C for 5 min.
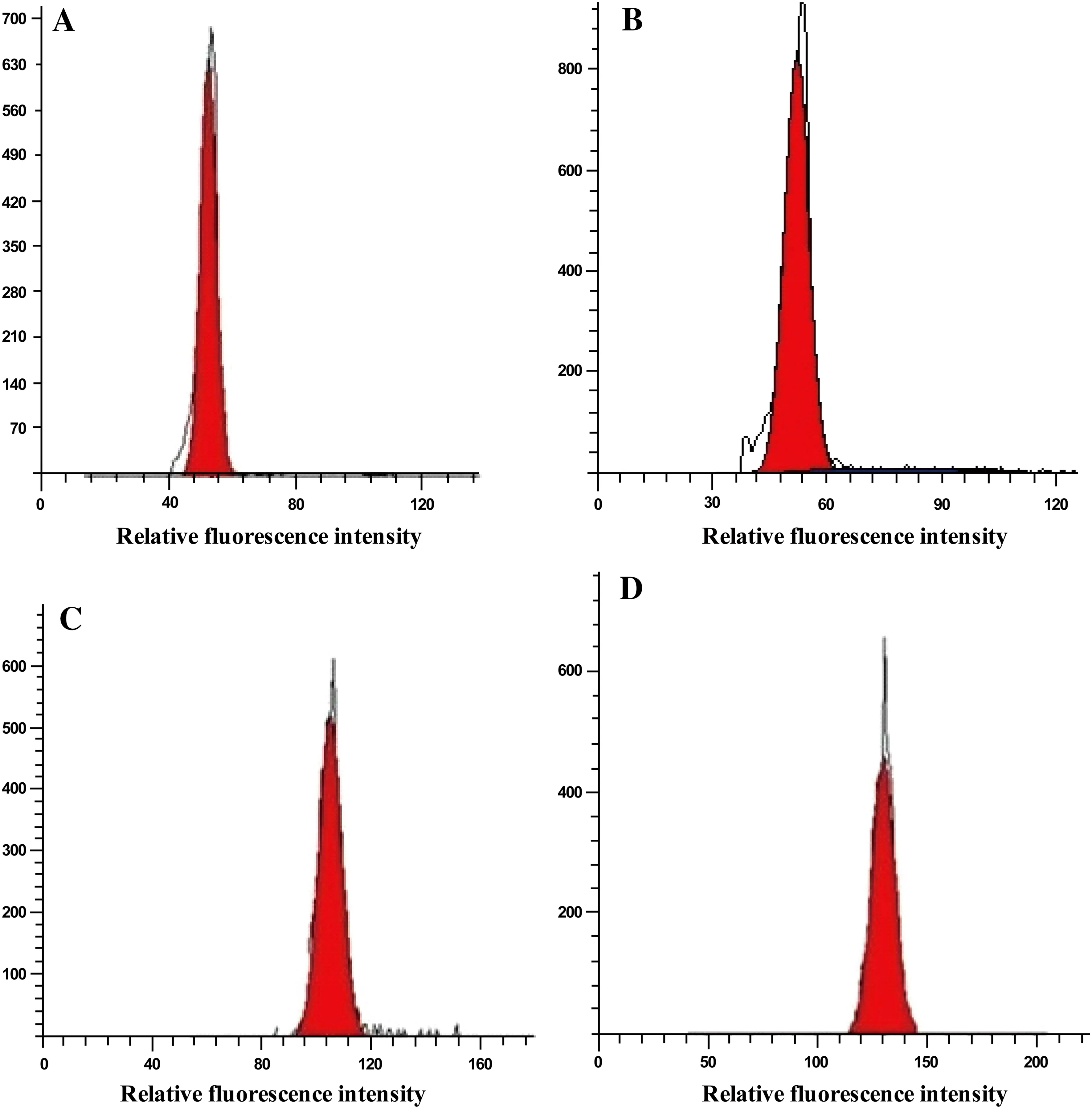
Fig.1 Examples of flow cytometric histograms showing the relative PI fluorescence intensity in nuclei from the leaves of different Chrysanthemum species.Abscissa:Relative fluorescence intensity (channel number);ordinate:number of nuclei.a is external control (2×),b is diploid(2×),c is tetraploid(4×),d is hexaploid(6×) plants
The purified nucleiwere stained with 250μL propidium iodide(PI;50μg·mL-1;Fluka,Buchs,Switzerland).RNase (50μg mL-1;Fluka)was then added to the nuclear suspension to preventstaining of double-stranded RNA.The samples were incubated on ice in the dark for 30 min before analysis by flow cytometry.All stages of extraction and staining were performed at 4°C.Control(diploid, 2n=2×=18)plants were used as an external reference standard(1000 nucleiwere analyzed per sample).
Flow cytometric analysis
The prepared material was analyzed on a standard FACS-can flow cytometer(Becton–Dickinson Immunocytometry Systems,San Jose,CA,USA)using a 488 nm laser at 15 mW.For each measurement,PI fluorescence area signals (relative fluorescence intensity,FL2-A)from 1000 nuclei were collected using CellQuest software(Becton–Dickinson).A live gate was set using the FL2-A and FSC parameters,allowing the fluorescence measurements from nuclei to generate a histogram of FL2-A.The mean positions of the G1/G2(nuclei)peak for the sample and the external standard were determined by analyzing the data using CellQuest software(Becton–Dickinson).Young leaf samples from Chrysanthemum nankingense L.(2n=18) (Ma et al.2013)were used as a diploid reference.To estimate ploidy level,the position of the G1 peak on the histogram obtained foreach individualplantwas compared with thatof C.nankingense.A software package(BD)wasused to calculate the coefficient of variation(CV).A minimum of three samples were analyzed for each accession.Three independent replicates were examined on successive days.

Table 2 Analysis of ploidy level in the genus Chrysanthemum

Fig.2 Non-flowering branches of C.chanetii.a The morphology of a non-flowering plant;b comparison of leaves of two branches.a1 and a2 (left)are flowering branches;b1 and b2(right)are non-flowering branches.Scale bar is 1 cm
Results and discussion
Excellent preparation of nuclei is crucial for flow cytometry analysis.However,obtaining nucleifrom different plantspecies requires differentextraction buffers suited to a particular plant’s tissue structure and metabolic components.We found thatmodified OTTO extraction buffer was suitable for most species examined,leading to the generation of clearly defined histograms.Only C.zawadskii required the use of Tris·MgCl2(Pfosser et al.1995)extraction buffer.LB01 and MgSO4extraction buffer did not generate histograms for any species.
After PI staining,nuclei isolated from young leaf tissue emitted fluorescence when irradiated with UV radiation, which was measured by the flow cytometer.For each sample,analysis of the relative fluorescence intensity of the isolated nuclei yielded a histogram showing a dominant peak corresponding to nuclei in the G1 phase of the cell cycle.The amount of debris in each sample was negligible. Clearly defined histograms for accurate determination of ploidy levels were obtained following flow cytometric analysis of intact leaf nuclei(Fig.1).Table 2 shows the ploidy level inference for each accession analyzed.The results implied thattwo of the accessions were diploid,three were tetraploid,and two were hexaploid species(Table 2).Most histograms showed a low CV(lessthan 5%),indicating that the results were reliable(Galbraith et al.2002).
Earlier chromosome counting identified different ploidy levels within the genus Chrysanthemum.Most ploidy levels determined in the current study were consistent with previous findings from chromosome counting.Here we found two morphologies in one species of C.chanetii:non-flowering branches had larger leaves than did flower branches (Fig.2a).The different ploidy levels encountered in thegermplasm collection examined in the currentstudy suggest that gamogenesis played an important role in the origin of modern chrysanthemum.
There were no significant differences among the same species of Chrysanthemum collected from different locations(such as C.nankingense and C.chanetii)with respect to ploidy level,which indicates that ploidy level is not associated with geographic origin.There were distinct differences in morphological and reproductive behavior in C.chanetii plants collected from Qinhuangdao.The leaves of non-flowering branches were nearly twice the size of flowering branches(Fig.2b).Itis interesting thatthese two morphologically different plants were derived from the same species.The reason for this requires further study.
In conclusion,we successfully used flow cytometry to determine ploidy levels in Chrysanthemum.The high sensitivity and accuracy of flow cytometry enabled us to rapidly estimate the ploidy levels of Chrysanthemum species.This efficienttechnique can be used to further clarify the genetic diversity of both wild and cultivated Chrysanthemum species.
AcknowledgmentsChrysanthemum chanetii plants from Hebei were supplied by Professor Liu Z.L.(College of Landscape Architecture,Hebei Normal University of Science and Technology).
Angeles M,Anto´n A,Olmos E,Pe´rez-Pe´rez JM,Acosta M(2013) Evaluation of ploidy level and endoreduplication in carnation (Dianthus spp.).Plant Sci 201–202:1–11
Bennett MD,Leitch IJ(2005)Plant genome size research:a field in focus.Ann Bot 95:1–6
Burson BL,Actkinson JM,Hussey MA,Jessup RW(2012)Ploidy determination of buffel grass accessions in the USDA National Plant Germplasm System collection by flow cytometry.S Afr J Bot 79:91–95
Dai SL,Wang WK,Xu YX(2005)Phylogenetic relationship of Dendranthema(DC.)Des Moul.revealed by fluorescent in situ hybridization.J Integr Plant Biol 7:783–791
Dolezˇel J,Dolezelova M,Novak FJ(1994)Flow cytometric estimation of nuclear-DNA amount in diploid bananas(Musa acuminata and M.balbisiana).Biol Plantarum 36:351–357
Dolezˇel J(1997)Application of flow cytometry for the study of plant genomes.J Appl Genet 38:285–302
Dolezˇel J,Greilhuber J,Lucretti S,Meister MA,Lysak MA,Nardi L, Obermayer R(1998)Plant genome size estimation by flow cytometry:inter-laboratory comparison.Ann Bot 82(Suppl A):17–26
DolezˇelJ,Kubala´kova´M,BartosˇJ,Macas J(2004)Flow cytogenetics and plant genome mapping.Chromosome Res 12:77–91
Dowrick GJ(1952)The chromosome of Chrysanthemum(I).Heredity 6:365–375
Dowrick GJ(1953)The chromosomes ofchrysanthemum(II).Garden varieties.Heredity 7:59–72
Fukai S,Zhang W,Goi M(1998)Some Dendranthema species native to Japan.Acta Hort 454:85–90
Galbraith DW,Lambert G,Macas J,Dolezel J(2002)Analysis of nuclear DNA contentand ploidy in higherplants.In:Robinson J, Darzynkiewicz Z,Dean P,Hibbs A,Orfao A,Rabinovitch P, Wheeless L(eds)Current protocols in cytometry.Wiley,New York,pp 761–862
Hamill SD,Smith MK,Dodd WA(1992)In vitro induction ofbanana autotetraploids by colchicine treatment of micropropagated diploids.Aust J Bot 40:887–896
Kiszczak W,Krzyzanowska D,Strycharczuk K,Kowalska U,Wolko B,Gorecka K(2011)Determination of ploidy and homozygosity ofcarrotplants obtained from anthercultures.Acta PhysiolPlant 33:401–407
Kondo K,Abd El-Twab MH,Idesawa R,Kimura S,Tanaka R(2003) Genome phylogenetics in Chrysanthemum sensu lato.In: Sharma AK,Sharma A(eds)Plant genome biodiversity and evolution,Part A.phanerogams,vol 1.Science Publishers, Enfield,pp 117–200
Kumar PP,Turner IM,Rao AN,Arumuganathan K(2011)Estimation of nuclear DNA content of various bamboo and rattan species. Plant Biotechnol Rep 5:317–322
Ma YP,Zhou YZ,Wang YZ,Wei JX,Yu ZY,Yang S,Wang Y,Dai SL(2013)CnFL,a FLORICAULA/LEAFY homologue in Chrysanthemum nankingense is dramatically up-regulated in induced shootapicalmeristems.Biochem Syst Ecol50:114–120
Moon YH,Cha YL,Choi YH,Yoon YM,Koo BC,Ahn JW,An GH, Kim JK,Park KG(2013)Diversity in ploidy levels and nuclear DNA amounts in Korean Miscanthus species.Euphytica 193:317–326
Nakata M(1987)Species of wild Chrysanthemum in Japan: cytologicaland cytogeneticalview on its entity.Acta Phytotaxon Geobot 38:241–259
Nakata M,Hong D,Qiu J,Uchiyama H,Tanaka R,Chen X(1992) Cytogenetic studies on wild Chrysanthemum sensu lato in China. II.A naturalhybrid between Chrysanthemum indicum(2n=36) D.vestitum(2n=54)from Hubei Province.J Jpn Bot 67:92–100
Obidiegwu J,Loureiro J,Ene-Obong E,Rodriguez E,Kolesnikova-Allen M,Santos C,Muoneke C,Asiedu R(2009)Ploidy level studies on the Dioscorea cayenensis/Dioscorea rotundata complex core set.Euphytica 3:319–326
Obidiegwu J,Rodriguez E,Ene-Obong E,Loureiro J(2010)Ploidy levels of Dioscorea alata L.germplasm determined by flow cytometry.Genet Resour Crop Evol 57:351–356
Ono S,Hosaka K(2010)Efficient chromosome number estimation using flow cytometry in the backcross of Solanum demissum (2n=6x=72)to S.tuberosum(2n=4x=48).Am J Potato Res 87:553–556
Pfosser M,Amon A,Lelley T(1995)Evaluation of sensitivity of flow cytometry in detecting aneuploidy in wheat using disomic and ditelosomic wheat rye addition lines.Cytometry 21:387–393
Scaldaferro M,Chiarini F,Santin˜aque FF,Bernardello G,Mosconen EA(2012)Geographical pattern and ploidy levels of the weed Solanum elaeagnifolium(Solanaceae)from Argentina.Genet Resour Crop Evol 59:1833–1847
Schlaepfer DR,Edwards PJ,Billete R(2010)Why only tetraploid Solidago gigantea(Asteraceae)became invasive:a common garden comparison of ploidy levels.Oecologia 163:661–673
Tahara M(1915)Cytological studies in Chrysanthemum(A preliminary note).Bot Mag Tokyo 29:48–50
Zhou SJ,Wang JW(1997)The cytological study on ten species of Dendranthema(in Chinese).J Wuhan Bot Res 15:289–292
10 October 2013/Accepted:28 July 2014/Published online:30 April 2015
©Northeast Forestry University and Springer-Verlag Berlin Heidelberg 2015
Project funding:This work was supported by the National Natural Science Foundation of China(No.31470699)and the Fundamental Research Funds for the Central Universities(No.130420003).
The online version is available athttp://www.springerlink.com
Corresponding editor:Zhu Hong
✉Yue-ping Ma mypluna@sina.com Si-lan Dai silandai@sina.com
1College of Life and Health Sciences,Northeastern University,Shenyang 110004,People’s Republic of China
2College of Landscape Architecture,Beijing Forestry University,Beijing 100083,People’s Republic of China
 Journal of Forestry Research2015年3期
Journal of Forestry Research2015年3期
- Journal of Forestry Research的其它文章
- Axial variations in anatomical properties and basic density of Eucalypturograndis hybrid(Eucalyptus grandis×E.urophylla) clones
- Nest site characteristics and nest loss of Marsh Grassbird at Zhalong National Nature Reserve,China
- Variations in electrical impedance and phase angle among seedlings of Pinus densata and parental species in Pinus tabuliformis habitat environment
- Optimization of corn-stalk skin flake-wood shaving composite technology
- Effects of salinity on the nail-holding power of dimension lumber used in light-frame wood building
- Spatial distribution of heavy metals(Cu,Pb,Zn,and Cd) in sediments of a coastal wetlands in eastern Fujian,China
