Entamoeba histolytica acetyl-CoA synthetase: biomarker of acute amoebic liver abscess
Lim Boon Huat, Alfonso Olivos Garcia, Tan Zi Ning, Wong Weng Kin, Rahmah Noordin, Siti Shafiqah Anaqi Azham, Lee Zhi Jie, Guee Cher Ching, Foo Phiaw Chong, Pim Chau Dam
1Biomedicine Programme, School of Health Sciences, Universiti Sains Malaysia, 16150 Kubang Kerian, Kelantan, Malaysia
2Departmento de Medicina Experimental, Facultad de Medicina, Universidad Nacional Autonoma de Mexico, 04510 Mexico D.F., Mexico
3Department of Medicine, Allianze University College of Medical Sciences, Pulau Pinang, Malaysia
4Institute for Research in Molecular Medicine, 11800 Universiti Sains Malaysia, Penang, Malaysia
Entamoeba histolytica acetyl-CoA synthetase: biomarker of acute amoebic liver abscess
Lim Boon Huat1*, Alfonso Olivos Garcia2, Tan Zi Ning3, Wong Weng Kin1, Rahmah Noordin4, Siti Shafiqah Anaqi Azham1, Lee Zhi Jie1, Guee Cher Ching1, Foo Phiaw Chong1, Pim Chau Dam1
1Biomedicine Programme, School of Health Sciences, Universiti Sains Malaysia, 16150 Kubang Kerian, Kelantan, Malaysia
2Departmento de Medicina Experimental, Facultad de Medicina, Universidad Nacional Autonoma de Mexico, 04510 Mexico D.F., Mexico
3Department of Medicine, Allianze University College of Medical Sciences, Pulau Pinang, Malaysia
4Institute for Research in Molecular Medicine, 11800 Universiti Sains Malaysia, Penang, Malaysia
PEER REVIEW
Peer reviewer
Professor Dr. Armando Acosta Domínguez, Department of Basic Research, Finlay Institute, Avenue 27 No. 19805, La Lisa, Havana City AP 16017, CP 11600, Cuba.
Tel: 5372716911
E-mail: aracosta2005@yahoo.es
Comments
This is a well-researched and clearly presented manuscript that will be useful to both fundamental and applied scientists who research on E. histolytica, the parasitic protozoan that causes high morbidity and mortality in developing countries endemic for the infection.
Details on Page 450
Objective:To characterize the Entamoeba histolytica (E. histolytica) antigen(s) recognized by moribound amoebic liver abscess hamsters.
Entamoeba histolytica, Amoebic liver abscess, Biomarker, Acetyl-CoA synthetase, Recombinant EhACS, Western blot
1. Introduction
Amoebic liver abscess (ALA) is a fatal manifestation of the enteric protozoanEntamoeba histolytica(E. histolytica). Human and some non-human primates are the main reservoirs for this protozoan[1]. In order to simulate human ALA to better understand the underlying pathogenesis, mouse, rat and rabbit models have been tested, but only hamster (Mesocricetusauratus) and gerbil (Meriones unguiculatus) were reported to be susceptible to the trophozoites[2]. The experimental infection routes include direct inoculation of viable, virulence trophozoites into the liver, hepatic portal vein or peritoneal cavity of the animal[3]. ALA infected rodent usually deteriorates to moribund state in approximately a week, which is accompanied by increase malaise, liver tenderness, lethargy and reduce appetite.
In rodents, antibodies produced against pathogen could be detected as early as 5-7 d, post infection[4]. Hence, the antibodies produced during the acute stage may recognise potentially important biomarker(s) pertinent to the understanding of pathogenesis, vaccine development and diagnosis of ALA. To date, there was no report on the analysis of antibodies produced in either hamster or gerbil post-infected with virulentE. histolyticatrophozoites.
The objective of this study was to identify and characterize potentialE. histolyticabiomarker(s) recognised by sera of moribund ALA hamsters. pulse-on and 0.5 seconds pulse-off. The lysate was then centrifuged at 10 000 × g (9020 r/min) for 10 min at 4 °C to collect the CSA supernatant.
2. Materials and methods
2.1.HM-1:IMSS E.histolytica culture
HM-1:IMSSE. histolyticatrophozoites were axenically cultured in Diamond TYI-S-33 medium[5]. The trophozoites were continuously passed through the liver of hamsters to maintain their virulence.
2.2. ExperimentalALAin hamster and serum collection
The animal experimentation was conducted in accordance with the requirements of Universiti Sains Malaysia Animal Ethics Committee, PPSG/07(A)/044/(2008)(40). A maximum of three 8-10 week old male hamsters were housed in each polycarbonate cage with regular changes of clean woodchips beddings. The animals were provided with alternate 12 h lighting and 12 h darkness, as well asad libitumfood pellets and water. ALA was induced by inoculating each hamster with 1×106trophozoites in 0.2 mL phosphate buffered saline via the portal vein[6]. In approximately 6-8 d post-inoculation, the hamster was euthanised with three-time overdose of sodium pentobarbital. Blood was immediately collected via cardiac puncture into a sterile micro-centrifuge tube and allowed to clot. Serum was separated, labelled as ‘hamster ALA serum’ and kept at -20 °C until used. In the control group, each healthy animal was inoculated with 0.2 mL sterile phosphate buffer saline (PBS) without the trophozoites. The serum collected was labelled as ‘hamster control serum’ and kept at -20 °C until used. Twenty microlitres of each ALA serum sample were pooled into a microfuge tube, mixed and labelled as ‘pooled hamster ALA sera’. Similarly, ‘pooled hamster control sera’were pooled from the control serum samples.
2.3. Preparation of crude soluble antigen (CSA)
Ten millionE. histolyticatrophozoites were mixed with 500 µL complete Lysis-M buffer added with protease inhibitor cocktail (Roche, Germany) and 20 µL of 0.5 mol/L iodoacetamide (Sigma, USA). The mixture was sonicated (Branson, UK) at 10% amplitude for three cycles of 1 min sonication with 0.5 seconds
2.4. 1D-PAGEandWestern blot analysis
Twenty micrograms of CSA per well were separated by 9% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and electro-blotted (Bio-Rad, USA) onto a 0.45 µm-pore-size nitrocellulose membrane. The membrane was blocked for 1 h at room temperature with 5% skimmed milk. Subsequently, the membrane was cut into multiple strips and each strip was incubated with Tris-buffered saline (TBS) control, pooled hamster control sera, individual hamster ALA serum samples, or pooled hamster ALA sera at dilution of 1:50 in TBS containing 0.1% Tween 20 (TBS-T) for 2 h at RT. After washing with TBS-T for 3×5 min, the strips were incubated with monoclonal mouse anti-hamster IgG-HRP (Sigma, USA) at a dilution of 1: 4 000 in TBS for 1 h at RT. The signals were developed using enhanced chemiluminescence (ECL) blotting reagent (Roche Diagnostics, Germany) and captured using X-ray film.
2.5. 2D-PAGEandWestern blot analysis
The proteins ofE. histolyticaCSA were first fractionated based on their isoelectric points (pI) using 3100 OFFGEL Fractionator (Agilent Technologies, Germany). ImmobilineTMDry Strip (GE Healthcare, UK) with linear pH range 3-10 in a 12-well setup was used. The fractionation was performed according to the manual provided by the manufacturer. Twenty microlitres of a fractionated protein sample were mixed with 5 µL of 5X sample buffer without boiling, and subsequently separated by 9% SDS-PAGE. Western blot analysis was performed on the 12 protein fractions using pooled hamster ALA sera and pooled hamster control sera to identify the antigenic fraction(s). For confirmation of the antigenic fraction, the Western blot experiment was repeated on the fraction using individual hamster ALA serum samples.
2.6. Mass spectrometry analysis and protein identification
The antigenic protein band was sent to the Australian Proteomic Service for tandem mass spectrometry (MS/MS) analysis. At the proteomic facility, the protein sample was digested with trypsin and the peptides were extracted and analysed by electro-spray ionisation mass spectrometry using the Ultimate 3000 nano high performance liquid chromatography system [Dionex] coupled to a 4000 Q TRAP mass spectrometer [Applied Biosystems]. The mass-spectrometry analysis was performed with two different gel slice samples to ensure reproducibility. According to peptides sequence BLAST via Mascot,E. histolyticaacetyl-CoA synthetase (UniProt accession number: Q9NAT4) was identified.
2.7. Cloning and expression of recombinant acetyl-CoA synthetase
Genomic DNA was isolated from 3×106E. histolyticatrophozoites using QIAamp DNA mini kit (QIAGEN GmbH, Germany). PCR primers targeting the amplification ofE. histolyticaacetyl-CoA synthetase (EhACS) gene from the genomic DNA were F-5’-GGA ATT CCA TAT GAT GCA ATT TGA GCC ACT and R-5’-CCG CTC GAG TTA TGG TTG GAT GAC GA. The PCR product (2 142 bp) was cloned into the pET-14b vector and confirmed via sequencing. Recombinant EhACS (rEhACS), was then propagated inEscherichia coliXL 1-blue and expressed inEscherichia coliBL21-AI. During expression, 1 mL of overnight starter culture was inoculated into 50 mL Luria-Bertani broth supplemented with 50 µg/mL ampicillin. At OD6000.6-0.8, overexpression of rEhACS protein was induced with 0.2% (w/v) L-arabinose at 32 °C, overnight. The recombinant protein was purified using HisPurTMNi-NTA Resin (Fisher Scientific, USA), according to the manufacturer’s protocol, and the concentration was estimated using the Bradford method.
2.8. Evaluation of rEhACSvia indirectELISA
The rEhACS identified as a potential biomarker in hamster experimentation was evaluated using hamster ALA serum samples in an indirect-ELISA format. The customised rEhACSELISA was optimized and performed based on modifications of the method described by Reen[7]. Each well of the flat-bottom microtiter plate (NUNC, Denmark) was coated with 100 µL of 25 µg/µL of the recombinant protein in 0.1 mol/L carbonate buffer, pH 9.6, overnight at 4 °C in a humid box. Each well was then blocked with 200 µL blocking reagent (Roche, Germany) for 1 h. Then, 100 µL of each hamster serum sample, followed by 100 µL of monoclonal mouse anti-hamster IgG-HRP (Sigma, USA) were incubated in each well for 1 h at serum dilutions of 1:100 and 1:1 000 in PBS, respectively. Each cycle of washing (3× 5 min) was performed using 200 µL per well of phosphate-buffer saline (PBS) containing 0.05% Tween 20. After a final incubation with 100 µL TMB substrate per well for 15 min in the dark, 100 µL of 1 N H2SO4was added as the stop solution. The absorbances were read at 450 nm using a Multiskan FC Microplate Reader (Fisher Scientific, USA). The cut-off value for the indirect ELISA was then determined based on the mean optical density (OD) readings plus 2 standard deviations (SD) obtained for the five hamster control serum samples.
3. Results
3.1. 1D-Western blot analysis ofCSAprobed with hamster serum samples
The antigenic profile of CSA probed with the hamster ALA serum samples revealed 5 antigenic bands,i.e.~100 kDa,~75 kDa, ~65 kDa, ~50 kDa and ~45 kDa, which did not react with the controls. Interestingly, only the ~75 kDa protein band was detected by all the hamster ALA serum samples (Figure 1).
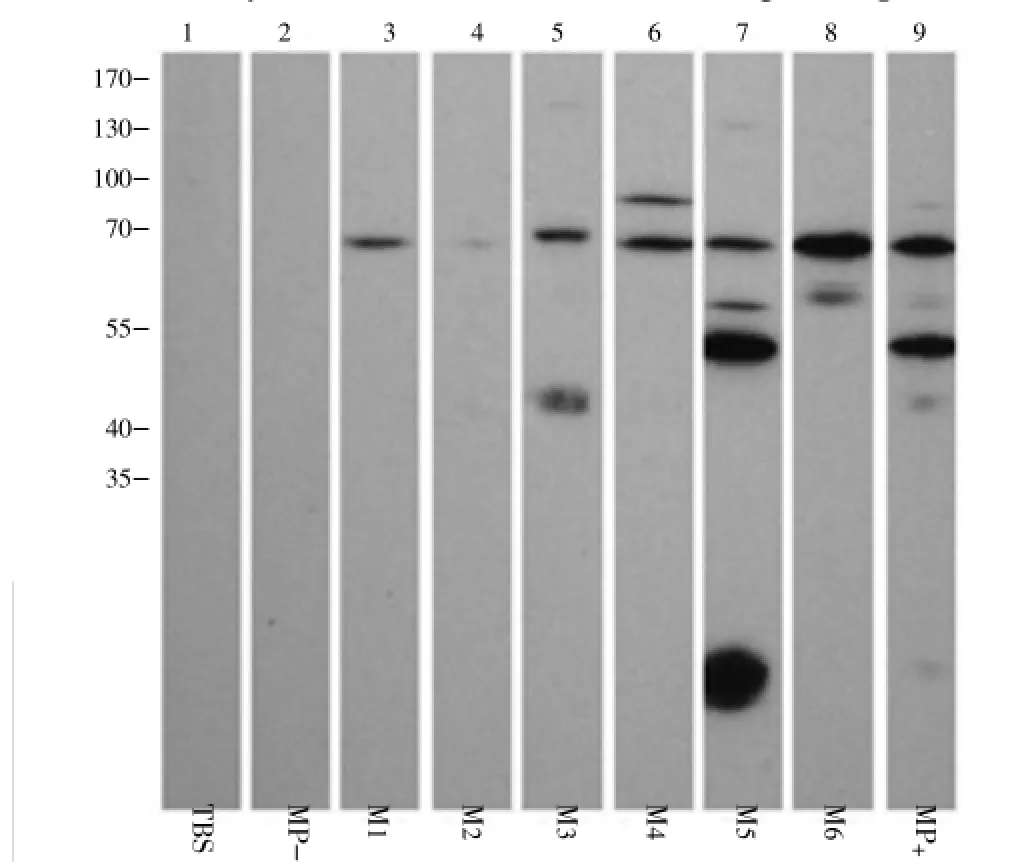
Figure 1. Western blot analysis of CSA probed with hamster serum samples (Representive data).MP+: Pooled hamster ALA sera; MP-: Pooled hamster healthy sera; M1-M6 represent individual hamster ALA serum samples.

Figure 2. Western blot analysis of fractionated CSA probed with hamster pooled control and ALA serum samples. *: Position of ~75 kDa protein band; M+: Pooled hamster ALA sera; M-: Pooled hamster healthy sera.
3.2. 2D protein separation and identification
CSA was fractionated into 12 fractions according to the pI values of its proteins. Western blot analysis of the fractions revealed that the expected ~75 kDa protein was located mostly in Fraction 6 with a pI range of 5.91-6.5 (Figure 2). Further Western blot analysis on this protein fraction indicated that the~75 kDa protein was recognized by all the individual hamster ALA serum samples (Figure 3). The targeted protein band was excised from SDS-PAGE gel and sent for MS/MS analysis. Mass spectrometric analysis identified the ~75 kDa antigenic protein as: tr|C4LUV9|acetyl-CoA synthetase, putative Tax_Id=5759 [E. histolytica]. The protein and peptide scores were 294 and 1 456 respectively, with sequence coverage of 44%. In the peptide report, there were 7 of 48 peptides which were above the cut-off ion score of 60 which indicated identity or extensive homology (P<0.05).

Figure 3. 2D-Western blot analysis of CSA probed with individual hamster ALA serum samples.*: Position of the ~75kDa protein band; M1+ to M5+: Individual hamster ALA serum samples; MP+: Pooled hamster ALA sera; M-: Pooled hamster control sera.
3.3. Examination rEhACSantigenicity viaELISA
In the customized rEhACS-ELISA, a cut-off value of 0.1 573 was determined based on mean OD+2SD of the five hamster control serum samples. The ELISA revealed 100% sensitivity and specificity when tested with serum samples from moribund hamster with ALA (n=31) and the control group (n=5), respectively (Figure 4).
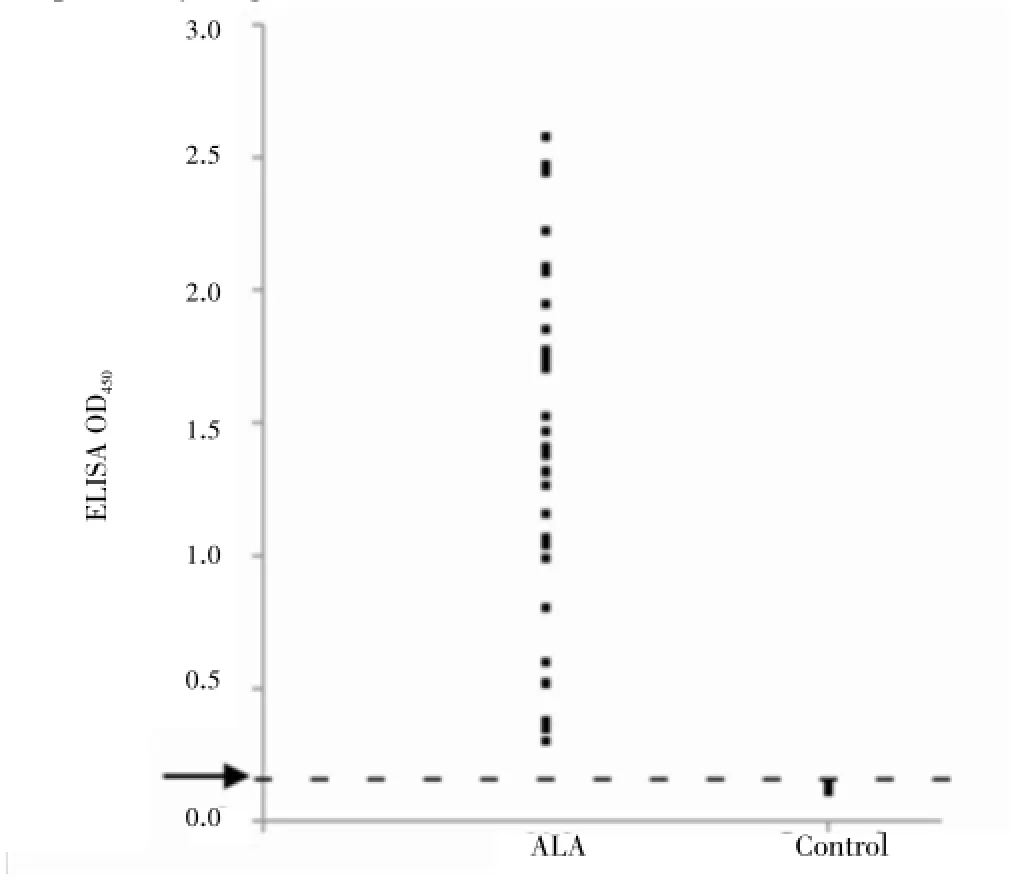
Figure 4. OD450distribution of hamster ALA and control serum samples in rEhACS-ELISA.
4. Discussion
Virulent strain of HM-1:IMSS ofE. histolyticawas maintained by routinely passing the trophozoites through hamster livers twice a month[6]. In this study, serum samples obtained from the euthanized animals were found to possess sufficient antibodies for immunoblot analysis. In the Western blot analyses, all the ALA hamster sera, but not the healthy control sera, were found to recognise the 77 kDaE. histolyticaADP-forming ACS. This is therefore a potential diagnostic marker during the acute and terminal stage of the infection. The detectable antibody level produced during this relatively short period of 7 d was in accordance with a previous report that revealed rodent antibodies were generally first detected between 5-7 d postinfection[4].
In experimentally induced hamster ALA, there was massive death of trophozoites during the first few hours post-infection[6]. It is highly probable thatE. histolyticaantigens, including EhACS, released from these dead amoebas triggered the host humoral responses during this early stage of infection. It was also pointed out that the 12 h post-infection period was the critical stage for successful invasion by the amoebas into the hamster liver, where the lowest number of trophozoites was observed in the infected liver[6]. Possibly the live amoebas continued to secrete or excrete EhACS that boosted the production of antibody, or antibody production was due to release of the highly immunogenic antigen during subsequent death of trophozoites.
In mitochondrial organisms, acetyl-CoA is the fuel for tricarboxylic acid cycle but this pathway is absent from the amitochondriateE. histolytica. The amoeba ADP-forming converts acetyl-CoA to acetate by hydrolysis to generate ATP from ADP and Pi[8]. However, it does not appear to synthesise ATP from AMP and Ppi[9]. Interestingly, it is also involved in degradation of amino acids by accepting propionyl-CoA as asubstrate to generate ATP[8,10]. Although the functions of EhACS inE. histolyticametabolism are relatively well reported, its potential roles in pathogenesis, vaccine and diagnosis are basically unexplored[11-13].
In conclusion, this study has successfully identified EhACS as a biomarker for ALA in moribund hamsters. The potential role(s) of EhACS in elucidating pathogenesis, developing vaccine and improving diagnosis of human amoebiasis should be further studied.
Conflict of interest statement
We declare that we have no conflict of interest.
Acknowledgements
We thank Dr. R. Perez Tamayo (Departmento de Medicina Experimental, Facultad de Medicina, Universidad Nacional Autonoma de Mexico) for the kind gift ofE. histolyticaaxenic culture and Dr. Graham C. Clark (London School of Hygiene and Tropical Medicine, LSHTM) for his valuable review on the manuscript and sharing his expertise with LBH during his three-month sabbatical leave at LSHTM. We also thank Dr. Rumaizi Shaari (Universiti Malaysia Kelantan) and Dr. See Too Wei Cun (School of Health Sciences, Health Campus of Universiti Sains Malaysia) for their contributions. This work was supported by Malaysia Ministry of Education Long-Term Research Grant Scheme (LRGS) No. 203/PSK/6722002.
Comments
Background
ALA is a fatal manifestation ofE. histolytica. To better understand the pathogenesis of human ALA, hamster (Mesocricetus auratus) has been used to maintain the virulence ofE. histolyticain liver of hamster. This study attempted to identify and characterize potential biomarker(s) ofE. histolyticarecognized by antibodies of hamsters with acute ALA.
Research frontiers
This is the first study that utilized a 5-7 d post-infected hamster sera in Western blotting to revealE. histolyticaacetyl-CoA synthetase as a potentially important biomarker in acute ALA infection.
Related reports
TheE. histolyticaacetyl-CoA synthetase recombinant protein was subsequently expressed and utilized in an indirect ELISA format to show that it was highly specific and sensitive in the detection of hamster antibodies during acute stage of ALA infection. There are no other reports and this is a new finding.
Innovations and breakthroughs
The innovative aspect of this paper is the utilization of 5-7 d sera obtained from hamsters with acute ALA to perform Western blotting to identify the biomarker. In comparison, other studies normally focused on the infected liver.
Applications
The potential roles of EhACS in elucidating pathogenesis, developing vaccine and improving diagnosis of human amoebiasis should be further studied. In addition, it would be interesting to ascertain the potential of rEhACS in determining antibodies from human with ALA.
Peer review
This is a well-researched and clearly presented manuscript that will be useful to both fundamental and applied scientists who research onE. histolytica, the parasitic protozoan that causes high morbidity and mortality in developing countries endemic for the infection.
[1] Anuar TS, Al-Mekhlafi HM, Abdul Ghani MK, Abu Bakar E, Azreen SN, Salleh FM, et al. Molecular epidemiology of amoebiasis in Malaysia: highlighting the different risk factors of Entamoeba histolytica and Entamoeba dispar infections among Orang Asli communities. Int J Parasitol 2013; 42(13-14): 1165-1175.
[2] Tsutsumi V, Shibayama M. Experimental amebiasis: a selected review of some in vivo models. Arch Med Res 2006; 37(2): 210-220.
[3] Villalba-Magdaleno JD, Pérez-Ishiwara G, Serrano-Luna J, Tsutsumi V, Shibayama M. In vivo programmed cell death of Entamoeba histolytica trophozoites in a hamster model of amoebic liver abscess. Microbiology 2011; 157(5): 1489-1499.
[4] Compton SR, Riley LK. Detection of infectious agents in laboratory rodents: traditional and molecular techniques. Comp Med 2001; 51(2): 113-119.
[5] Pires-Santos GM, Santana-Anjos KG, Vannier-Santos MA. Optimization of Entamoeba histolytica culturing in vitro. Exp Parasitol 2012; 132(4): 561-565.
[6] Faust DM, Guillen N. Virulence and virulence factors in Entamoeba histolytica, the agent of human amoebiasis. Microbes Infect 2012; 14(15): 1428-1441.
[7] Wild D. The immunoassay handbook: theory and applications of ligand binding, ELISA and related techniques. Amsterdam, Netherlands: Elsevier; 2013.
[8] Pineda E, Encalada R, Olivos-García A, Néquiz M, Moreno-Sánchez R, Saavedra E. The bifunctional aldehyde-alcohol dehydrogenase controls ethanol and acetate production in Entamoeba histolytica under aerobic conditions. FEBS Letters 2013; 587(2): 178-184.
[9] Clark CG, Alsmark UCM, Tazreiter M, Saito-Nakano Y, Ali V, Marion S, et al. Structure and content of the Entamoeba histolytica genome. Adv Parasitol 2007; 65: 51-190.
[10] Atteia A, van Lis R, Tielens AG, Martin WF. Anaerobic energy metabolism in unicellular photosynthetic eukaryotes. Biochim Biophys Acta 2013; 1827(2): 210-223.
[11] Beaumier CM, Gillespie PM, Hotez PJ, Bottazzi ME. New vaccines for neglected parasitic diseases and dengue. Transl Res 2013; 162(3): 144-155.
[12] Singh A, Houpt E, Petri WA. Rapid diagnosis of intestinal parasitic protozoa, with a focus on Entamoeba histolytica. Interdiscip Perspect Infect Dis 2009; doi: 10.1155/2009/547090.
[13] Carranza-Rosales P, Santiago-Mauricio MG, Guzmán-Delgado NE, Vargas-Villarreal J, Lozano-Garza G, Viveros-Valdez E, et al. Induction of virulence factors, apoptosis, and cytokines in precision-cut hamster liver slices infected with Entamoeba histolytica. Exp Parasitol 2012; 132(4): 424-433.
10.12980/APJTB.4.2014C1169
*Corresponding author: Lim Boon Huat, (PhD), Senior Lecturer in Biomedicine Programme, School of Health Sciences, Health Campus, Universiti Sains Malaysia, 16150 Kubang Kerian, Kelantan, Malaysia.
Tel: (+6)097677619
Fax: (+6)097677515
E-mail: limbh493@gmail.com, limbh@kb.usm.my
Foundation Project: Supported by Malaysia Ministry of Education Long-Term Research Grant Scheme (LRGS) No. 203/PSK/6722002.
Article history:
Received 1 Apr 2014
Received in revised form 8 Apr, 2nd revised form 14 Apr, 3rd revised form 20 Apr 2014
Accepted 29 Apr 2014
Available online 28 Apr 2014
Methods:Crude soluble antigen of E. histolytica was probed with sera of moribund hamsters in 1D- and 2D-Western blot analyses. The antigenic protein was then sent for tandem mass spectrometry analysis. The corresponding gene was cloned and expressed in Escherichia coli BL21-AI to produce the recombinant E. histolytica ADP-forming acetyl-CoA synthetase (EhACS) protein. A customised ELISA was developed to evaluate the sensitivity and specificity of the recombinant protein.
Results:A ~75 kDa protein band with a pI value of 5.91-6.5 was found to be antigenic; and not detected by sera of hamsters in the control group. Tandem mass spectrometry analysis revealed the protein to be the 77 kDa E. histolytica ADP-forming acetyl-CoA synthetase (EhACS). The customised ELISA results revealed 100% sensitivity and 100% specificity when tested against infected (n=31) and control group hamsters (n=5) serum samples, respectively.
Conclusions:This finding suggested the significant role of EhACS as a biomarker for moribund hamsters with acute amoebic liver abscess (ALA) infection. It is deemed pertinent that future studies explore the potential roles of EhACS in better understanding the pathogenesis of ALA; and in the development of vaccine and diagnostic tests to control ALA in human populations.
 Asian Pacific Journal of Tropical Biomedicine2014年6期
Asian Pacific Journal of Tropical Biomedicine2014年6期
- Asian Pacific Journal of Tropical Biomedicine的其它文章
- A retrospective evaluation of the quality of malaria case management at twelve health facilities in four districts in Zambia
- Study of the efficacy of a Wheaton coated bottle with permethrin and deltamethrin in laboratory conditions and a WHO impregnated paper with bendiocarb in field conditions
- Pharmacognostical study and establishment of quality parameters of aerial parts of Costus speciosus-a well known tropical folklore medicine
- Hepatocurative potential of Vitex doniana root bark, stem bark and leaves extracts against CCl4-induced liver damage in rats
- In vitro α-amylase inhibitory activity and in vivo hypoglycemic effect of methanol extract of Citrus macroptera Montr. fruit
- Antimicrobial activity of some essential oils against oral multidrugresistant Enterococcus faecalis in both planktonic and biofilm state
