高等植物适应盐逆境研究进展
张金林,李惠茹,郭姝媛,王锁民,施华中,韩庆庆,包爱科,马清
(1.草地农业生态系统国家重点实验室,兰州大学草地农业科技学院,甘肃 兰州 730020;2.美国德克萨斯理工大学化学与生物化学系,德克萨斯 拉伯克 TX79409,美国)
高等植物适应盐逆境研究进展
张金林1*,李惠茹1,郭姝媛1,王锁民1,施华中2,韩庆庆1,包爱科1,马清1
(1.草地农业生态系统国家重点实验室,兰州大学草地农业科技学院,甘肃 兰州 730020;2.美国德克萨斯理工大学化学与生物化学系,德克萨斯 拉伯克 TX79409,美国)
土壤盐碱化已经成为制约农作物生长及产量的重要因子之一,寻求将盐碱化对植物的危害降低到最小程度的策略势在必行。关于植物对盐逆境适应能力的研究已成为全球关注的热点, 如何提高植物的耐盐能力也已成为研究的重中之重。深入探究高等植物适应盐逆境的机制,有助于提高植物耐盐性,增加作物产量和保护生态环境。本文就高等植物适应盐逆境的重点研究进展,综述了盐胁迫对植物的危害;植物耐盐的生理机制,包括渗透调节、营养元素平衡和增强抗氧化胁迫等;植物耐盐相关基因研究进展,包括离子转运蛋白基因、渗透调节相关基因、信号传导相关基因和细胞抗氧化相关基因等;提高植物耐盐性的途径。最后针对今后植物适应盐逆境方面的研究方向进行了展望。
高等植物;盐胁迫;耐盐性;耐盐基因
地球表面70%的面积被海洋覆盖,海水中Na+浓度大约在500 mmol/L以上,而K+浓度仅为9 mmol/L[1];地壳当中钠元素含量为2.8%,而钾元素为2.6%[2]。因此,地球被称为“咸行星”[1]。在中世纪,盐(氯化钠,NaCl)被称为“白色金子”,因为它是和黄金一样昂贵的商品[3]。盐碱土在陆地生态系统上分布广泛,全世界盐渍土面积约l0亿hm2[4]。在全球的干旱和半干旱地区,约有50%的灌溉土地受到盐碱化的影响,区域内的非灌溉土地同样会发生盐碱化[4]。中国土地盐渍化大约占到全球盐渍土面积的1/10[5],从滨海到内陆,从低地到高原都分布着不同类型的盐渍化土地[6]。在长期的自然进化过程中,海洋植物保留了对高浓度盐分的耐受性,然而,大多数陆生高等植物在进化过程中丧失了这种耐受性而采取了一种甜土植物的生活方式[7]。土壤中可溶性盐分过多,会对植物造成伤害[4,8]。因此,土壤盐碱化已经成为影响农作物生长及产量的重要因子之一,关于植物对盐逆境适应能力的研究已成为全球关注的热点。如何提高植物的耐盐能力已成为研究的重中之重。高等植物对盐逆境的适应是一个综合的生物调节过程,需要各种生理生化过程的协同作用,而非某种单一的过程就能够使植物成功地抵御盐逆境。本文综述了盐胁迫对植物的危害,植物适应盐逆境的方式,提高植物耐盐性的途径和植物耐盐的相关基因研究进展,最后针对植物适应盐逆境方面的研究进行了展望。
1 盐逆境对植物的危害
盐逆境对植物造成的直接危害首先表现为渗透胁迫,并且持续存在;紧接着表现为离子失调引起的毒害和营养元素的亏缺;最后引起的氧化胁迫导致膜透性的改变、生理生化代谢的紊乱和有毒物质的积累,进而引起植物生长发育和形态建成的改变(图1)[1,7-10]。
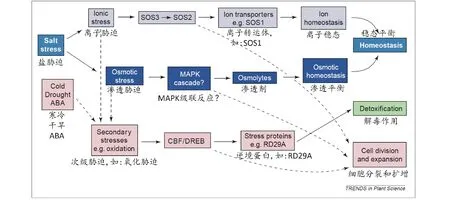
图1 盐胁迫对植物的危害以及植物耐盐的主要生理机制[9]
1.1渗透胁迫
首先,在盐胁迫下,植物种子的萌发会受到影响,一般分为渗透效应和离子效应。渗透效应引起溶液渗透势降低而使种子吸水受阻,从而影响种子萌发;离子效应通过盐离子(Na+、Cl-、SO42-等)直接毒害而抑制种子萌发[11]。在对一年生植物种子耐盐机制进行研究时,发现高盐浓度会对种子产生渗透抑制,只有胁迫减轻时才能消除这种抑制作用[12]。李昀等[13]通过对碱茅(Puccinelliadistans)、黑麦草(Loliumperenne)、野大麦(Hordeumbrevisubulatum)、盐爪爪(Kalidiumcapsicum)、碱蓬(Suaedaglauca)五种牧草研究发现,盐浓度升高,种子的发芽率会降低。其次,对于整株植物而言,若土壤中的盐分过多,则会导致土壤中的水势下降,植物细胞的水势相对过高,导致植物吸水困难,严重的还会引起植物失水,引起生理干旱[7]。
1.2离子失调
土壤中的盐分多以离子形式存在,虽然高等植物对土壤中的离子具有选择吸收作用,但是在吸水的同时,也必定会吸收大量的盐离子[1,7-10]。K+在细胞内可作为60多种酶的活化剂,能促进蛋白质的合成,促进糖类的合成与运输;K+也是构成细胞渗透势的重要成分[14]。而在根内K+可以从薄壁细胞转运至导管,从而降低导管中的水势,使水分能从根系表面转运到木质部中去[15]。K+对气孔开放有直接作用,有研究表明,K+可以维持细胞的渗透平衡,在气孔的关闭中起作用,还可以作为许多酶的辅助因子行使生理功效[16]。土壤中K+的浓度范围在0.2~10.0 mmol/L之间,而K+和Na+的水合半径相似,因此高浓度的Na+会阻碍植物对K+的吸收,造成K+匮缺,从而抑制了以上提到的依赖于K+的生理生化反应的正常进行[4,7]。同时,大量Na+、Cl-进入细胞可以破坏Ca2+平衡,细胞质中游离Ca2+急剧增加,使Ca2+介导的钙调蛋白(calmodulin,CaM)调节系统和磷酸肌醇调节系统失调,细胞代谢紊乱甚至伤害死亡[17]。此外,土壤中高浓度的盐分还会抑制植物对NO3-和NH4+的吸收,而对NO3-吸收的抑制作用更大[18]。
1.3氧化胁迫
细胞膜是植物细胞重要的保护屏障,对物质运输、能量传递、信号转导有重要作用。细胞膜本身具有选择透过性,因而可以调节细胞内的离子平衡,同时满足植物生理活动的需要。然而,盐胁迫导致的氧化胁迫会使膜的透性发生改变,一方面对离子的选择性、流速、运输等产生影响;另一方面,也造成了磷和有机物质的外渗,从而使得细胞的生命活动受到影响[7]。丙二醛(malondialdehyde,MDA)含量的高低和细胞质膜的透性变化是反映膜脂过氧化作用强弱和质膜破坏程度的重要指标,其含量可说明植物遭受逆境伤害的程度[19]。活性氧的增加还会破坏细胞中具有膜结构细胞器的结构,如引起线粒体DNA的突变,造成细胞衰老,导致内质网部分膨胀、线粒体数目减少而体积膨胀、液泡膜破碎、胞质降解等。
1.4光合作用受挫
盐逆境会使得植物的光合速率下降,叶绿素是表征光合利用率的最重要的指标之一。随着盐浓度升高,叶绿素a、b和类胡萝卜素含量均显著降低,表明高盐胁迫抑制或破坏了光系统Ⅱ的部分功能,光合作用的能量及电子传递受到抑制[20]。同时,盐分过多可使磷酸烯醇式丙酮酸羧化酶(phosphoenolpyruvate carboxylase,PEPC)与1,5-二磷酸核酮糖羧化酶(ribulose-1,5-bisphosphate,Rubisco)活性降低,引起胁迫初期光合作用明显下降[21]。
1.5有毒物质的积累
盐胁迫下,由于植物细胞结构的损伤、活性氧的积累、生理代谢的破坏,植株体内蛋白质的合成速率降低,水解加速,造成植株体内氨基酸积累,会产生许多的有毒物质,如大量氮代谢的中间产物,包括NH3和某些游离氨基酸(异亮氨酸、鸟氨酸和精氨酸)转化成有一定毒性的腐胺(如丁二胺、戊二胺等),而腐胺又可被氧化成NH3和H2O2,当它们达到一定浓度时,细胞会中毒死亡[22]。这些物质的积累,会抑制植物内相关物质的合成,使得植物生长受抑。
2 植物耐盐的生理机制
植物受到盐害胁迫时,会采取两种方式来减轻盐胁迫造成的危害:躲避盐离子的伤害和增强对盐胁迫的耐受性[6]。植物避盐的方式主要分为4种:泌盐、稀盐、积盐和拒盐。泌盐(salt excretion)是指植物在吸收盐分后,不在体内存储,而是通过体内的盐腺等器官或机制排出体外,再通过雨水冲刷等方式脱盐,从而维持植物体内的离子稳态。泌盐被认为是一个有助于盐生植物抗盐的重要机制[5,23-24]。稀盐(salt-dilution)是指植物通过吸收大量的水分、使细胞加速生长(增加薄壁细胞组织,使细胞质膨胀,增大细胞壁伸展度等)、改变形态等方法,稀释体内盐分,降低盐浓度[7,23]。积盐(salt-accumulation)是指植物体内的原生质特化后,将根部吸收的盐分区隔化至液泡,同时,抑制盐分从液泡内溢出,即将盐分储存在液泡内,作为廉价的渗透调节剂[7,23]。拒盐(salt-exclusion)是指植物本身排斥盐离子进入细胞,依靠其对盐的不透性,阻止盐分进入植物体;另一方面,植物根部通过阻止Na+向木质部的装载和加大对Na+外排来减少植株对Na+积累。绝大多数栽培植物都属于拒盐能力弱的甜土植物,拒盐能力强的盐生植物有芦苇、碱茅[7,23,25]。除了通过以上4种方式来躲避盐离子的伤害,所有高等植物都可以通过以下几个方面的生理调节过程来增强对盐胁迫的耐受性,即渗透调节、营养元素平衡和增强抗氧化胁迫(图1)。
2.1渗透调节
在盐逆境胁迫下,高等植物通常会采用两种渗透调节方式,一是在植物体内合成有机调节物质;二是积累更多的无机离子。
通常有机溶质大体可分为3类:1) 游离氨基酸(如脯氨酸),具有很大的水溶性,其疏水端可和蛋白质结合,亲水端可与水分子结合,蛋白质可借助脯氨酸束缚更多的水,从而防止渗透胁迫条件下蛋白质的脱水变性;它可以维持细胞内外渗透平衡,防止水分散失[26]。2)甜菜碱,作为一种无毒的渗透调节剂和酶的保护剂,它的积累使植物细胞在盐胁迫下保持膜的完整性,在渗透胁迫下仍能保持正常的功能。许多高等植物,尤其是藜科和禾本科植物,在受到水/盐胁迫时积累大量甜菜碱[27]。甜菜碱对类囊体膜也有稳定作用,并显著提高光系统Ⅱ光合放氧的稳定性;甜菜碱能保护抗氧化酶系统[超氧化物歧化酶(superoxide dismutase,SOD)、抗坏血酸过氧化物酶(ascorbate peroxidase,APX)、过氧化氢酶(catalase,CAT)等]的活性[28];甜菜碱还可以稳定生物大分子的结构与功能,解除高浓度盐对酶的毒害作用,防止脱水诱导的蛋白质热动力学干扰[29]。3)可溶性糖和多元醇,也可以作为渗透调节物质,调节植物细胞的渗透势,从而增强植物的耐盐机制[30]。
无机离子(K+,Na+和Cl-)作为渗透调节剂具有很多优点,也日益受到重视。首先,由于离子的大量存在,无需消耗物质和能量来大量合成,所以较为廉价;第二,无机离子的调节作用可以在短时间内迅速完成;第三,它们的作用也显著高效[31]。无机离子在整株植物渗透调节中的作用可以很简易地通过直接测量茎或根的渗透压而容易的验证,并且可以与它们在组织液中的浓度进行比较。无机离子主要是 K+、Na+、Cl-,无论是在盐生植物还是在非盐生双子叶植物中,这3种离子提供了80%至95%的细胞液渗透压[31]。K+是循环丰富的离子,同时K+是植物生长的必需元素,在维持细胞的基本功能中扮演了重要角色,并且在保持低水平的蛋白酶和核酸内切酶活性以及防止植物在盐胁迫下细胞损伤和死亡中的作用也是不言而喻的[32]。有研究表明,碱茅与更敏感的长穗偃麦草(Elytrigiaelongata)相比,在严重的高盐或缺氧条件下,长穗偃麦草将更严重地丢失K+,从而证明了K+保留在根中对高盐缺氧土地耐受性的作用[33]。而对积盐型盐生植物而言,对Na+的吸收远远大于K+的吸收[34]。绝大部分被植物细胞吸收的Na+并非存在于细胞质中,而是区隔化在液泡中作为廉价的渗透调节物质来维持正常细胞膨压,从而增强植物抵御盐逆境的能力[35-37]。研究表明,在介质当中添加适量的NaCl可以增强荒漠植物霸王(Zygophyllumxanthoxylum)的抗旱性,从而促进其生长[38-39];进一步分析表明,干旱胁迫下霸王叶中Na+浓度显著增加了64%,Na+对叶渗透势的贡献由8%增至13%;50 mmol/L NaCl处理使干旱胁迫下霸王叶中Na+显著增加了2.3倍,Na+对叶渗透势的贡献增至28%,从而提高了植株的渗透调节能力[38]。
2.2营养元素平衡
在正常生理状态下,植物细胞内的离子保持均衡状态,而在盐胁迫下,细胞质中过多的离子尤其是Na+对植物细胞的代谢活动会有伤害。大多数植物在盐胁迫下,组织内的K+含量会降低[4]。王锁民等[40]在对植物Na+、K+的选择性吸收及运输的研究基础上,首次提出植物根系对土壤中Na+、K+的选择性吸收(selective absorption,SA)能力以及植株不同部位对Na+、K+选择性运输(selective transport,ST)能力的计算公式。研究表明,小花碱茅(Puccinelliatenuiflora)可以在高Na+环境下生存,且其体内维持很低的Na+浓度,主要依靠其对K+/Na+强大的选择性吸收能力和限制Na+的吸收[25]。这一结果在对盐敏感和耐盐品种的长穗偃麦草的比较研究中得到了进一步证实,即耐盐品种的SA和ST值均显著大于盐敏感品种[41]。海滨碱蓬(Suaedamaritima)体内的K+含量变化是随NaCl浓度的升高而呈升高趋势,这样既可以保持一定的K+营养,还可以保持相对稳定的K+/Na+,这对植物本身生长有利[42]。在盐胁迫的条件下,提供某些微量元素,可有效地提高植物的含水量,促进植物的光合作用,有利于植物生长。研究表明,在含有Na+的土壤中添加硅时,植株的含水量提高了7.3%,同时,植物的光合效率、CO2同化效率等也得到了显著地提高;在黄瓜(Cucumissativus)[43]、大麦(Hordeumvulgare)[44]、水稻(Oryzasativa)[45-46]、紫花苜蓿(Medicagosativa)[47]、小麦[48]和草地早熟禾(Poapratensis)[49]等植物研究中也得到了同样的结果。
2.3增强抗氧化胁迫
在正常的生理条件下,植物体内的活性氧自由基和自身的抗氧化系统对活性氧的清除是动态平衡的,可以保持体内正常的代谢过程。在干旱、盐渍等胁迫下,膜脂过氧化作用加剧,植物体内活性氧含量上升,随之超氧化物歧化酶、过氧化物酶(peroxidase,POD)、过氧化氢酶和抗坏血酸(ascorbic acid,ASA)等保护酶的活性也相应增加,从而防止膜脂的过氧化作用,以此来增强植物对逆境的耐受性[50]。管博等[51]在盐地碱蓬(Suaedasalsa)的研究发现,随着盐胁迫的增强,SOD作为防御活性氧自由基(reactive oxygen species,ROS)的第一道防线,其活性显著增加,CAT活性变化趋势与SOD相似。此发现在对梭梭(Haloxylonammodendron)[52]和沟叶结缕草(Zoysiamatrella)[53]的研究中也得到证实。有研究表明,高盐和干旱处理下,西藏野生大麦(Hordeumvulgarevar.trifurcatum)体内的ROS水平会显著升高来抵御伤害[54]。然而在重度盐胁迫下,植物体内的这些活性氧去除剂的结构发生破坏,植物清除活性氧的防御能力下降,使膜脂的过氧化作用加剧,破坏细胞膜的透性[4,7,55]。
3 高等植物耐盐相关基因研究进展
随着分子生物学的发展,人们能够在基因组成、表达调控及信号转导等分子水平上认识植物的耐盐机理;并且已经通过对植物耐盐相关基因的研究,来进一步提高植物的耐盐性。目前,高等植物耐盐相关基因的研究主要集中在离子转运蛋白基因、渗透调节相关基因、信号传导相关基因、细胞抗氧化相关基因等。
3.1离子转运蛋白基因
3.1.1质膜上离子转运蛋白基因 有研究表明,环核苷酸调控通道(cyclic-nucleotide-gated channel,CNGCs)和非选择性阳离子通道/电压非依赖型通道(non-selective cation channel/voltage-independent channel,NSCCs/VICs)是不同类型的通道蛋白,在一些植物中CNGCs涉及Ca2+的信号转导,而NSCCs/VICs涉及Na+的摄入[56]。而NSCCs/VICs[57]是根部Na+进入细胞膜的通道蛋白,当NSCCs通道被抑制时,就可缓解细胞的盐胁迫(图2)。Tyerman和Skerrett[58]在研究小麦根部的NSCCs通道时发现,涌入的Na+主要分布于根部的各个区室及表皮的原生质内。低亲和性阳离子转运体(low-affinity cation transporter,LCT1)是从小麦中发现的一类能够介导低亲和性阳离子吸收的蛋白[59]。研究表明,小麦中的LCT1不仅可以维持Rb+与Na+的浓度的平衡,吸收少量的Ca2+,也可维持Na+/K+的浓度[60];但由于受到土壤中Ca2+浓度的影响,LCT1并不是Na+流入的主要通道[4,61]。
高亲和K+转运载体(high-affinity K+transporter,HKT)是一种与植物耐盐性密切相关的Na+或Na+-K+转运蛋白,能将植物木质部中过多的Na+卸载到其周围薄壁细胞中,降低地上部Na+含量,并维持体内K+稳态平衡[4,62](图2)。根据在异源表达系统中对Na+和K+运输的不同,HKT蛋白可被分成两种[63]:HKT1作用于外部K+缺乏时,主要用于特异性Na+的运输和介导Na+的吸收[64];HKT2则是有K+-Na+协同转运蛋白及其同系物的功能[4]。Ren等[65]绘制了一个水稻SKC1基因的QTL图谱,并发现SKC1基因编码一种HKT型K+/Na+通道蛋白,在植物盐胁迫下调节K+/Na+离子平衡,维持细胞内外的渗透压,从而提高植物耐盐能力,后来证明SKC1为水稻HKT1;5基因。Laurie等[66]研究表明HKT2基因的低表达可以抑制Na+的进入,从而降低组织内Na+的浓度。作为高亲和性Na+转运的转运体,HKT转运蛋白在草类以及其他植物中可能都在控制整体植株中Na+的转运起重要作用[2]。Kader等[67]在研究水稻时发现,转运蛋白基因OsHKT1、OsHKT2和OsVHA在盐胁迫下诱导表达,可以通过调节Na+与K+的比例而降低Na+的浓度。研究表明,SsHKT1在盐地碱蓬的离子平衡和耐盐方面有重要作用[68]。
众多研究表明,高亲和K+转运载体(high-affinity K+transporter,HAK)基因对Na+的吸收也有重要作用[69]。盐对HvHAK1积累影响的分析结果表明,在转录水平上,一个强而短暂NaCl胁迫触发HvHAK1的上调,说明HvHAK1是一个高盐应答的快速反应基因。还有研究表明,在盐逆境胁迫下,HvHAK1 mRNA的积累和对K+摄取的控制是高度相关的[70],对Na+吸收的控制则对维持细胞内的pH重要[71]。研究表明,PhaHAK5、PhaHAK2-n、PhaHAK2-e、PhaHAK2-u等基因均可调节盐生植物中Na+的浓度[72]。
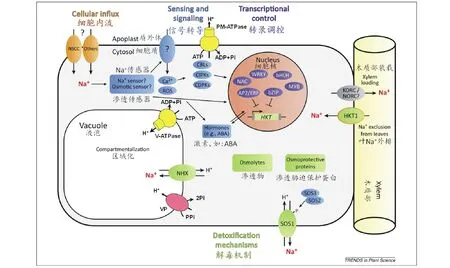
图2 植物细胞Na+运输机制及响应盐逆境的重要调控网络(在Deinlein等[62]基础上修订)
拟南芥K+转运体(ArabidopsisK+transporter,AKT1)类的通道蛋白是在植物中广泛表达的一类内整流K+通道。作为一条对NH4+不敏感的K+吸收途径,AKT1的主要功能是与对NH4+敏感的K+吸收途径并行存在,使植物能够适应多样的外界环境。研究表明,小麦中K+缺乏时,TaAKT1通道会增强对Na+的吸收。众多研究显示,OsAKT1的表达并不影响植物中K+的浓度,但是Na+的积累却依靠此基因的表达[73]。KUP基因家族对细胞内K+浓度的调节有多种类型[74],它们的表达可以表现出对K+的高吸收性,但是在拟南芥(Arabidopsisthaliana)中,NaCl的加入会抑制K+的吸收[75]。Wang等[34]研究表明,积盐型盐生植物海滨碱蓬存在两条低亲和性Na+吸收途径,即在低浓度NaCl(25 mmol/L)条件下主要由HKT介导Na+的吸收,在高浓度NaCl(150 mmol/L)条件下主要由AKT1介导Na+的吸收。Zhang等[76]进一步研究发现两条途径的外界Na+浓度介于95~100 mmol/L NaCl之间。
盐敏感(salt overly sensitive,SOS)基因是从一类超盐敏感突变体中发现的,遗传分析表明,这些突变体可分为5种,即sos1、sos2、sos3、sos4和sos5[77-78]。目前筛选出的一些SOS基因家族编码重要的植物离子载体蛋白、信号转导蛋白等,它们的存在对提高植物的耐盐性有重要作用。质膜Na+/H+逆向转运蛋白就是由SOS1编码的,该蛋白的主要功能就是将Na+排到细胞外部,从而减少细胞内Na+的积累[62,79-80](图2)。Guo等[81]研究表明,SOS1在小花碱茅拒Na+当中发挥着重要作用。Ma等[82]对霸王ZxSOS1的研究表明,ZxSOS1参与调控霸王体内Na+、K+转运和空间分配,进而影响植株生长。Feki等[83]将硬质小麦(Triticumdurum)TdSOS1转入拟南芥显著提高了转基因植物的耐盐性。Nie等[84]通过将大豆(Glycinemax)GmsSOS1基因转到缺失SOS1的拟南芥中,证明了其互补性,即验证了SOS1的功能。SOS2和SOS3都是编码信号转导途径中的蛋白,在信号转导过程中,彼此有着千丝万缕的联系。同时,有研究表明二者功能的行使与Ca2+有密切的关系[85]。目前研究的SOS信号通路就是:高浓度的Na+引起细胞内Ca2+浓度上升,Ca2+与SOS3结合,结合Ca2+后的SOS3与SOS2直接互作而激活了SOS2,之后通过SOS3氨基末端的酰化(amino terminus myristoylation)使SOS3-SOS2复合体会铆钉到细胞质薄膜上,该复合体中被激活的SOS2磷酸化质膜上的SOS1,从而提高SOS1对Na+的转运活性而促进高浓度Na+的外排[7]。但在拟南芥和水稻之间SOS信号通路似乎具有保守型,在水稻中,功能性同系物SOS1,SOS2,SOS3也构成一个重要的耐盐的信号通路[86]。SOS5则在细胞壁形成、细胞伸展、植物繁殖等过程中起重要作用[77]。这一系列研究表明,在盐胁迫下SOS基因家族对植物的耐盐性有重要作用。
3.1.2液泡膜上离子转运蛋白基因 成熟植物细胞的液泡为Na+储存提供弄了一个很大的空间。将Na+区域化在液泡中是减少细胞溶质之中Na+的有效方式,从而减少胞液中Na+的毒害作用。钠和氯离子在细胞溶质中的浓度是由穿过质膜和液泡膜的净通量决定的。液泡膜蛋白中,Na+/H+逆向转运蛋白(Na+/H+transporters,NHX)参与运输Na+,该转运蛋白是由液泡膜H+-ATP酶和H+-焦磷酸酶的作用所形成的质子梯度来驱动的(图2)[34,62]。NHX基因被证明可以将细胞质中的Na+区域化在液泡中。第一个从拟南芥克隆出的NHX型转运体基因被命名为AtNHX1[87],它属于AtNHX1-6亚科六成员之一。这组转运体运用质子驱动力来运输Na+和K+[88-89]。除了在耐盐性中的作用,NHX转运体还涉及许多其他细胞过程,包括细胞内pH值调节[90],囊泡运输和蛋白定位[91]。从发现拟南芥中NHX转运体后,已在许多其他植物中发现了此类型的转运体,包括水稻OsNHX1[92]、细叶海滨藜(Atriplexgmelini)AgNHX1[93]、大麦HvNHX1[94]、棉花(Gossypiumspp.)GhNHX1[95]、玉米ZmNHX[96]、小麦TaNHX1[97]和TaNHX2[98]、苜蓿MsNHX1[99]、大豆(Glycinemax)GmNHX1[100]、长穗偃麦草(Elytrigiaelongata)AeNHX1[101]、珍珠狼尾草(Pennisetumglaucum)PgNHX1[102]、灰绿藜(Chenopodiumglaucum)CgNHX1[103]、胡杨(Populuseuphratica)PeNHX1-6[104]、盐穗木(Halostachyscaspica)HcNHX1[105]、霸王ZxNHX1[37]、海蓬子(Salicorniabigelovii)SbNHX1[106]和花花柴(Kareliniacaspia)KcNHX1和KcNHX2[107]等。 Wu等[37]发现在盐或干旱条件下,霸王ZxNHX的上调与Na+的积累呈正相关关系。进一步研究表明,霸王ZxNHX在控制Na+、K+的吸收,长距离运输和整株内离子稳态方面具有重要作用[108]。自从报道AtNHX基因超表达可以使拟南芥具有耐盐性后[87],AtNHX或其他NHX基因在植物中超表达以使之具有抗逆性的成功例子也越来越多。例如将拟南芥AtNHX1分别导入玉米[109]、小麦[110]、棉花[111]和花生(Arachishypogaea)[112],将细叶海滨藜AgNHX1导入水稻[93],将棉花GhNHX1导入烟草[95],将短芒大麦草(Hordeumbrevisubulatum)HbNHX1导入烟草[98],将狼尾草(Pennisetumalopecuroides)PgNHX1导入芥菜(Brassicajuncea)[113]和水稻[102],将长穗冰草(Agropyronelongatum)AeNHX1导入拟南芥和高羊茅(Festucaelata)[101],将拟南芥AtNHX5导入兰猪耳(Toreniafournieri)[114],将獐茅(Aeluropussinensis)AlNHX导入烟草[115],将小麦TaNHX1导入紫花苜蓿[116],将北美海蓬子(SalicorniaBigelovii)SbNHX1导入麻疯树(Jatrophacurcas)[117],将绿豆(Vignaradiata)VrNHX1导入拟南芥[118],均显著提高了转基因植物的耐盐性。
NHX转运体通过跨液泡膜H+梯度驱动来转运Na+至液泡,因此通过H+-ATP酶和H+-焦磷酸酶的超表达增加H+驱动力,从而有可能提高抗盐性。然而,由于其多亚基的特征,液泡H-ATP酶并不是超表达的理想候选者,因为不大可能将所有的亚基在相似的水平过量表达而去创建一个功能性的超表达转基因植物。液泡膜H+-焦磷酸酶(vacuolar pyrophosphatase,VP)是用于此目的的一个更好的选择,因为它是一个单一的多肽蛋白[119-120],并且,它产生H+梯度的能力可与H-ATP酶相媲美(图2)[62,121]。H-焦磷酸酶基因也首先在拟南芥中被克隆出来,命名为AVP1[119]。VP基因也已从其他许多高等植物中克隆得到,如盐芥TsVP[121]和盐地碱蓬SsVP等[122]。拟南芥AVP1的过量表达赋予转基因植物耐旱和耐盐性,在许多植物中得到了证实,如拟南芥[120,123]、番茄[124]、紫花苜蓿[125]、匍匐翦股颖(Agrostisstolonifera)[126]、大麦[127]、甘蔗(Saccharumofficinarum)[128]。同时,其他VP基因导入植物后,植物抗逆性也得到了提高,例如将盐地碱蓬SsVP1转入拟南芥[122],将盐芥(Thellungiellahalophila)TsVP转入烟草[129]、棉花[130]和玉米[130],将盐爪爪KfVP1转入拟南芥[131],将小麦TaVP1转入烟草[132],将小麦TaVP基因转入烟草[133]。
近年来,一些学者将NHX和VP两种转运蛋白基因构建双价表达载体同时超表达进行了转基因研究,如在水稻中超表达盐地碱蓬SsNHX1和拟南芥AVP1[134],水稻OsNHX1和OsVP1[135];在拟南芥中超表达小麦TaNHX1和TaTVP1[136];在西红柿中超表达狼尾草PgNHX1和拟南芥AVP1[137];在烟草中超表达小麦TaNHX1和TaTVP1[138]以及在百脉根(Lotuscorniculatus)中超表达霸王ZxNHX和ZxVP1-1[139]。
3.2渗透调节相关基因
渗透胁迫是植物盐胁迫的主要方面,渗透调节相关基因在植物耐盐性中的作用也进行了较多研究。多元醇、脯氨酸、海藻糖、甜菜碱等均是植物耐盐的重要渗透调节物质,因而合成这些渗透调节物质的一些关键基因在耐盐中起到重要作用。如mtlD基因和gutD基因,它们分别编码合成甘露醇和山梨醇的酶。超表达mtlD基因增加了甘露醇在毛白杨(Populustomentosa)体内的积累从而增强了其耐盐性[140];转gutD基因可使转基因植株产生山梨醇,转基因玉米可耐受1.17% NaCl的浓度[141];在水稻中超表达mtlD和gutD基因提高了其耐盐性[142]。Kishor等[143]对P5CS基因进行研究,获得P5CS转基因烟草,其中脯氨酸的含量明显提高,与对照相比,耐盐性有提高。晚期胚胎发生富集蛋白(late embriogenesis abundant protein,LEA)是Dure等[144]首次在棉花种子发育晚期胚胎中发现的一类蛋白。Xu 等[145]将LEA基因转入水稻悬浮细胞中,得到的转基因植株经过繁殖,在第二代中表现出耐盐的能力。张宁等[146]从菠菜(Spinaciaoleracea)叶片中分离了甜菜碱醛脱氢酶(betaine-aldehyde dehydrogenase,BADH)基因,并将该基因与其他植物的BADH序列作了同源性分析,同时,证实了菠菜BADH基因的转录与表达受干旱和盐胁迫的诱导。Jia等[147]将山菠菜(Atriplexhortensis)的BADH基因转入烟草,提高了烟草的耐盐性。在水稻中过量表达缺失D结构域的组成型活性突变形式OsbZIP46CA1可以显著增强耐旱性和抵抗渗透胁迫的能力,并可显著上调已知的逆境应答基因(包括ABF或AREB类成员的下游基因)的表达[148]。
3.3信号传导相关基因
在盐胁迫下,植物可以关闭或开启某些基因的表达,从而维持自身的营养平衡。与植物耐盐性相关的信号转导途径包括:SOS途径(见3.1.1)、脱落酸(abscisic acid,ABA)信号和蛋白激酶途径。
ABA应答基因可分为ABA依赖型和ABA非依赖型。目前,已发现多种转录因子与盐胁迫有关,这些基因的表达分为4条途径:两条依赖ABA途径,两条不依赖ABA途径[149]。十字花科植物盐芥是拟南芥的近亲,它耐盐,耐低温和氧化胁迫,盐芥基因组相对较小,因此成为研究植物耐受逆境分子机理的理想材料[150]。在盐芥中研究发现,所有涉及ABA生物合成途径的基因家族在盐胁迫下基因数目增加,这种增加会调节复杂的ABA生物合成过程[45,151]。
蛋白激酶在调节植物对非生物胁迫的响应中发挥了重要作用。蛋白激酶是信号转导的重要元件,信号转导中丝裂原活化蛋白激酶(mitogen-activated protein kinase,MAPK)途径是重要途径之一。目前已分离到多种能被逆境诱导的MAPK基因:拟南芥AtMEKK1、AtMEK1/AtMEK2、ATMPK3、AtMPK4、ANP1、AtMKK2以及烟草NPK1、AtMPK3和AtMPK6等[152-153]。III型亚家族SnRK2蛋白(SnRK2.6/2.3/2.2)是拟南芥脱落酸信号转导过程中关键的正调控因子。这些激酶受脱落酸或渗透胁迫激活后,可磷酸化与胁迫相关的转录因子和离子通道,最终使植物免受高盐的危害[154]。有研究表明,野大麦的CIPK蛋白(CBL-interacting protein kinase,CIPK)HbCIPK2对盐和渗透胁迫耐受性起正调节作用,HbCIPK2有助于阻止根中K+的丢失和Na+的积累,以便维持K+/Na+的动态平衡和细胞免于死亡[155]。有研究结果表明,PLDα1(磷脂酶)D衍生的磷脂酸(phosphatidic acid,PA)与微管结合蛋白(microtubule-associated protein,MAP)MAP65-1结合后,可调控微管的稳定及对盐胁迫的耐受性。MAP65-1是PA的一个靶蛋白,该研究揭示了在环境胁迫引起的信号转导过程中膜脂与细胞骨架之间的功能性联系[156]。这一研究结果提出了植物中磷脂酸通过调控微管结合蛋白活性参与盐胁迫反应的新机制。
3.4细胞抗氧化相关基因
膜脂的过氧化导致细胞膜透性的增加是盐胁迫的主要危害之一。细胞膜本身具有选择透过性,表面具有多种离子运输与传递的蛋白,内部有多种抗氧化机制,因而可以抵制膜内不饱和脂肪酸的过氧化作用。目前发现,SOD、POD、GSH等是细胞内主要的抗氧化物质。它们本身可抑制氧自由基的产生,清除体内的活性氧。众多研究表明,植物体内某些基因的存在,对植物的抗氧化有重要作用。如:NtGST/GPX基因编码的酶既有谷胱苷肽-S-转移酶活性,又有谷光苷肽过氧化物酶活性,将该基因在烟草中过量表达,可以增强植物的耐盐性和耐寒性[157]。Kovtun等[158]将一种MAPKKK基因——ANP1基因转入烟草中发现,它可通过MAPK级联反应激活谷胱苷肽-S-转移酶基因(GST6)的表达,从而使植株表现出耐盐性。S-腺苷甲硫氨酸脱羧酶(S-adenosylmethionine decarboxylase,SAMDC)是多胺合成中的一个关键酶,SAMDC基因在烟草中过量表达可以增强植物对盐和其他非生物胁迫的抗性。维生素C是植物体必需的维生素之一,在光合电子传递、活性氧清除、氧化还原平衡调控、细胞分裂与生长、衰老与凋亡等生理过程中具有重要作用。在对拟南芥的研究中表明,拟南芥AtERF98是维生素C合成的关键转录调控因子,盐可以诱导AtERF98表达,而AtERF98突变显著抑制盐诱导的维生素C合成,并降低清除活性氧的能力,表现为盐敏感。这些结果表明,ERF类转录因子可通过调控抗氧化物质的合成来调节植物对盐胁迫的应答反应[159]。
4 提高植物耐盐性的途径
提高植物耐盐性的途径有很多,其中包括抗盐锻炼,加入生长调节剂和培育耐盐新品种等。
4.1抗盐锻炼
植物耐盐能力常随生长发育时期的不同而异,且对盐分的抵抗力有一个适应锻炼过程。种子在一定浓度的盐溶液中吸水膨胀,然后再播种萌发,可提高作物生育期的耐盐能力。因此,在对植物进行抗盐锻炼时,可逐渐升高Na+浓度,从而提高植物的耐盐能力。用CaCl2浸种的玉米在盐胁迫下的叶绿素含量、细胞膜透性和根系活力的变化程度均小于水浸种,脯氨酸含量、干物质重高于水浸种,水势低于水浸种,提高了三叶期玉米的耐盐能力[160]。在低浓度钠盐处理下,夏枯草(Prunellavulgaris)种子的发芽率、发芽势、活力指数、根长、苗高以及鲜重都得到了显著提高[161]。
4.2加入生长调节剂
在很多情况下,植物对逆境胁迫的响应是通过改变内源激素的水平来实现的。如植物处于非生物逆境胁迫条件下会产生大量的ABA和乙烯(ethylene,Eth)等。生长素不仅调控植物的生长发育,也广泛参与逆境胁迫反应[162]。植物使用不同的策略来应对高盐土壤,植物可以调整自身的根系结构和根生长的方向来避免局部盐浓度过高的情况[163]。有一种模式就是通过盐度影响了植物激素在根系的分布,从而影响根生长。除了植物生长素(indole-3-acetic acid,IAA),细胞分裂素(cytokinin,CTK),乙烯和脱落酸也有相应的作用。植物生长素的运用可以通过促进侧根形成从而缓解渗透胁迫并不受ABA的影响,这表明这两种激素的行为独立地确定侧根原基的命运[164]。用植物激素处理植株,是常用的提高植物耐盐性的途径之一。例如,在含0.15% Na2SO4土壤中的小麦生长不良,但在播前用IAA浸种,可以抵消Na2SO4抑制小麦根系生长的作用,使小麦生长良好[165]。ABA是一种逆境激素,能诱导气孔关闭,减少蒸腾作用、减少盐的吸收,提高作物的耐盐能力,它普遍存在于高等植物中,在植物对逆境的适应中有着重要的作用[166]。此外,NO在植物耐受逆境中的作用尤其引人注意。如NO能显著缓解盐对盐地碱蓬种子萌发的抑制,外源NO供体硝普钠(sodium nitroprusside,SNP)能通过促进种子吸水以缓解盐的渗透胁迫来缓解盐胁迫下盐地碱蓬种子萌发[167]。并且SNP还可以缓解盐胁迫对蒺藜苜蓿(Medicagotruncatula)种子萌发的抑制作用[168]。
4.3培育耐盐品种
随着植物分子生物学研究的深入、植物基因工程的发展、育种学的进步,培育耐盐的植物品种已经从普通的生理生化等表面的研究深入到了分子领域。现今,人们可以通过改造植物内部的基因、构建耐盐基因的载体和遗传转化等方法进一步改良植株。如上文所述,利用高等植物耐盐相关基因培育耐盐作物品种将会成为未来研究的主要方向。此外有研究表明,植物的多倍体化同样可以使植物具有耐盐性,同时增强植物钾的积累[169]。
5 展望
近年来对于作物的耐盐性研究发展迅速,但在很大程度上仍然停留在通过鉴定和利用拟南芥中已知的相应基因来进行耐盐性调控的水平。对水稻[64,170]和小麦[171-172]的研究已经指出了一条阐明作物特别是禾谷类作物耐盐机制的研究方向,但对作物的系统性研究仍然需要使用正向和反向遗传工具以及植物生理学、生物化学和分子生物学等手段。用正向遗传筛选作物盐敏感突变体从而确定作物中重要的耐盐基因仍然没有广泛开展,这种方法将有助于找到新基因或特定的作物耐盐机制。可以相信,拟南芥中未确定的某些机制,可能存在于作物中,特别是具有不同体系结构和组织构架的单子叶植物组织中。
由3种类型的Na+转运体调控的3种细胞水平耐盐机制(控制Na+吸收,增强Na+外排,提高Na+区域化)可以共同协调来提高植物的抗盐能力。然而,虽然在细胞水平上这3种Na+转运机制易于理解,但在整体植物中这些转运体在特定位置上的协调作用机制仍然没有完全搞清楚。因此,还需要在生理和分子水平上进一步研究来确定这些转运体的组织和细胞定位以阐明其在抗盐中的相互协调功能。除此之外,这些转运体在其他生物过程中的作用也有待于进一步研究。显而易见的是,这些转运体不仅仅对植物耐盐性很重要,而且还涉及其他的细胞生理学过程,这在拟南芥中已经得到证实;然而,除了耐盐性以外,这些转运体在作物中的其他作用还不是很清楚。对这些方面研究将会有助于我们估计过量表达这些转运体来增强植物耐盐性后所产生的潜在副作用。
盐生植物为阐明高等植物耐盐机制的研究提供了天然资源。利用亲缘关系密切的盐生植物和甜土植物以及比较基因组学研究方法将有助于阐明盐生植物耐盐性的遗传基础。Wu等[150]的研究证明了这一结论,即利用比较基因组学方法来确定盐芥基因组和拟南芥基因组之间的差异以及其与盐胁迫反应和抗盐的关系,最终鉴定其耐盐的分子机制。虽然比较基因组学的研究需要完整的基因组序列,但可以预见的是,随着测序技术的迅速发展和相关成本的减少,可供利用的基因组序列将会更加丰富。这种方法也可以用于鉴定自然变异种群中耐盐变种中的耐盐基因的变化,这会在分子育种方面为作物的耐盐性遗传改良提供有价值的信息。此外,转录组学、蛋白质组学和表观遗传学等手段将加速盐生植物耐盐机制的研究和耐盐基因的大量挖掘。
[1] Flowers T J.Improving crop salt tolerance.Journal of Experimental Botany, 2004, 55:307-319.
[2] Kronzucker H J, Coskun D, Schulze L M,etal.Sodium as nutrient and toxicant.Plant and Soil, 2013, 369:1-23.
[3] Janz D, Polle A.Harnessing salt for woody biomass production.Tree Physiology, 2012, 32:1-3.
[4] Zhang J L, Flowers T J, Wang S M.Mechanisms of sodium uptake by roots of higher plants.Plant and Soil, 2010, 326:45-60.
[5] Zhao K F, Li F Z, Fan S J,etal.Halophytes in China.Chinese Bulletin of Botany, 1999, 16(3):201-207.
[6] Zhao K F.Plants adapt to salt adversity.Bulletin of Biology, 2002, 37(6):7-10.
[7] Zhang J L, Shi H Z.Physiological and molecular mechanisms of plant salt tolerance.Photosynthesis Research, 2013, 115:1-22.
[8] Munns R, Tester M.Mechanisms of salinity tolerance.Annual Review of Plant Biology, 2008, 59:651-681.
[9] Zhu J K.Plant salt tolerance.Trends in Plant Science, 2001, 6:66-71.
[10] Kronzucker H J, Britto D T.Sodium transport in plants:a critical review.New Phytologist, 2011, 189:54-81.
[11] Gorai M, El A W, Yang X,etal.Toward understanding the ecological role of mucilage in seed germination of a desert shrubHenophytondeserti:interactive effects of temperature, salinity and osmotic stress.Plant and Soil, 2014, 374:727-738.
[12] Wei Y, Dong M, Huang Z Y,etal.Factors influencingseed germination ofSalsolaaffinis(Chenopodiaceae), a dominant annual halophyte inhabiting the deserts of Xinjiang.Flora of China, 2008, 203:134-140.
[13] Li Y, Shen Y Y, Yan S G.Comparative studies of effect of NaCl stress on the seed germination of 5 forage species.Pratacultural Science, 1997, 14(2):50-53.
[14] Liang Y C.Effects of silicon on enzyme activity and sodium, potassium and calcium concentration in barley under salt stress.Plant and Soil, 1999, 209:217-224.
[15] Trono D, Flagella Z, Laus M N,etal.The uncoupling protein and the potassium channel are activated by hyperosmotic stress in mitochondria from durum wheat seedlings.Plant Cell and Environment, 2004, 27:437-448.
[16] Becker D, Hoth S, Ache P,etal.Regulation of the ABA-sensitiveArabidopsispotassium channel gene GORK in response to water stress.Febs Letters, 2003, 554:119-126.
[17] Ottow E A, Brinker M, Teichmann T,etal.Populuseuphraticadisplays apoplastic sodium accumulation, osmotic adjustment by decreases in calcium and soluble carbohydrates, and develops leaf succulence under salt stress.Plant Physiology, 2005, 139:1762-1772.
[18] Song J, Ding X D, Feng G,etal.Nutritional and osmotic roles of nitrate in a euhalophyte and a xerophyte in saline conditions.New Phytologist, 2006, 171:357-366.
[19] Liu J, Cai H, Liu Y,etal.A study on physiological characteristics and cmparison of salt resistance of twoMedicagosativaat the seeding stage.Acta Prataculturae Sinica, 2013, 22(2):250-256.
[20] Jarunee J, Kenjiusui, Hiroshi M.Differences in physiological responses to NaCl between salt-tolerantSesbaniarostrataBrem.and Obem.And non-tolerantPhaseolusvulgarisL.Weed Biology and Management, 2003, 3:21-27.
[21] Makela P, Karkkainen J, Somersalo S.Effect of glycinebetaine on chloroplast ultrastructure, chlorophyll and protein content, and RuBPCO activities in tomato grown under drought or salinity.Biologia Plantarum, 2000, 43:471-475.
[22] Leshem Y, Melamed B N, Cagnac O,etal.Suppression ofArabidopsisvesicle-SNARE expression inhibited fusion of H2O2containing vesicles with tonoplast and increased salt tolerance.Proceedings of the National Academy of Sciences of the United States of America, 2006, 103:18008-18013.
[23] Flowers T J, Colmer T D.Salinity tolerance in halophytes.New Phytologist, 2008, 179:945-963.
[24] Shabala S, Bose J, Hedrich R.Salt bladders:do they matter.Trends in Plant Science, 2014, 19(11):687-691.
[25] Wang C M, Zhang J L, Liu X S,etal.Puccinelliatenuifloramaintains a low Na+level under salinity by limiting unidirectional Na+influx resulting in a high selectivity for K+over Na+.Plant, Cell and Environment, 2009, 32:486-496.
[26] Ueda A, Yamamoto-Yamane Y, Takabe T.Salt stress enhances proline utilization in the apical region of barley roots.Biochemical and Biophysical Research Communications, 2007, 355:61-66.
[27] Wang C Q, Zhao J Q, Chen M,etal.Identification of betacyanin and effects of environmental factors on its accumulation in halophyteSuaedasalsa.Journal of Plant Physiology and Molecular Biology, 2006, 32(2):195-201.
[28] Md A H, Mst N A B, Yoshimasa N,etal.Proline and glycinebetaine enhance antioxidant defense and methylglyoxal detoxification systems and reduce NaCl-induced damage in cultured tobacco cells.Journal of Plant Physiology, 2008, 165:813-824.
[29] Incharoensakd A, Takabe T, A kazawa T.Effect of bteaine on enzyme activity and subunit internation of ribulose-1,5 -bisphosphate carboxylase/oxygenas fromAphnothecehalophytica.Plant Physiology, 1986, 81:1044-1049.
[30] Dubey R S, Singh A K.Salinity induces accumulation of soluble sugars and alters the activity of sugar metabolising enzymes in rice plants.Biologia Plantarum, 1999, 42:233-239.
[31] Shabala S, Shabala L.Ion transport and osmotic adjustment in plants and bacteria.Biomolecular Concepts, 2011, 2:407-419.
[32] Demidchik V, Cuin T A, Svistunenko D,etal.Arabidopsisroot K+-efflux conductance activated by hydroxyl radicals:single-channel properties, genetic basis and involvement in stress-induced cell death.Journal of Cell Science, 2010, 123:1468-1479.
[33] Teaklea N L, Bazihizina N, Shabala S,etal.Differential tolerance to combined salinity and O2deficiency in the halophytic grassesPuccinelliaciliataandThinopyrumponticum:The importance of K+retention in roots.Environmental and Experimental Botany, 2013, 87:69-78.
[34] Wang S M, Zhang J L, Flowers T J.Low-affinity Na+uptake in the halophyteSuaedamaritima.Plant Physiology, 2007, 145(2):559-571.
[35] Cuin T A, Bose J, Stefano G,etal.Assessing the role of root plasma membrane and tonoplast Na+/H+exchangers in salinity tolerance in wheat:in planta quantification methods.Plant, Cell and Environment, 2009, 34:947-961.
[36] Schmidt U G, Endler A, Schelbert S,etal.Novel tonoplast transporters identified using a proteomic approach with vacuoles isolated from cauliflower buds.Plant Physiology, 2007, 145:216-229.
[37] Wu G Q, Xi J J, Wang Q,etal.TheZxNHXgene encoding tonoplast Na+/H+antiporter from the xerophyteZygophyllumxanthoxylumplays important roles in response to salt and drought.Journal of Plant Physiology, 2011, 168:758-767.
[38] Ma Q, Yue L J, Zhang J L,etal.Sodium chloride improves photosynthesis and water status in the succulent xerophyteZygophyllumxanthoxylum.Tree Physiology, 2012, 32(1):4-13.
[39] Yue L J, Li S X, Ma Q,etal.NaCl stimulates growth and alleviates water stress in the xerophyteZygophyllumxanthoxylum.Journal of Arid Environments, 2012, 87:153-160.
[40] Wang S M, Zhu X Y, Shu X X.Studies on the characteristics of ion absorption anddistribution inPuccinelliatenuiflora.Acta Prataculturae Sinica, 1994, 3(1):39-43.
[41] Guo Q, Meng L, Mao P C,etal.Salt tolerance in two tall wheatgrass species is associated with selective capacity for K+over Na+.Acta Physiologiae Plantarum, 2014, 37:1708.
[42] Zhang J L, Flowers T J, Wang S M.Differentiation of low-affinity Na+uptake pathways and kinetics of the effects of K+on Na+uptake in the halophyteSuaedamaritime.Plant and Soil, 2013, 368(1-2):629-640.
[43] Zhu Z J, Wei G Q, Li J,etal.Silicon alleviates salt stress and increases antioxidant enzymes activity in leaves of salt-stressed cucumber (CucumissativusL.).Plant Science, 2004, 167:527-533.
[44] Liang Y C, Zhang W H, Chen Q,etal.Effects of silicon on H+-ATPase and H+-PPase activity, fatty acid composition and fluidity of tonoplast vesicles from roots of salt-stressed barley (HordeumvulgareL.).Environmental and Experimental Botany, 2005, 53:29-37.
[45] Gong H J, Randall D P, Flowers T J.Silicon deposition in the root reduces sodium uptake in rice (OryzasativaL.) seedlings by reducing bypass flow.Plant Cell and Environment, 2006, 29:1970-1979.
[46] Ma J F, Yamaji N, Mitani N,etal.An efflux transporter of silicon in rice.Nature, 2007, 448:209-212.
[47] Wang X S, Han J G.Effects of NaCl and silicon on ion distribution in the roots, shoots and leaves of two alfalfa cultivars with different salt tolerance.Soil Science and Plant Nutrition, 2007, 53:278-285.
[48] Tuna A L, Kaya C, Higgs D,etal.Silicon improves salinity tolerance in wheat plants.Environmental and Experimental Botany, 2008, 62:10-16.
[49] Chai Q, Shao X, Zhang J.Silicon effects onPoapratensisresponses to salinity.HortScience, 2010, 45:1876-1881.
[50] Bose J, Rodrigo-Moreno A, Shabala S.ROS homeostasis in halophytes in the context of salinity stress tolerance.Journal of Experimental Botany, 2014, 65(5):1241-1257.
[51] Guan B, Yv J B, Lu Z H,etal.Effects of water-salt stresses on seeding growth and activities of antioxidative enzyme ofSuaedasalsain coastal wetlands of the yellow river delta.Environmental Science, 2011, 32(8):2422-2429.
[52] Lu Y, Lei J Q, Zeng F J,etal.Effects of salt treatments on the growth and ecophysiological characteristies.Acta Prataculturae Sinica, 2014, 23(3):152-159.
[53] Xue X D, Dong X Y, Duan Y X,etal.A comparison of salt resistance of three kinds ofZoysiaat different salt concentrations.Acta Prataculturae Sinica, 2013, 22(6):315-320.
[54] Ahmed I M, Nadira U A, Bibi N,etal.Secondary metabolism and antioxidants are involved in the tolerance to drought and salinity, separately and combined, in Tibetan wild barley.Environmental and Experimental Botany, 2015, 111:1-12.
[55] Gao H J, Yang H Y, Bai J P,etal.Ultrastructural and physiological responses of potato (SolanumtuberosumL.) plantlets to gradient saline stress.Frontiers in Plant Science, 2014, 5:787.
[56] Talke I N, Blaudez D, Maathuis F J M,etal.CNGCs:prime targets of plant cyclic nucleotide signalling.Trends in Plant Science, 2003, 8:286-293.
[57] Amtmann A, Sanders D.Mechanism of Na+uptake by plant cells.Advances in Botanical Research, 1999, 29:75-112.
[58] Tyerman S D, Skerrett I M.Root ion channels and salinity.Scientia Horticulturae, 1999, 78:175-235.
[59] Zhang H F, Wang S M.Advances in study of Na+uptake and transport in higher plants and Na+homeostasis in the cell.Chinese Bulletin of Botany, 2007, 24(5):561-571.
[60] Amtmann A, Fischer M, Marsh E L,etal.The wheat cDNA LCT1 generates hypersensitivity to sodium in a salt-sensitive yeast strain.Plant Physiology, 2001, 126:1061-1071.
[61] Kronzucker H J, Szczerba M W, Schulze L M,etal.Non-reciprocal interactions between K+and Na+ions in barley (HordeumvulgareL.).Journal of Experimental Botany, 2008, 59:2793-2801.
[62] Deinlein U, Stephan A B, Horie T,etal.Plant salt-tolerance mechanisms.Trends in Plant Science, 2014, 19(6):371-379.
[63] Zamani B M, Ebrahimie E, Niazi A.In silico analysis of high affinity potassium transporter (HKT) isoforms in different plants.Aquatic Biosystems, 2014, 10:9.
[64] Wang Q, Guan C, Wang P,etal.AtHKT1;1 andAtHAK5 mediate low-affinity Na+uptake inArabidopsisthalianaunder mild salt stress.Plant Growth Regulation, 2015, 75(3):615-623.
[65] Ren Z H, Gao J P, Li L G,etal.A rice quantitative trait locus for salt tolerance encodes a sodium transporter.Nature Genetics, 2005, 37:1141-1146.
[66] Laurie S, Feeney K A, Maathuis F J M,etal.A role forHKT1 in sodium uptake by wheat roots.Plant Journal, 2002, 32:139-149.
[67] Kader M A, Seidel T, Golldack D,etal.Expressions ofOsHKT1,OsHKT2, andOsVHAare differentially regulated under NaCl stress in salt-sensitive and salt-tolerant rice (OryzasativaL.) cultivars.Journal of Experimental Botany, 2006, 57(15):4257-4268.
[68] Shao Q, Zhao C, Han N,etal.Cloning and expression pattern ofSsHKT1 encoding a putative cation transporter from halophyteSuaedasalsa.DNA sequence, 2008, 19(2):106-114.
[69] Senn M E, Rubio F, Banuelos M A,etal.Comparative functional features of plant potassiumHvHAK1 andHvHAK2 transporters.Journal of Biological Chemistry, 2001, 30:44563-44569.
[70] Fulgenzi F R, Peralta M L, Mangano S,etal.The ionic environment controls the contribution of the barleyHvHAK1 transporter to potassium acquisition.Plant Physiology, 2008, 147:252-262.
[71] Carden D E, Walker D J, Flowers T J,etal.Single-cell measurements of the contributions of cytosolic Na+and K+to salt tolerance.Plant Physiology, 2003, 131:676-683.
[72] Takahashi R, Nishio T, Ichizen N,etal.Cloning and functional analysis of the K+transporter PhaHAK2 from salt-sensitive and salt-tolerant reed plants.Biotechnol Letters, 2007, 29:501-506.
[73] Golldack D, Quigley F, Michalowski C B,etal.Salinity stress-tolerant and -sensitive rice (OryzasativaL.) regulate AKT1-type potassium channel transcripts differently.Plant Molecular Biology, 2003, 51:71-81.
[74] Kim E J, Kwak J M, Uozumi N,etal.AtKUP1:AnArabidopsisgene encoding high-affinity potassium transport activity.Plant Cell, 1998, 10:51-62.
[75] Fu H H, Luan S.AtKUP1:a dual-affinity K+transporter fromArabidopsis.Plant Cell, 1998, 10:63-73.
[76] Zhang J L.Low-Affinity Na+Uptake and Accumulation in the Halophyte Suaeda maritina[D].Lanzhou: Lanzhou University, 2008.
[77] Shi H, Kim Y, Guo Y,etal.TheArabidopsisSOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion.Plant Cell, 2003, 15(1):19-32.
[78] Shi H Z, Quintero F J, Pardo J M,etal.The putative plasma membrane Na+/H+antiporterSOS1 controls long-distance Na+transport in plants.Plant Cell, 2002, 14:465-477.
[79] Wu G Q, Wang P, Ma Q,etal.Selective transport capacity for K+over Na+is linked to the expression levels ofPtSOS1 in halophytePuccinelliatenuiflora.Functional Plant Biology, 2012, 39:1047-1057.
[80] Liu M, Wang T Z, Zhang W H.Sodium extrusion associated with enhanced expression ofSOS1 underlies different salt tolerance betweenMedicagofalcataandMedicagotruncatulaseedlings.Environmental and Experimental Botany, 2015, 110:46-55.
[81] Guo Q, Wang P, Ma Q,etal.Selective transport capacity for K+over Na+is linked to the expression levels ofPtSOS1 in halophytePuccinelliatenuiflora.Functional Plant Biology, 2012, 39:1047-1057.
[82] Ma Q, Li Y X, Yuan H J,etal.ZxSOS1 is essential for long-distance transport and spatial distribution of Na+and K+in the xerophyteZygophyllumxanthoxylum.Plant and Soil, 2014, 374:661-676.
[83] Feki K, Quintero F J, Khoudi H,etal.A constitutively active form of a durum wheat Na+/H+antiporterSOS1 confers high salt tolerance to transgenicArabidopsis.Plant Cell Reports, 2014, 33(2):277-288.
[84] Nie W X, Xu L, Yu B J.A putative soybeanGmsSOS1 confers enhanced salt tolerance to transgenicArabidopsissos1-1 mutant.Protoplasma, 2015, 252(1):127-134.
[85] Ishitani M, Liu J, Halfter U,etal.SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding.Plant Cell, 2000, 12(9):1667-1678.
[86] Martinez-Atienza J, Jiang X, Garciadeblas B,etal.Conservation of the salt overly sensitive pathway in rice.Plant Physiology, 2007, 143:1001-1012.
[87] Gaxiola R A, Rao R, Sherman A,etal.TheArabidopsisthalianaproton transporters, AtNHX1 and AVP1, can function in cation detoxification in yeast.Proceedings of the National Academy of Sciences of the United States of America, 1999, 96:1480-1485.
[88] Apse M P, Aharon G S, Snedden W A,etal.Salt tolerance confermi by over expression of a vacuolar NaCMC antiport inArabidopsis.Science, 1999, 285(12):1256-1258.
[89] Venema K, Quintero F J, Pardo J M,etal.TheArabidopsisNa+/H+exchanger catalyzes low affinity Na+and K+transport in reconstituted vesicles.Journal of Biological Chemistry, 2002, 277:2413-2418.
[90] Yamaguchi T, Fukuda-Tanaka S, Inagaki Y,etal.Genes encoding the vacuolar Na+/H+exchanger and flower coloration.Plant Cell Physiology, 2001, 142:451-461.
[91] Sottosanto J B, Gelli A, Blumwald E.DNA array analyses ofArabidopsisthalianalacking a vacuolar Na+/H+antiporter:impact of AtNHX1 on gene expression.Plant Journal, 2004, 40:752-771.
[92] Fukuda A, Nakamura A, Tanaka Y.Molecular cloning and expression of the Na+/H+exchanger gene inOryzasativa.BBA-Gene Structure and Expression, 1999, 1446:149-155.
[93] Ohta M, Hayashi Y, Nakashima A,etal.Introduction of a Na+/H+antiporter gene fromAtriplexgmeliniconfers salt tolerance to rice.FEBS Letters, 2002, 532:279-282.
[94] Fukuda A, Chiba K, Maeda M,etal.Effect of salt and osmotic stresses on the expression of genes for the vacuolar H+-pyrophosphatase, H+-ATPase subunitA, and Na+/H+antiporter from barley.Journal of Experimental Botany, 2004, 55:585-594.
[95] Wu C A, Yang G D, Meng Q W,etal.The cottonGhNHX1 gene encoding a novel putative tonoplast Na+/H+antiporter plays an important role in salt stress.Plant Cell Physiology, 2004, 45:600-607.
[96] Zorb C, Noll A, Karl S,etal.Molecular characterization of Na+/H+antiporters (ZmNHX) of maize (ZeamaysL.) and their expression under salt stress.Journal of Plant Physiology, 2005, 162:55-66.
[97] Brini F, Gaxiola R A, Berkowitz G A,etal.Cloning and characterization of a wheat vacuolar cation/proton antiporter and pyrophosphatase proton pump.Plant Physiology and Biochemistry, 2005, 43:347-354.
[98] Yu J N, Huang J, Wang Z M,etal.An Na+/H+antiporter gene from wheat plays an important role in stress tolerance.Journal of Biosciences, 2007, 32:1153-1161.
[99] Yang Q C, Wu M S, Wang P Q,etal.Cloning and expression analysis of a vacuolar Na+/H+antiporter gene from alfalfa.DNA sequece, 2005, 16:352-357.
[100] Li W Y, Wong F L, Tsai S N,etal.Tonoplast-locatedGmCLC1 andGmNHX1 from soybean enhance NaCl tolerance in transgenic bright yellow (BY)-2cells.Plant, Cell and Environment, 2006, 29:1122-1137.
[101] Qiao W H, Zhao X Y, Li W,etal.Overexpression ofAeNHX1, a root-specific vacuolar Na+/H+antiporter fromAgropyronelongatum, confers salt tolerance toArabidopsisandFestucaplants.Plant Cell Reports, 2007, 26:1663-1672.
[102] Verma D, Singla-Pareek S L, Rajagopal D,etal.Functional validation of a novel isoform of Na+/H+antiporter fromPennisetumglaucumfor enhancing salinity tolerance in rice.Journal of Biosciences, 2007, 32:621-628.
[103] Li J Y, He X W, Xu L,etal.Molecular and functional comparisons of the vacuolar Na+/H+exchangers originated from glycophytic and halophytic species.Journal of Zhejiang University Science, 2008, 9:132-140.
[104] Ye C Y, Zhang H C, Chen J H,etal.Molecular characterization of putative vacuolar NHX-type Na+/H+exchanger genes from the salt-resistant treePopuluseuphratica.Physiologia Plantarum, 2009, 137:166-174.
[105] Guan B, Hu Y, Zeng Y,etal.Molecular characterization and functional analysis of a vacuolar Na+/H+antiporter gene (HcNHX1) fromHalostachyscaspica.Molecular Biology Reports, 2010, 38:1889-1899.
[106] Jha A, Joshi M, Yadav N,etal.Cloning and characterization of theSalicorniabrachiataNa+/H+antiporter geneSbNHX1 and its expression by abiotic stress.Molecular Biology Reports, 2011, 38:1965-1973.
[107] Liu L, Zeng Y, Pan X,etal.Isolation, molecular characterization, and functional analysis of the vacuolar Na+/ H+antiporter genes from the halophyteKareliniacaspica.Molecular Biology Reports, 2012, 39:7193-7202.
[108] Yuan H J, Ma Q, Wu G Q,etal.ZxNHXcontrols Na+and K+homeostasis at the whole-plant level inZygophyllumxanthoxylumthrough feedbackregulation of the expression of genes involved in their transport.Annals of Botany, 2015, 115(3):495-507.
[109] Yin X Y, Yang A F, Zhang K W,etal.Production and analysis of transgenic maize with improved salt tolerance by the introduction ofAtNHX1 gene.Acta Botanica Sinica, 2004, 46:854-861.
[110] Xue Z Y, Zhi D Y, Xue G,etal.Enhanced salt tolerance of transgenic wheat (TritivumaestivumL.) expressing a vacuolar Na+/H+antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na+.Plant Science, 2004, 167:849-859.
[111] He C, Yan J, Shen G,etal.Expression of anArabidopsisvacuolar sodium/proton antiporter gene in cotton improves photosynthetic performance under salt conditions and increases fiber yield in the field.Plant Cell Physiology, 2005, 46:1848-1854.
[112] Banjara M, Zhu L, Shen G,etal.Expression of anArabidopsissodium/proton antiporter gene (AtNHX1) in peanut to improve salt tolerance.Plant Biotechnology Reports, 2012, 6:59-67.
[113] Rajagopal D, Agarwal P, Tyagi W,etal.PennisetumglaucumNa+/H+antiporter confers high level of salinity tolerance in transgenicBrassicaJuncea.Molecular Breeding, 2007, 19:137-151.
[114] Shi L Y, Li H Q, Pan X P,etal.Improvement ofToreniafournierisalinity tolerance by expression ofArabidopsisAtNHX5.Functional Plant Biology, 2008, 35:185-192.
[115] Zhang G H, Su Q, An L J,etal.Characterization and expression of a vacuolar Na+/H+antiporter gene from the monocot halophyteAeluropuslittoralis.Plant Physiology and Biochemistry, 2008, 46:117-126.
[116] Zhang Y M, Liu Z H, Wen Z Y,etal.The vacuolar Na+-H+antiport geneTaNHX2 confers salt tolerance on transgenic alfalfa (Medicagosativa).Functional Plant Biology, 2012, 39:708-716.
[117] Joshi M, Jha A, Mishra A,etal.Developing transgenic Jatropha using theSbNHX1 gene from an extreme halophyte for cultivation in saline wasteland.PLoS One, 2013, 8(8):e71136.
[118] Mishra S, Alavilli H, Lee B H,etal.Cloning and functional characterization of a vacuolar Na+/H+antiporter gene from mungbean (VrNHX1) and its ectopic expression enhanced salt tolerance inArabidopsisthaliana.PLoS One, 2014, 9(10):e106678.
[119] Sarafian V, Kim Y, Poole R J,etal.Molecular cloning and sequence of cDNA encoding the pyrophosphate-energized vacuolar membrance proton pump ofArabidopsisthaliana.Proceedings of the National Academy of Sciences of the United States of America, 1992, 89:1775-1779.
[120] Gaxiola R A, Palmgren M G, Schumacher K.Plant proton pumps.FEBS Letters, 2007, 581:2204-2214.
[121] Gao F, Gao Q, Duan X G,etal.Cloning of an H+-PPase gene fromThellungiellahalophilaand its heterologous expression to improve tobacco salt tolerance.Journal of Experimental Botany, 2006, 57:3259-3270.
[122] Guo S L, Yin H B, Zhang X,etal.Molecular cloning and characterization of a vacuolar H+-pyrophosphatase gene, SsVP, from the halophyteSuaedasalsaand its overexpression increases salt and drought tolerance ofArabidopsis.Plant Molecular Biology, 2006, 60:41-50.
[123] Li J, Yang H, Peer W A,etal.ArabidopsisH+-PPaseAVP1 regulates auxin-mediated organ development.Science, 2005, 310:121-125.
[124] Park S, Li J, Pittman J K,etal.Up-regulation of a H+- pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants.Proceedings of the National Academy of Sciences of the United States of America, 2005, 102:18830-18835.
[125] Bao A K, Wang S M, Wu G Q,etal.Overexpression of theArabidopsisH+-PPase enhanced resistance to salt and drought stress in transgenic alfalfa (MedicagosativaL.).Plant Science, 2009, 176:232-240.
[126] Li Z G, Baldwin M, Hu Q,etal.Heterologous expression ofArabidopsisH+-pyrophosphatase enhances salt tolerance in transgenic creeping bentgrass (AgrostisstoloniferaL.).Plant Cell Environment, 2010, 33:272-289.
[127] Schilling R K, Marschner P, Shavrukov Y,etal.Expression of theArabidopsisvacuolar H+-pyrophosphatase gene (AVP1) improves the shoot biomass of transgenic barley and increases grain yield in a saline field.Plant Biotechnology Journal, 2014, 12(3):378-386.
[128] Kumar T, Uzma, Khan M R,etal.Genetic improvement of sugarcane for drought and salinity stress tolerance usingArabidopsisvacuolarpyrophosphatase (AVP1) gene.Molecular Biotechnology, 2014, 56(3):199-209.
[129] Lv S L, Lian L J, Tao P L,etal.Overexpression ofThellungiellahalophilaH+-PPase (TsVP) in cotton enhances drought stress resistance of plants.Planta, 2009, 229:899-910.
[130] Pei L, Wang J, Li K,etal.Overexpression ofThellungiellahalophilaH+-pyrophosphatase gene improves low phosphate tolerance in maize.PLOS One, 2012, 7(8):e43501.
[131] Yao M, Zeng Y, Liu L,etal.Overexpression of the halophyteKalidiumfoliatumH+-pyrophosphatase gene confers salt and drought tolerance inArabidopsisthaliana.Molecular Biology Reports, 2012, 39:7989-7996.
[132] Khoudi H, Maatar Y, Gouiaa S,etal.Transgenic tobacco plants expressing ectopically wheat H+-pyrophosphatase (H+-PPase) geneTaVP1 show enhanced accumulation and tolerance to cadmium.Journal of Plant Physiology, 2012, 169:98-103.
[133] Li X, Guo C, Gu J,etal.Overexpression of VP, a vacuolar H+-pyrophosphatase gene in wheat (TriticumaestivumL.), improves tobacco plant growth under Pi and N deprivation, high salinity, and drought.The Journal of Experimental Botany, 2014, 65(2):683-696.
[134] Zhao F Y, Zhang X J, Li P H,etal.Co-expression of theSuaedasalsaSsNHX1 andArabidopsisAVP1 confer greater salt tolerance to transgenic rice than the singleSsNHX1.Molecular Breeding, 2006, 17:341-353.
[135] Liu S P, Zheng L Q, Xue Y H,etal.Overexpression ofOsVP1 andOsNHX1 increases tolerance to drought and salinity in rice.Journal of Integrative Plant Biology, 2010, 53:444-452.
[136] Brini F, Hanin M, Mezghani I,etal.Overexpression of wheat Na+/H+antiporterTNHX1 and H+- pyrophosphataseTVP1 improve salt- and drought-stress tolerance inArabidopsisthalianaplants.Journal of Experimental Botany, 2007, 58:301-308.
[137] Bhaskaran S, Savithramma D L.Co-expression ofPennisetumglaucumvacuolar Na+/H+antiporter andArabidopsisH+- pyrophosphatase enhances salt tolerance in transgenic tomato.Journal of Experimental Botany, 2011, 62:5561-5570.
[138] Gouiaa S, Khoudi H, Leidi E O,etal.Expression of wheat Na+/H+antiporterTNHXS1 and H+- pyrophosphataseTVP1 genes in tobacco from a bicistronictranscriptional unit improves salt tolerance.Plant Molecular Biology, 2012, 79:137-155.
[139] Bao A K, Wang Y W, Xi J J,etal.Co-expression of xerophyteZygophyllumxanthoxylumZxNHXandZxVP1-1 enhances salt and drought tolerance in transgenicLotuscorniculatusby increasing cations accumulation.Functional Plant Biology, 2014, 41:203-214.
[140] Hu L, Lu H, Liu Q L,etal.Overexpression ofmtlDgene in transgenicPopulustomentosaimproves salt tolerance through accumulation of mannitol.Tree Physiology, 2005, 25:1273-1281.
[141] Liu Y, Wang G Y, Liu J J,etal.Transfer ofE.coligutDgene into maize and regeneration of salt-tolerant transgenic plants.Science in China Series C-Life Science, 1999, 42(1):90-95.
[142] Wang H Z, Huang D N, Lu R F,etal.Salt tolerance of transgenic rice (OryzasativaL.) withmtlDgene andgutDgene.Chinese Science Bulletin, 2000, 45:1685-1690.
[143] Kishor P B K, Hong Z, Mian G H.Over expression of △’-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants.Plant Physiology, 1995, 108:1387-1394.
[144] Dure L, Greenway S C, Galau G A.Developmental biochemistry of cottonseed embryogenesis and germination-changing messenger ribonucleic-acid populations as shown byinvitroandinvivoprotein-synthesis.Biochemistry, 1981, 20(14):4162-4168.
[145] Xu D, Duan X, Wang B,etal.Expression of a late embryogenesis abundant protein geneHVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice.Plant Physiology, 1996, 110:249-257.
[146] Zhang N, Wang D, Si H J.Isolation and induced expression of betaine aldehyde dehydrogenase genefrom spinach.Journal of Agricultural Biotechnology, 2004, 12(5):612-613.
[147] Jia G X, Zhu Z Q, Chang F Q,etal.Transformation of tomato with theBADHgene fromAtripleximproves salt tolerance.Plant Cell Reports, 2002, 21(2):141-146.
[148] Tang N, Zhang H, Li X H,etal.Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice.Plant Physiology, 2012, 158:1755-1768.
[149] Shinozaki K, Yamaguchi-Shinozaki K.Gene expression and signal transduction in water stress response.Plant Physiology, 1997, 115:327-334.
[150] Wu H J, Zhang Z H, Wang J Y,etal.Insights into salt tolerance from the genome ofThellungiellasalsuginea.Proceedings of the National Academy of Sciences, 2012, 109(30):12219-12224.
[151] Taji T, Seki M, Satou M,etal.Comparative genomics in salt tolerance betweenArabidopsisand a Rabidopsis-related halophyte salt cress usingArabidopsismicroarray.Plant Physiology, 2004, 135:1697-1709.
[152] Sheen J.Signal transduction in maize andArabidopsismesophyll protoplasts.Plant Physiology, 2001, 127:1466-1475.
[153] Moon H, Lee B, Choi G,etal.NDP kinase 2 interacts with two oxidative stress-activated MAPKs to regulate cellular reduxstate and enhances multiple stress tolerance in transgenic plants.Proceedings of the National Academy of Sciences, 2003, 100(1):358-363.
[154] Xie T, Ren R, Zhang Y Y,etal.Molecular mechanism for inhibition of a critical component in theArabidopsisthalianaabscisic acid signal transduction pathways, SnRK2.6, by protein phosphataseABI1.Journal of Biological Chemistry, 2012, 287:794-802.
[155] Li R F, Zhang J W, Wu G Y,etal.HbCIPK2, a novel CBL-interacting protein kinase from halophyteHordeumbrevisubulatum, confers salt and osmotic stress tolerance.Plant, Cell and Environment, 2012, 35:1582-1600.
[156] Zhang Q, Lin F, Mao T,etal.Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress inArabidopsis.Plant Cell, 2012, 24:4555-4576.
[157] Roxas V P, Lodhi S A, Garrett D K,etal.Stress tolerance in transgenic tobacco seedlings that overexpress glutathione S-transferase/glutathione peroxidase.Plant & Cell Physiology, 2000, 41(11):1229-1234.
[158] Kovtun Y, Chiu W L, Tena G,etal.Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants.Proceedings of the National Academy of Sciences of the United States of America, 2000, 97:2940-2945.
[159] Zhang Z, Wang J, Zhang R X,etal.The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis inArabidopsis.The Plant Journal, 2012, 71:273-287.
[160] Ge Y, Gao P, Xia J Z,etal.The effects of calcium chloride on improving te salt resistance ofZeamaysL.Journal of Northeast Agicultural University, 2004, 35(3):281-284.
[161] Zhang L X, Chang Q S, Hou X G,etal.Effects of sodium salt stress on seed germination ofPrunellavulgaris.Acta Prataculturae Sinica, 2015, 24(3):177-186.
[162] Qian Q, Qu L J, Yuan M,etal.Research advances on plant science in China in 2012.Chinese Bulletin of Botany, 2013, 48:231-287.
[163] Galvan-Ampudia C S, Testerink C.Salt stress signals shape the plant root.Current Opinion in Plant Biology, 2011, 14:296-302.
[164] Brady S M, Sarkar S F, Bonetta D,etal.The abscisic acid insensitive 3(ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development inArabidopsis.Plant Journal, 2003, 34:67-75.
[165] An J P, Chen K S.The relations between the injury of plasma membrane and the increase of aba content in wheat leaves.Journal of Lanzhou University (Natural Science), 1994, 3:127-131.
[166] Iraudat J, Parcy F, Gosti F.Current advances in abseisic acid action and signaling.Plant Molecular Biology, 1994, 26:1557-1577.
[167] Yuan F, Yang J C, Chen M,etal.Effect of no donor sodium nitroprusside (SNP) on seed germination ofSuaedasalsaL.under NaCl Stress.Plant Physiology Journa, 2010, 46(1):24-28.
[168] Liu W Y, Yang H W, Wei X H,etal.Effects of exogenous nitric oxide on seed germination, physiological characteristics and active oxygen metabolism ofMedicagotruncatula.Acta Prataculturae Sinica, 2015, 24(2):85-95.
[169] Chao D Y, Dilkes B, Luo H,etal.Polyploids exhibit higher potassium uptake and salinity tolerance inArabidopsis.Science, 2013, 341:658-659.
[170] Horie T, Costa A, Kim T H,etal.RiceOsHKT2;1 transporter mediates large Na+influx component into K+-starved roots for growth.EMBO Journal, 2007, 26(12):3013-3014.
[171] James R A, Blake C, Byrt C S,etal.Major genes for Na+exclusion,Nax1 andNax2 (wheatHKT1;4 andHKT1;5), decrease Na+accumulation in bread wheat leaves under saline and waterlogged conditions.Journal of Experimental Botany, 2011, 62:2939-2947.
[172] Byrt C S, Platten J D, Spielmeyer W,etal.HKT1; 5-like cation transporter linked to Na+exclusion loci in wheat,Nax2 andKna1.Plant Physiology, 2007, 143:1918-1928.
参考文献:
[5] 赵可夫, 李法曾, 樊守金, 等.中国的盐生植物.植物学通报, 1999, 16(3):201-207.
[6] 赵可夫.植物对盐渍逆境的适应.生物学通报, 2002, 37(6):7-10.
[13] 李昀, 沈禹颖, 阎顺国.NaCl胁迫下5种牧草种子萌发的比较研究.草业科学, 1997, 14(2):50-53.
[19] 刘晶, 才华, 刘莹, 等.两种紫花苜蓿苗期耐盐生理特性的初步研究及其耐盐性比较.草业学报, 2013, 22(2):250-256.
[40] 王锁民, 朱兴运, 舒孝喜.碱茅离子吸收与分配特性研究.草业学报, 1994, 3(1):39-43.
[51] 管博, 于君宝, 陆兆华, 等.黄河三角洲滨海湿地水盐胁迫对盐地碱蓬幼苗生长和抗氧化酶活性的影响.环境科学, 2011, 32(8):2422-2429.
[52] 鲁艳, 雷加强, 曾凡江, 等.NaCl处理对梭梭生长及生理生态特征的影响.草业学报, 2014, 23(3):152-159.
[53] 薛秀栋, 董晓颖, 段艳欣, 等.不同盐浓度下3种结缕草的耐盐性比较研究.草业学报, 2013, 22(6):315-320.
[59] 张宏飞, 王锁民.高等植物Na+吸收、转运及细胞内Na+稳态平衡研究进展.植物学通报, 2007, 24(5):561-571.
[76] 张金林.盐生植物海滨碱蓬Na+吸收和积累的研究[D].兰州: 兰州大学, 2008.
[146] 张宁, 王蒂, 司怀军.菠菜甜菜碱醛脱氢酶基因的分离和诱导表达.农业生物技术学报, 2004, 12(5):612-613.
[160] 葛瑛, 高鹏, 夏激宗, 等.氯化钙在提高玉米抗盐性方面的作用.东北农业大学学报, 2004, 35(3):281-284.
[161] 张利霞, 常青山, 侯小改, 等.不同钠盐胁迫对夏枯草种子萌发特性的影响.草业学报, 2015, 24(3):177-186.
[162] 钱前, 瞿礼嘉, 袁明, 等.2012年中国植物科学若干领域重要研究进展.植物学报, 2013, 48:231-287.
[165] 安建平, 陈靠山.渗透胁迫下小麦的膜损伤与ABA增高的关系.兰州大学学报, 1994, (3):127-131.
[167] 袁芳, 杨剑超, 陈敏, 等.NaCl 胁迫下外源NO供体硝普钠(SNP)对盐地碱蓬种子萌发的影响.植物生理学通讯, 2010, 46(1):24-28.
[168] 刘文瑜, 杨宏伟, 魏小红, 等.外源NO调控盐胁迫下蒺藜苜蓿种子萌发生理特性及抗氧化酶的研究.草业学报, 2015, 24(2):85-95.
Research advances in higher plant adaptation to salt stress
ZHANG Jin-Lin1*, LI Hui-Ru1, GUO Shu-Yuan1, WANG Suo-Min1, SHI Hua-Zhong2, HAN Qing-Qing1, BAO Ai-Ke1, MA Qing1
1.StateKeyLaboratoryofGrasslandAgro-ecosystems,CollegeofPastoralAgricultureScienceandTechnology,LanzhouUniversity,Lanzhou730020,China; 2.DepartmentofChemistryandBiochemistry,TexasTechUniversity,LubbockTX79409,USA
Soil salinity is a serious worldwide problem causing reduction in crop growth and agricultural output potential.Consequently, finding new ways to minimize the adverse effects of soil salinization on agriculture is globally important.Understanding the adaptation mechanisms of higher plants to salt stress is critical for enhancing salt tolerance and yields of crop plants as well as protecting ecological environments.In this paper, we reviewed the key progresses in salt stress adaptation of higher plants, including the effects of salt stress in plants; physiological mechanism of plant salt tolerance (osmotic adjustment, nutrient balance and the antioxidant system); the diversity of genes relevant to salt tolerance (ion transporting protein genes, osmotic regulation-related genes, signal transduction-related genes and cellular antioxidant-related genes and so on); and the approaches for crop improvement in salt tolerance.Prospects for developing crop plants tolerant to salinity are also discussed.
higher plants; salt stress; salt tolerance; salt tolerant genes
10.11686/cyxb2015233
http://cyxb.lzu.edu.cn
2015-05-07;改回日期:2015-07-14
国家自然科学基金项目(31222053,31170431和31172256),教育部“长江学者和创新团队发展计划”(IRT13019)和中央高校基本科研业务费项目(lzujbky-2014-m01和lzujbky-2015-194)资助。
张金林(1975-),男,甘肃泾川人,教授,博士生导师,博士。
*通信作者Corresponding author.E-mail:jlzhang@lzu.edu.cn
张金林, 李惠茹, 郭姝媛, 王锁民, 施华中, 韩庆庆, 包爱科, 马清.高等植物适应盐逆境研究进展.草业学报, 2015, 24(12):220-236.
ZHANG Jin-Lin, LI Hui-Ru, GUO Shu-Yuan, WANG Suo-Min, SHI Hua-Zhong, HAN Qing-Qing, BAO Ai-Ke, MA Qing.Research advances in higher plant adaptation to salt stress.Acta Prataculturae Sinica, 2015, 24(12):220-236.

