Plasmid-mediated quinolone resistance determinants qnrA1and qnrA3 in Escherichia coli isolatesfrom pigs containing the aac(6')-Ib-cr gene
CHEN Xiang,ZHANG Ning,ZHANG Wei-qiu,LI Xiu,WANG Yan-hong,JIAO Xin-an
(Jiangsu Key Laboratory of Zoonosis / Jiangsu Co-Innovation Center for Prevention and Control of ImportantAnimal Infectious Diseases and Zoonoses, Yangzhou University, Yangzhou 225009, China)
Quinolone resistance was thought to be mediated only by chromosomal mutations, until plasmid-mediated quinolone resistance (PMQR) was described in 1998[1]. Since then, several PMQR mechanisms have been described: 1) the pentapeptide repeat family Qnr proteins (QnrA, QnrB, QnrS, QnrC, and QnrD); 2) AAC(6')-Ib-cr, a variant aminoglycoside acetyltransferase that is responsible for reduced susceptibility to ciprofloxacin by modifying ciprofloxacin; 3) QepA, an efflux pump belonging to the major facilitator subfamily, and OqxAB, a multidrug efflux pump that confers resistance to multiple agents, which has been recently reported to reduce susceptibility to ciprofloxacin and nalidixic acid. These mechanisms provide the low-level quinolone resistance showninvitroto facilitate the emergence of high-level resistance in the presence of quinolones at therapeutic levels[1].
The 218-amino-acid protein QnrA, which belongs to the pentapeptide repeat family, protects DNA gyrase and topoisomerase Ⅳ from the inhibitory activity of quinolones.QnrAhas been well known for its worldwide distribution and is located in complexsul1-type class 1 integrons. Nowadays, sevenqnrAalleles (QnrA1-7) have been described (http://www.lahey.org/qnrStudies/). To monitor the prevalence ofqnrAand other PMQR genes, a total of 198E.colifrom pigs were identified.
Materials and methods
Bacterial isolates
A total of 198E.coliisolates were collected from 9 provinces between 1998 and 2007. Among them, 188 strains were isolated from clinically affected pigs, and the rest were from pigs at slaughter. The isolates were identified using biochemical procedures[2]. Each isolate was derived from a different specimen. After isolation, the purified isolates were stored at -70 ℃ in Luria-Bertani (LB) broth with 20% glycerol.
Detection of PMQR genes
PMQR genes includingqnrA,qnrB,qnrS,qnrC,qnrD,aac(6')-Ib-cr,qepAamong the 198 clinicalE.coliisolates were analyzed[3].qnrAgene was determined by polymerase chain reaction (PCR), using the primers 5’-AGA GGA TTT CTC ACG CCA GG and 5’- GCA GCA CTA TKA CTC CCA AGG (K=A or C). The conditions were as follows: an initial step of 94 ℃ for 3 min, followed by 30 cycles of 30 sec at 94 ℃, 30 sec at 57 ℃ annealing temperature, 40 sec at 72 ℃, and a final extension step of 7 min at 72 ℃. To sequence differentqnrgenes, the primers 5’-GGG TAT GGA TAT TAT TGA TAA AG and 5’-CTA ATC CGG CAG CAC TAT TA were used[4-5].
Conjugation experiment
Isolates harboringqnrAgene were selected for conjugation experiments by the broth-mating method usingE.coliJ53 as the recipient[4]. Gentamicin and tetracycline were used with sodium azide to select the transconjugants. The transconjugants harboringqnrAgene and other PMQR genes were confirmed by PCR as previously described.
Antimicrobial susceptibility testing
Susceptibilities to ampicillin, cefotaxime, ciprofloxacin, chloramphenicol, enoxacin, fleroxacin, gatifloxacin, gentamicin, kanamycin, nalidixic acid, norfloxacin, tetracycline of the transconju-gants, recipient, and donors were assayed by the broth dilution method, according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI)[6].E.coliJ53 AzR was used as the control strain[4].
Location of PMQR genes
To analyze the location of PMQR genes in donors and transconjugants, S1 nuclease-PFGE and Southern blot analysis were also performed[7]. Briefly, whole-cell DNA of the donors and transconjugants embedded in agarose gel plugs was treated with S1 nuclease and separated by PFGE alongside the marker,Salmonellaserotype Braenderup reference standard (H9812), restricted withXbaI. Subsequently, Southern blot hybridization was performed with DNA probes generated from PCR products as previously described usingqnrAandaac(6')-Ib-crand a digoxigenin nucleic acid labeling and detection system (Roche Diagnostics, Mannheim, Germany).
Results
Prevalence of PMQR genes
There were four strains being positive forqnrA, in which one strain isolated in 2004 in Liao-ning Province was positive forqnrA1 and the other three isolates wereqnrA3 which derived on the same pig farm in 2005 in Jiangsu Province. The fourE.colistrains also carriedaac(6')-Ib-cr, which were confirmed by digestion withFokⅠ and direct sequencing. All the isolates were negative forqnrB,qnrC,qnrDorqepA.
Conjugation experiment and antimicrobial susceptibility testing
By conjugation experiment, 4 transconjugants carryingqnrAandaac(6')-Ib-crwere obtained from theqnrAandaac(6')-Ib-crpositive strains. The susceptibility testing results showed that the ciprofloxacin MICs of the four transconjugants ranged from 0.25 to 1 μg/mL, an increase of 32- to 128- fold over that of the recipient strain (Table 1). TheqnrA1 andaac(6')-Ib-crpositive transconjugant was resistance to gentamicin and kanamycin, and the threeqnrA3 andaac(6')-Ib-crpositive transconjugants were resistance to tetracycline.
Location of PMQR genes
The results of S1 nuclease-PFGE and Sou-thern blot analysis revealed that the plasmid transfered to J53 AzR, andqnrAandaac(6')-Ib-crwere both located on it. The location of the two genes in donor strain was the same. So as shown in Figure 1,qnrAandaac(6')-Ib-crgenes were located on the same plasmid (about 110 kb) in both the strain 23 and its transconjugant (Figure 1).
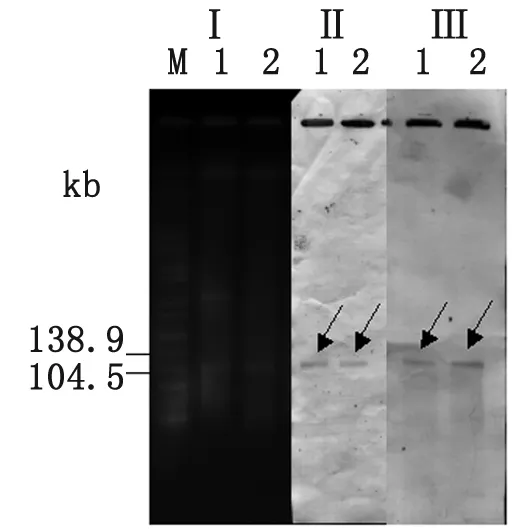
Fig.1PanelIshowsgelelectrophoresisoftheplasmidsofE.coli23(Lane1)anditstransconjugantstrain(Lane2)
Lane M isSalmonellaserotype Braenderup reference standard (H9812) restricted withXbaI. Panels II, III show the results of Southern blot analysis of uncut plasmids hybridized withqnrA(II) oraac(6')-Ib-cr(III) probe, respectively.
Tab.1CharacteristicsofdonorstrainsandE.coliJ53AzRtransconjugants

Note:aNo. 40, 41, 42 isolates were from the same farm.btransconjugant.cMIC--minimum inhibitory concentration; NA--nalidixic acid; CIP--ciprofloxacin; NOR--norfloxacin; ENX--enoxacin; FLX--fleroxacin; GAT--gatifloxacin; Amp--ampicillin; CTX--cefotaxime; Gen--gentamicin; Km--kanamycin; Tet--tetracycline; Cm--chloramphenicol.
Discussion
Until 1998, it was believed that quinolone resis-tance could be acquired only through chromosomal mutation. Since then, it has become clear that the plasmid-borne geneqnrAcan generate inEntero-bacteriaceaean 8- to 32-fold increase in MICs of quinolones by protecting DNA gyrase directly from quinolone inhibition[1]. The occurrence of theqnrA-positive enterobacterial isolates has been reported in the United States and then in Asia, Europe and Australia.
In this study, a total of 198E.coliisolates were collected in 9 provinces from 1998 to 2007. The occurrence ofqnrAgene in all theE.colistrains was investigated by PCR. DifferentqnrAgenes were also sequenced. The results showed that one was positive forqnrA1 and the other three isolates wereqnrA3, all of which were positive foraac(6')-Ib-cr. And then conjugation experiments were carried out to obtain the transconjugants. Four transconjugants carryingqnrAandaac(6')-Ib-crwere obtained using the liquid mating-out assay. The ciprofloxacin MICs of the four transconjugants ranged from 0.25 to 1 μg/mL, an increase of 32- to 128- fold over that of the recipient strain. Various determinants for resistance to other antimicrobial agents were also transferred with the plasmid. All of the strains were resistant to ampicillin. TheqnrA1 andaac(6')-Ib-crpositive transconjugant was resistant to gentamicin and kanamycin, and the threeqnrA3 andaac(6')-Ib-crpositive transconjugants were resistant to tetracycline (Table 1). The coexistence ofqnrAandaac(6')-Ib-crin a single plasmid and increasedqnrAexpression can account for the different levels of ciprofloxacin resistance shown in the transconju-gants[8].
It has been hypothesised thatqnrAemerged among clinical strains because of the selective pressure of quinolones[1,4,9]. Our study demonstrated thatqnrAcould transfer horizontally with the plasmid. It led to various determinants for resistance to other antimicrobial agents. It might explain why multidrug resistance, including resistance to quinolones, is rising worldwide.
In the present work,qnrAandaac(6')-Ib-crwere found to be located on the same plasmid, which was in accordance with a previous report thatqnralleles were frequently collocated withaac(6')-Ib-cron the same plasmid[1,9]. The association of these resistance determinants is worrisome, because it may facilitate the selection of high-level multidrug-resistant strains, which pose a potential threat to public health.
[1]Strahilevitz J, Jacoby GA, Hooper DC, et al. Plasmid-mediated quinolone resistance: a multifaceted threat[J]. Clin Microbiol Rev, 2009, 22(4): 664-689. DOI: 10.1128/CMR.00016-09
[2]Chen X, Gao S, Jiao XA, et al. Prevalence of serogroups and virulence factors ofEscherichiacolistrains isolated from pigs with postweaning diarrhea in eastern China[J]. Vet Microbiol, 2004, 103(1-2): 13-20. DOI: 10.1016/j.vetmic.2004.06.014
[3]Chen X, Zhang WQ, Pan WJ, et al. Prevalence ofqnr,aac(6')-Ib-cr,qepA, andoqxABinEscherichiacoliisolates from humans, animals, and the environment[J]. Antimicrob Agents Chemother, 2012, 56(6): 3423-3427. DOI: 10.1128/AAC.06191-11
[4]Wang M, Tran JH, Jacoby GA, et al. Plasmid-mediated quino-lone resistance in clinical isolates ofEscherichiacolifrom Shanghai, China[J]. Antimicrob Agents Chemother, 2003, 47(7): 2242-2248. DOI: 10.1128/AAC.47.7.2242-2248.2003
[5]Jacoby G, Cattoir V, Hooper D, et al.qnrgene nomenclature[J]. Antimicrob Agents Chemother, 2008, 52(7): 2297-2299. DOI: 10.1128/AAC.00147-08
[6]Clinical and Laboratory Standards Institute. Performance stan-dards for antimicrobial susceptibility testing; 23rd informational supplement[S]. In: CLSI Document M100-S23. Wayne: Clinical and Laboratory Standards Institute, 2013.
[7]Liu BT, Yang QE, Li L, et al. Dissemination and characterization of plasmids carryingoqxAB-blaCTX-Mgenes inEscherichiacoliisolates in food-producing animals[J]. PLoS ONE, 8(9): e73947. DOI: 10.1371/journal.pone.0073947
[8]Martinez-Martinez L, Pascual A, Jacoby GA. Quinolone resis-tance from a transferable plasmid[J]. Lancet, 1998, 351(9105): 797-799. DOI: 10.1016/S0140-6736(97)07322-4
[9]Rodríguez-Martínez JM, Cano ME, Velasco C, et al. Plasmid-mediated quinolone resistance: an update[J]. J Infect Chemo-ther, 2011, 17(2): 149-182. DOI: 10.1007/s10156-010-0120-2

