猕猴桃溃疡病菌漆酶基因的克隆与功能分析
刘 普, 胡家勇, 何 蓉, 朱立武
(安徽农业大学 园艺学院果树学重点实验室, 合肥 230036)
猕猴桃溃疡病菌漆酶基因的克隆与功能分析
刘 普, 胡家勇, 何 蓉, 朱立武
(安徽农业大学 园艺学院果树学重点实验室, 合肥 230036)
为探明猕猴桃溃疡病病菌漆酶基因的序列特征与功能,研究从12株溃疡病病菌丁香假单孢杆菌猕猴桃致病变种(Pseudomonassyringaepv.actinidae)中扩增获得漆酶基因,分别命名为PsalacJF、PsalacHY、PsalacGC、PsalacHWD、PsalacINS、Psalac7285、PsalacLT12、PsalacLT16、PsalacLT26、Psalac349、PsalacKw30和PsalacK3。序列分析结果显示上述序列之间只有少许核酸位点有差异,基因编码全长都为1374 bp,BLAST预测基因编码有457个氨基酸序列(包括N-端20个氨基酸的信号肽),启动子位于基因上游175 bp。生物信息学分析发现该基因具有漆酶特有的4个Cu2+活性位点,且活性位点高度保守。系统进化分析结果显示该基因编码序列与多种细菌的漆酶基因同源关系很近,其中与P.fluorescensPf0-1的同源性最高。研究结果表明溃疡病病菌中普遍含有具备细菌漆酶基因家族的序列特性,推测该基因可能参与调控溃疡病病菌致病性和Cu2+耐受性。
猕猴桃;溃疡病;漆酶
猕猴桃溃疡病是一种毁灭性病害,由丁香假单孢杆菌猕猴桃致病变种(Pseudomonassyringaepv.actinidae, Psa)引起,主要危害猕猴桃的主干、枝干和叶片。在枝干上表现为病部溢出大量初为乳白色、后变红褐色的粘液,随后上部枝条萎蔫死亡。在叶片上则表现为产生中间为褐色、边缘为黄色晕圈的近圆形病斑[1]。
近年研究发现,世界猕猴桃Psa存在4种不同的致病基因型(Genotype),即Psa-J(含有编码菜豆毒素基因簇),Psa-K(含有编码冠菌素基因簇),Psa-V(缺乏编码菜豆毒素和冠菌素基因簇)和Psa-LV(缺少部分effector基因簇)[2-3],不同基因型菌株存在着明显的致病差异。其中,Psa-LV致病力较弱,仅在叶片上形成病斑;而Psa-V致病力最强,危害也最为严重[2,4]。
漆酶(Laccase, EC 1.10.3.2)是一种含铜的多酚氧化酶,能有效地降解木质素物质,与植物病原菌的致病性和色素形成密切相关。漆酶按照来源可以分为植物漆酶、真菌漆酶和细菌漆酶。铜制剂是猕猴桃溃疡病生产中比较常用的一类药剂,如硫酸铜。1988年,丁香假单胞菌番茄致病变种中分离获得一种漆酶的类似物CopA,它表现出Cu2+抗性[5]。CopA和CopB是丁香假单孢杆菌猕猴桃致病种常见的两个耐铜基因。由于铜制剂的长期使用,最近在有些病菌中发现携带有新型的耐铜基因CopR和CopS的丁香假单孢杆菌猕猴桃致病[6-8]。
酚类物质(phenolic compounds)是植物体内一类具有防御功能的次生代谢物质,主要由类黄酮、酚酸和单宁等组成[9],它们广泛参与植物生长、发育和防御等生理过程,尤其在抵御病菌侵染方面具有关键作用[10-11]。植物在同病菌相互作用的过程中,会产生大量的酚类物质,调节植物抗病能力,保护自身免受伤害[12-15]。猕猴桃植物组织中,含有丰富的没食子酸、酒石酸、绿原酸、槲皮素和p-对香豆酸等酚类物质[16-18];在Psa侵染过程中,猕猴桃叶片中与酚类物质代谢相关的蛋白种类、含量变化明显[19]。由此可见,漆酶可能在病菌铜离子耐受(抗铜)、猕猴桃酚类物质脱毒等方面起到关键作用。为此,本研究拟根据丁香假单孢杆菌猕猴桃致病种基因组序列设计引物克隆漆酶基因的全长,通过生物信息学的方法分析不同基因间的关系,预测基因功能,为进一步分析溃疡病菌漆酶基因的功能提供理论基础。
1 材料与方法
1.1 试验材料
猕猴桃溃疡病病菌丁香假单孢杆菌猕猴桃致病变种(Pseudomonassyringaepv.actinidae)由课题组从中国和意大利、日本、韩国等国家共分离或收集(表1),其中Kw30为Psa-J,K3为Psa-K,其他均为Psa-V。
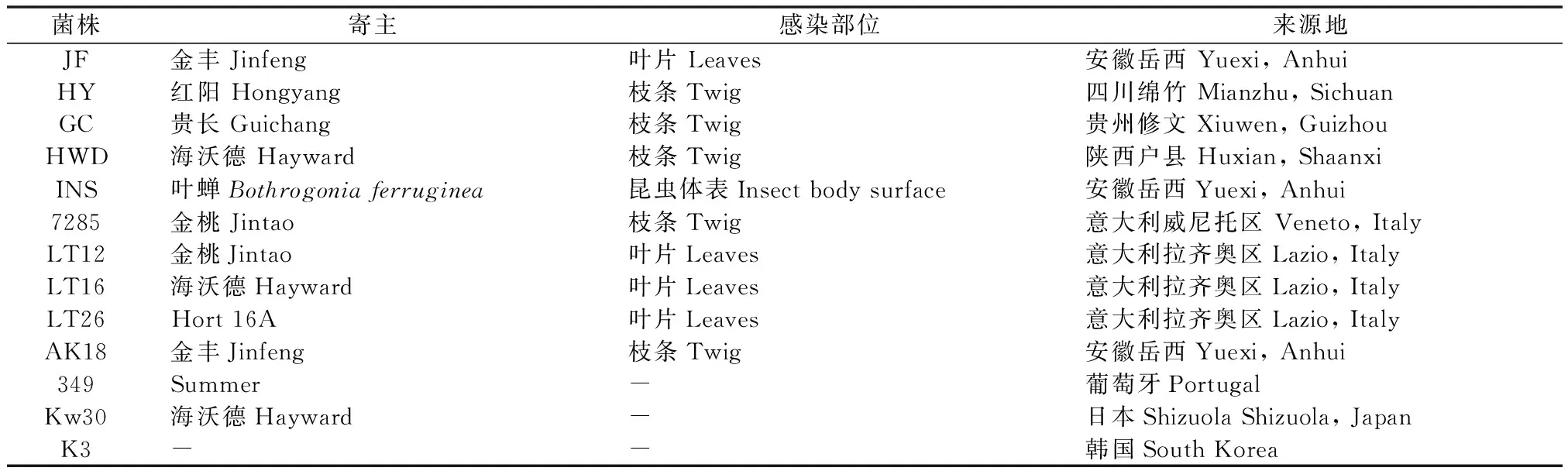
表1试验采用的溃疡病菌株
1.2 实验方法
1.2.1 猕猴桃溃疡病病菌漆酶基因全长克隆
根据丁香假单孢杆菌猕猴桃致病变种基因组序列设计,Cu3F CTTTTACCTGCCTCTTTTTCCG,Cu3R TGCCTCCCATTTTCGTTTCCTG。以猕猴桃溃疡病病菌基因组DNA为模板,扩增体系反应条件为:95℃预变性5 min;94℃ 1 min,61℃ 2 min,72℃ 2 min,32个循环;72℃ 10 min;4℃ 保温。取6 μL PCR扩增产物,经1.2%琼脂糖凝胶,1.5~2 V/cm 电泳检测,照相。回收目的条带并克隆测序。利用BLAST软件将测序结果与GenBank中的已知基因进行同源性分析。
1.2.2 猕猴桃溃疡病菌漆酶基因结构与功能预测
利用BLAST、DNAMAN、Swissplot、MEGA3.1和Clustal X2.0等软件对漆酶基因进行结构和功能分析。
1.2.3 大肠杆菌异源表达分析
利用引物MeiQCuF GGGATTACATATGTCACGACACCTCGATGGCGGCCATG和MeiQCuF GTTCTCGAGATGTCCTTCACCCGTCGTCAGATCC引入酶切位点Nde I和 Xho I并构建表达载体pET22b-Psalac转化大肠杆菌BL21(DE3),IPTG诱导表达。细胞破碎(30%功率,开4 s、关6 s)后上清液SDS-PAGE蛋白电泳分析。同时,提取物利用丁香醛连氮为底物检测漆酶的活性。
2 结果与分析
2.1 漆酶基因的克隆
从不同危害时期和国家(地区)的溃疡病病菌中克隆得到了不同类型病菌的Psalac全长,分别命名为Psalac-JF/HY/HWD等。PsalacJF编码全长1374 bp,BLAST预测该基因编码有457个氨基酸序列(包括N-端20个氨基酸的信号肽)。生物信息学分析发现启动子位于基因上游175 bp。同时该基因编码的氨基酸序列与NCBI进行BLASTP同源比对,分析结果显示该漆酶基因与基因组漆酶基因序列相同。
Psalac序列比较表明,不同类型病菌之间在部分核酸区域有差异。聚类分析发现在假单胞杆菌属其他种基因组中同样存在Psalac类似序列,表明Psalac在假单胞杆菌属中普遍存在(图1)。我们对Psalac与其他细菌漆酶进行多序列比对,结果显示Psalac 与其他细菌漆酶一样在铜离子结合位点区域表现保守(图2),因此可以认定Psalac是溃疡病病菌所编码的漆酶基因。

图1 分离自不同时期和国家(地区)的溃疡病菌psalac
AE016853.1 (P.syringaepv.tomatoDC3000), CP000058.1 (P.syringaepv.phaseolicola1448A),CP000075.1 (P.syringaepv.syringaeB728a),CP000094.2 (P.fluorescensPf0-1),CP002585.1 (P.brassicacearumsubsp.brassicacearumNFM421),CP004045.1 (P.poaeRE 1-1-14),CP003880.1 (Pseudomonassp. UW4),CP003150.1 (P.fluorescensF113),AF326404.1 (P.chlororaphis)、AF326399.1 (Pseudomonassp. GB13),AF326405.1 (P.fluorescen),AF086638.1 (P.putida),AF326403.1 (Pseudomonassp. MG1),AF326401. (Pseudomonas sp. ISO1),AF326398.1 (Pseudomonassp. ADP) and AF326407.1 (P.putidaMnB1)
2.2 漆酶基因的异源表达
通过构建pET22b-PsalacJF载体和大肠杆菌异源表达,粗酶SDS-PAGE分析(图3),结果显示PsalacJF蛋白质分子质量约为50 ku,与生物信息学预测的蛋白质分子质量大小一致。利用丁香醛连氮为底物,纯化的酶蛋白在50 mmol/L Na2HPO4-KH2PO4缓冲液(pH值7.5)环境中可以有效地使底物氧化成红色(图3),显示纯化酶蛋白具有漆酶活性。

图2 Clustal X2.0 分析Psalac及其他细菌漆酶蛋白质序列
CAA17652(Mycobacteriumtuberculosis), NP854527 (M.bovisAF2122/97), BAB05801(BacillushaloduransC-125), AAP57087(B.halodurans), AAD24211(P.putidaGB-1), ADM87301(uncultured bacterium) and AAC16140(RhodobactercapsulatusSB 1003). Four histidine-rich copper binding domains were indicated by full-length vertical boxes

图3 Psalac大肠杆菌异源表达SDS-PAGE和酶活显色
A—SDS-PAGE; B—酶活显色; The final concentration of IPTG screening with 0、0.1、0.2、0.5和0.75 mmol/L; temperature screening by using 16℃ and 37℃
3 讨 论
漆酶是一种含铜的多酚氧化酶,能够催化上百种酚类物质氧化。近年来研究发现漆酶在细菌中广泛存在[20],Ausec等[21]通过对2200种细菌的1200种漆酶进行分析后发现76% 漆酶具有可将蛋白分泌至胞外的信号肽。不同物种之间漆酶的核酸和氨基酸序列相似度低,只有在铜离子结合区域比较保守,漆酶可能在不同细菌中参与了不同的细胞功能。目前,漆酶已报道在病原菌抵御外界不良环境,如孢子形成、毒性金属离子耐受(如铜离子)及致病性等生物学过程中发挥关键作用[22]。
针对漆酶,另一个尤为关注的是漆酶作为多铜蛋白,具有多个铜离子结合位点,可以增强病菌铜离子耐受性。猕猴桃溃疡病是树体枝干型病害,常规的化学药剂防治效果不明显,目前防治溃疡病主要依靠铜制剂(如氢氧化铜、硫酸铜和氧氯化铜等)和链霉素。因此,病菌的铜抗性机制研究已成为一个热点。人们对分离自不同危害时期和国家的溃疡病病菌进行分析后发现病菌原先仅有抗铜基因copA(已被证实具备漆酶活性)和copB, 现在部分病菌已出现了copR和copS等其他抗铜基因[23]。
试验利用猕猴桃溃疡病病菌的基因组序列设计引物扩增获得的漆酶基因序列,预测其氨基酸序列。对PsalacJF与其他细菌漆酶进行多序列比对,结果显示PsalacJF 与其他细菌漆酶一样在铜离子结合位点区域表现保守。大肠杆菌异源表达结果显示克隆获得的漆酶基因具有漆酶活性,可以认定该扩增产物为漆酶基因编码序列。下一步验证漆酶基因与病菌的铜制剂耐药性和致病性的关系。
[1]赵利娜,胡家勇,叶振风,等. 猕猴桃溃疡病病原茵的分子鉴定和致病力测定[J].华中农业大学学报,2012, 31(5): 604-608.
[2]Scortichini M, Marcelletti S, Ferrante P, et al.Pseudomonassyringaepv. actinidiae: a re-emerging, multi-faceted, pandemic pathogen[J]. Molecular Plant Pathology, 2012, 13: 631-640.
[3]Serizawa S, Ichikawa T, Takikawa Y, et al. Occurence of bacterial canker of kiwifruit in Japan: description of symptoms, isolation of the pathogen and screening of bactericides[J]. Annals of the Phytopathological Society of Japan, 1989, 55: 427-436.
[4]Chapman J R, Taylor R K, Weir B S, et al. Phylogenetic relationships among global populations ofPseudomonassyringaepv. Actinidiae[J]. Phytopathology, 2012, 102:1034-1044.
[5]Mellano M A, Cooksey D A. Nucleotide sequence and organization of copper resistance genes fromPseudomonassyringaepv. tomato[J]. Journal of Bacteriology, 1988, 170(6):2879-2883.
[6]Mellano M A, Cooksey D A. Nucleotide sequence and organization of copper resistance genes fromPseudomonassyringaepv. tomato[J]. Journal of Bacteriology, 1988, 170(6):2879-2883.
[7]Held C, Kandelbauer A, Schroeder M, et al. Biotransformation of phenolics with laccase containing bacterial spores[J].Environmental Chemistry Letters, 2005, 3(2):74-77.
[8]Cha J S, Cooksey D A. Copper resistance inPseudomonassyringaemediated by periplasmic and outer membrane proteins[J]. Proceedings of the National Academy of Sciences of the United States of America, 1991, 88(20):8915-8919.
[9]Cheynier V, Comte G, Davies K M, et al. Plant phenolics: recent advances on their biosynthesis, genetics, and ecophysiology[J]. Plant Physiology and Biochemistry, 2013, 72: 1-20.
[10]Terrier N, Poncet-Legrand C, Cheynier V. Flavanols, flavonols and dihydroflavonols[J]. Wine Chemistry and Biochemistry. Springer New York, 2009, 463-507.
[11]Haminiuk C W I, Maciel G M, Plata-Oviedo M S V, et al. Phenolic compounds in fruits-an overview[J]. International Journal of Food Science & Technology, 2012, 47: 2023-2044.
[12]Ruelas C, Tiznado-Hernandez M E, Sanchez-Estrada A, et al. Changes in phenolic acid content duringAlternariaalternatainfection in tomato fruit[J]. Journal of Phytopathology, 2006, 154: 236-244.
[13]Kim H G, Kim G S, Lee J H, et al. Determination of the change of flavonoid components as the defence materials ofCitrusunshiuMarc. fruit peel againstPenicilliumdigitatumby liquid chromatography coupled with tandem mass spectrometry[J]. Food Chemistry, 2011, 128: 49-54.
[14]Ballester A R, Lafuente M T, Forment J, et al. Transcriptomic profiling of citrus fruit peel tissues reveals fundamental effects of phenylpropanoids and ethylene on induced resistance[J]. Molecular Plant Pathology, 2011, 12: 879-897.
[15]Daayf F, El-Hadrami A, El-Bebany A F, et al. Phenolic compounds in plant defense and pathogen counter-defense mechanisms[J]. Recent Advances in Polyphenol Research, 2012(3): 191.
[16]Park Y S, Leontowicz H, Leontowicz M, et al. Comparison of the contents of bioactive compounds and the level of antioxidant activity in different kiwifruit cultivars[J]. Journal of Food Composition and Analysis, 2011, 24: 963-970.
[17]Webby R F, Wilson R D, Ferguson A R. Leaf flavonoids of actinidia[J]. Biochemical Systematics and Ecology, 1994, 22: 277-286.
[18]Lee D E, Shin B J, Hur H J, et al. Quercetin, the active phenolic component in kiwifruit, prevents hydrogen peroxide-induced inhibition of gap-junction intercellular communication[J]. British Journal of Nutrition, 2010, 104: 164-170.
[19]Petriccione M, Di Cecco I, Arena S, et al. Proteomic changes inActinidiachinensisshoot during systemic infection with a pandemicPseudomonassyringaepv. actinidiae strain[J]. Journal of Proteomics 2013, 78: 461-476.
[20]Fang Z M, Li T L, Wang Q, et al. A bacterial laccase from marine microbial metagenome exhibiting chloride tolerance and dye decolorization ability[J]. Applied Microbiology and Biotechnology,2011, 89: 1103-1110.
[21]Ausec L, Zakrzewski M, Goesmann A, et al. Bioinformatic analysis reveals high diversity of bacterial genes for laccase-like enzymes[J]. PLoS ONE, 2011, 6, e25724.
[22]Galai S, Lucas-Elio P, Marzouki M N, et al. Molecular cloning of a copper-dependent laccase from the dye-decolorizing strainStenotrophomonasmaltophiliaAAP56[J]. Journal of Applied Microbiology, 2011, 111:1394-1405.
[23]Nakajima M, Goto M, Hibi, T. Similarity between copper resistance genes fromPseudomonassyringaepv. actinidiae andP.syringaepv. Tomato[J]. Journal of General Plant Pathology, 2002, 68:68-74.
Cloning and analysis of the expression of laccase genes inPseudomonassyringaepv.actinidae
LIU Pu, HU Jia-yong, HE Rong, ZHU Li-wu
(College of Horticulture, the Key Laboratory of Pomology, Anhui Agricultural University, Hefei 230036, China)
The objective of this study was to reveal sequence features of laccase, which would be used in further study of pathogenesis of kiwifruit bacterial canker caused byPseudomonassyringaepv.actinidae. 12 laccase genes homologsPsalacJF,PsalacHY,PsalacGC,PsalacHWD,PsalacINS,Psalac7285,PsalacLT12,PsalacLT16,PsalacLT26,Psalac349,PsalacKw30 andPsalacK3 were cloned from differentPseudomonassyringaepv.actinidaestrains isolated from diverse regions and countries. The results showed that the complete DNA sequence of laccase gene were 1374 bp in length, encoding a putative proteins of 457-amino acid. All canker strains share the same amino acid sequences and small nucleic acid sequence differences, containing four highly conservative Cu2+active sites. Promoter is located in the upstream gene with 175 bp. Phylogenetic analysis indicated that putative laccase sequence was homologous toP.fluorescensPf0-1 laccase CP000094.2. The studies suggested that laccase gene had the sequence characteristics of laccase gene family in bacteria, which meant that laccase might involve in the pathogenicity and copper-resistant ofPseudomonassyringaepv.actinidae.
kiwifruit; kiwifruit bacterial canker; laccase
2014-04-17;
2014-04-25
安徽省自然科学基金(1408085QC62)
刘 普,博士,讲师,主要从事果树病害分子机制,E-mail:puliu@ahau.edu.cn;
朱立武,教授,主要从事果树种质资源利用与创新,E-mail: zhuliwu@ahau.edu.cn。
S663.4;Q78
A
2095-1736(2015)01-0048-04
doi∶10.3969/j.issn.2095-1736.2015.01.048

