Genetic dissection of tetraploid cotton resistant to Verticillium wilt using interspecific chromosome segment introgression lines
Peng Wng,Zhiyun Ning,Ling Lin,Hong Chen,Hongxin Mei,Jun Zho,Bingling Liu,Xin Zhng,Wngzhen Guo,Tinzhen Zhng,*
aNational Key Laboratory of Crop Genetics& Germplasm Enhancement,Cotton Research Institute,Nanjing Agricultural University,Nanjing 210095,China
bInstitute of Plant Protection,Jiangsu Academy of Agricultural Sciences,Nanjing 210014,China
cCotton Research Institute,Xinjiang Academy of Agriculture and Reclamation Sciences,Xinjiang 832000,China
1.Introduction
Cotton (Gossypium spp.) is one of the most important fiber crops in the world and serves as a source of oil and biofuel[1].Verticillium wilt has worldwide distribution and causes serious economic losses [2].The disease is caused by the soilborne fungus Verticillium dahliae Kleb.The fungus infects the roots of the cotton plant in the soil by entering through cortical cells.Once inside,the spores and mycelia of the pathogen block the vessels of the plant.V.dahliae toxins and acidic glycoproteins are also important pathogenicity factors that can rapidly induce wilting[3].Strains of the pathogen in China can be divided into two types according to their virulence: defoliating and nondefoliating [4].An early symptom seen in the host plant after infection by a defoliating pathogen is downward curling and epinasty of the terminal leaf,followed by epinasty of most of the other leaves.These epinastic leaves then exhibit general chlorosis,which eventually leads to defoliation.If cotton plants are infected with a non-defoliating pathogen,the lower leaves exhibit interveinal chlorosis that leads to necrosis,but there is little or no epinasty and any dead leaves usually remain attached to the plant [4].The main factors affecting the virulence of Verticillium wilt in cotton are the V.dahliae pathotype and the inoculum density of the fungus[5].
To date,the most effective and feasible way to control Verticillium wilt disease is the development of cotton cultivars with resistance to the pathogen using conventional breeding and transgenic technologies [6–9].There are approximately 50 species in the Gossypium genus,of which four are cultivated,including two allotetraploids(Gossypium hirsutum and Gossypium barbadense)and two diploids(Gossypium herbaceum and Gossypium arboreum)[10,11].G.hirsutum,also known as upland cotton,is the most widely planted of the four cultivated Gossypium spp.,and has been the subject of most genetic studies and breeding efforts.It produces more than 95% of the annual cotton crop worldwide (National Cotton Council,http://www.cotton.org/,2006),but most of the commercial cultivars of the species are susceptible or only tolerant to Verticillium wilt.G.barbadense,another important cultivated species of cotton,is characterized by its extra-long-staple cotton compared to upland cotton.Of the four cultivated cotton species,G.barbadense is the most resistant to Verticillium wilt.For this reason,breeders have tried to introgress resistance gene(s) from G.barbadense to G.hirsutum.However,linkage drag between the resistance and undesired agronomic traits and distortion in segregation of the interspecific hybrid has severely hampered the exploitation of these lines.As a result,little progress has been made toward the selective breeding of cotton for resistance to Verticillium wilt,and the needs of the cotton industry are far from being achieved[2].
Quantitative trait loci (QTL)/genes resistant to Verticillium wilt have been detected in G.barbadense and G.hirsutum cultivars.A random amplified polymorphic DNA marker linked with a resistance gene at a distance of 12.4 cM was identified.This marker was associated with a phenotypic variance (PV) of 12.1% [12].Two QTL clusters with high contributions were detected on chromosome (Chr.) D7 and Chr.D9 by composite interval mapping [13].With the use of an F2population (from a cross between a G.barbadense cultivar and a G.hirsutum cultivar) and a single isolate of V.dahliae,three large-effect QTL(CM12,STS1,and BNL3147-2)conferring resistance to Verticillium wilt were detected on Chr.A11 [14].Several QTL showing resistance to the disease have been also detected in various studies[4,15,16].However,differences in markers,isolates,and developmental stages among these studies and the unavailability of chromosome tagging data make comparisons of results obtained from these studies difficult.
Chromosomal segment introgression lines (CSILs) carrying introgressed chromosomal segments in the same genetic background offer great advantages for studying the genetic functions of chromosomal segments.Moreover,CSILs are a unique system for mapping purposes,thanks to the marked reduction in confounding interactive effects between segregating loci in the genetic background,such as the ones that occur in other types of mapping populations [17].We developed a CSIL population using the cotton genetic standard G.hirsutum cv.TM-1 as the recipient parent and the long-staple cotton G.barbadense cv.Hai 7124 as the donor parent,and employed our 330 simple sequence repeat (SSR) anchored markers for molecular marker-assisted selection (MAS) in the BC5S1–4and BC4S1–3generations.The CSIL population comprised 174 lines containing 298 introgressed segments,of which 86 lines(49.4%)contained a single introgressed segment.The introgressed segments covered a total length of 2948.7 cM(with an average length of 16.7 cM),representing 83.3%of the cotton genome[18].In the present study,we used these CSILs to identity QTL affecting resistance to Verticillium wilt.Our major objectives were to conduct genome wide screening of chromosome regions containing resistance gene(s),identify the genetic mechanisms of tetraploid cotton resistance to Verticillium wilt,and find markers linked to QTL conferring resistance to multiple V.dahliae isolates during the seedling stage in order to facilitate improved cotton breeding programs.
2.Materials and methods
2.1.Pant materials
G.hirsutum cv.TM-1,the genetic standard upland cotton,was obtained from the Southern Plains Agricultural Research Center,USDA-ARS,College Station,Texas,U.S.A.[19].G.barbadense cv.Hai 7124,grown extensively in China,is the offspring of an individual plant selected during earlier studies of inherited resistance to V.dahliae in our laboratory[20,21].G.hirsutum cv.Junmian 1 is distributed widely in Xinjiang municipality and is highly sensitive to Verticillium wilt,and was selected as a control.One set of CSILs was developed using MAS in the genetic standard G.hirsutum cv.TM-1 background(the recipient parent) and the G.barbadense cv.Hai 7124 (the donor parent)which is resistant to Verticillium wilt.
2.2.Inoculation and phenotyping of CSILs in the greenhouse
Two defoliating V.dahliae isolates found commonly in the Yangtze River cotton-growing region of China,V991 and V07DF2,were selected to represent isolates with strong and extrastrong virulence.The defoliating isolate D8092,from the Yellow River cotton-growing region,was selected to represent isolates of intermediate virulence.V.dahliae isolates were grown on potato dextrose agar plates at 25 °C for 10–14 d.Inocula for experiments were prepared by spreading a conidial suspension on agar plates that were then incubated at 25 °C for 6–7 d.Conidia were then collected and diluted to 1 × 107cells mL-1.
The 166 CSILs were grown in paper cups of 7.3 × 5.1 × 8.3 cm in the greenhouse at Nanjing Agriculture University (Nanjing,China) from 2009 to 2011.These CSILs were planted in a randomized block design with two replicates.V.dahliae V991(in 2009),V07DF2 (in 2010) and D8092 (in 2011) were used to inoculate CSIL individuals (after the emergence of two true leaves) by watering with 15 mL of conidial suspension.Disease symptoms were scored at 25,30,and 35 days post inoculation (dpi),according to a standard National Grade Criteria for leaf disease symptoms at the seedling stage[22,23]as follows:grade 0,healthy with no disease symptoms;1,one or two cotyledons showing disease symptoms; 2,one true leaf showing disease symptoms or defoliated by disease;3,two true leaves showing disease symptoms or defoliated by disease;and 4,all leaves showing disease symptoms or killed by disease.Single plant numbers and grades were recorded at every time point,and the disease index (DI) and relative DI (RDI) of the tested canopy were calculated according to the following formulae[24]:
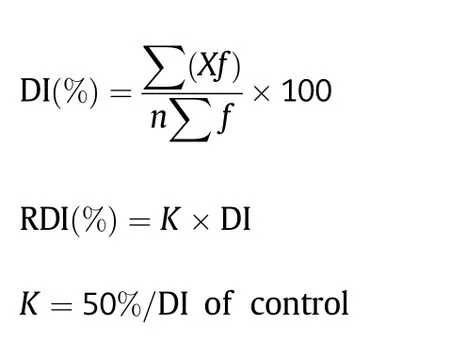
where X denotes the grade of disease severity according to the National Grade Criteria above,n is the value of the greatest severity among all tested canopies,and f is the number of plants in each grade.The RDI values were used to divide disease severity of the test canopies to Verticillium wilt into five grades:immunity,RDI = 0;high resistance,RDI <10.0%;resistance,RDI(%) = 10.1–20.0;tolerance,RDI(%) = 20.1–35.0;and susceptibility,RDI >35.0%.
2.3.Data analysis and QTL mapping
Trait means were calculated using SPSS 17.0 (SPSS,Chicago,Illinois,U.S.).Wang et al.[25,26]proposed a likelihood ratio test method based on stepwise regression (RSTEP-LRT) to detect QTL of non-idealized CSIL,because the t-test is unsuitable.QTL IciMapping 3.0(http://www.isbreeding.net/)was used to detect the additive effects of QTL and the epistatic QTL of nonidealized CSIL[25,26].A log-of-odds(LOD)score >3.0 was used to identify the additive effects of QTL.
The QTL nomenclature was adapted from the method established for rice [27].Thus,names start with “q” and this is followed by an abbreviation of the trait name,the name of the chromosome,and the number of the QTL affecting the trait on that chromosome.To identify resistance QTL from the resistant parent Hai7124 and pyramid different resistant QTL to breed cotton cultivars with broad-spectrum resistance,we defined QTL identified in this study as resistance or susceptibility QTL depending on whether the resistance-increasing alleles were from the resistance donor Hai 7124.

Table 1-Relative disease indices of parent plants and CSILs after challenge by three V.dahliae isolates.
3.Results
3.1.Resistance of CSILs inoculated with the three V.dahliae isolates in the greenhouse
The RDI of G.barbadense cv.Hai 7124 ranged from 12.22% for V.dahliae D8092 to 17.51% for V.dahliae V07DF2 (Table 1),indicating that this cultivar is resistant to these pathogen isolates.The RDI of G.hirsutum cv.TM-1 ranged from 33.54%for V.dahliae D8092 to 40.81% for V.dahliae V07DF2 (Table 1),suggesting that some resistance or tolerance genes are present in this cultivar.The mean RDIs of the CSILs were 31.35%(9.09–49.68%) for V.dahliae V991,34.46% (19.23–53.54%) for V.dahliae V07DF2,and 31.36% (7.83–49.63%) for V.dahliae D8092.Although the average RDIs of the CSILs were closer to the values observed for G.hirsutum cv.TM-1 than to those of G.barbadense cv.Hai 7124,there were individual CSILs with RDIs that fell on both sides of the values observed for the V.dahliae V991 and D8092 isolates(Table 1).
According to the RDIs,no CSIL line was immune to all three V.dahliae isolates(Fig.1).Only one CSIL showed high resistance to both the V.dahliae V991 and D8092 isolates.Respectively 16,3,and 11 CSILs were resistant;73,78,and 79 were tolerant;and 75,84,and 74 were susceptible to V.dahliae V991,V07DF2 and D8092 isolates.These results indicated that fewer than 10% of the CSILs showed resistance to Verticillium wilt.
3.2.Mapping of QTL associated with resistance to three defoliating V.dahliae isolates
A total of 42 QTL were identified and mapped on 18 chromosomes with LOD values ranging from 3.00 to 9.29(Tables 2 and 3; Fig.2).Of these QTL,23 showed resistanceincreasing effects and the remaining 19 showed susceptibilityincreasing effects in response to the three V.dahliae isolates.Interestingly,most of QTL responded to different isolates.
3.2.1.QTL associated with resistance to V.dahliae V991
Based on RDIs obtained in the greenhouse experiment in 2009,10 QTL showed resistance to V.dahliae V991 and were mapped on eight chromosomes,Chrs.A3,A7,A8,A9,A13,D4,D5,and D12,of which Chr.A3 and Chr.A7 contained two QTL each.The additive effect on increasing resistance to V.dahliae V991 ranged from-11.04 to-7.59 for a single QTL,and the phenotypic variation explained ranged from 1.7% to 3.7%.Eight susceptibility QTL were detected on Chrs.A1,A3,A5,A12,D1,D2,and D3.Among these eight,two were located on Chr.D1.The additive effect of the decrease in G.hirsutum cv.TM-1 resistance to V.dahliae V991 ranged from 6.80 to 9.12,indicating that some resistance and/or tolerance QTL presenting G.hirsutum cv.TM-1 were substituted by susceptible chromosome segments from G.barbadense cv.Hai 7124,resulting in greater susceptibility of these CSILs than of G.hirsutum cv.TM-1.The percentage of PV ranged from 1.6 to 2.8%.
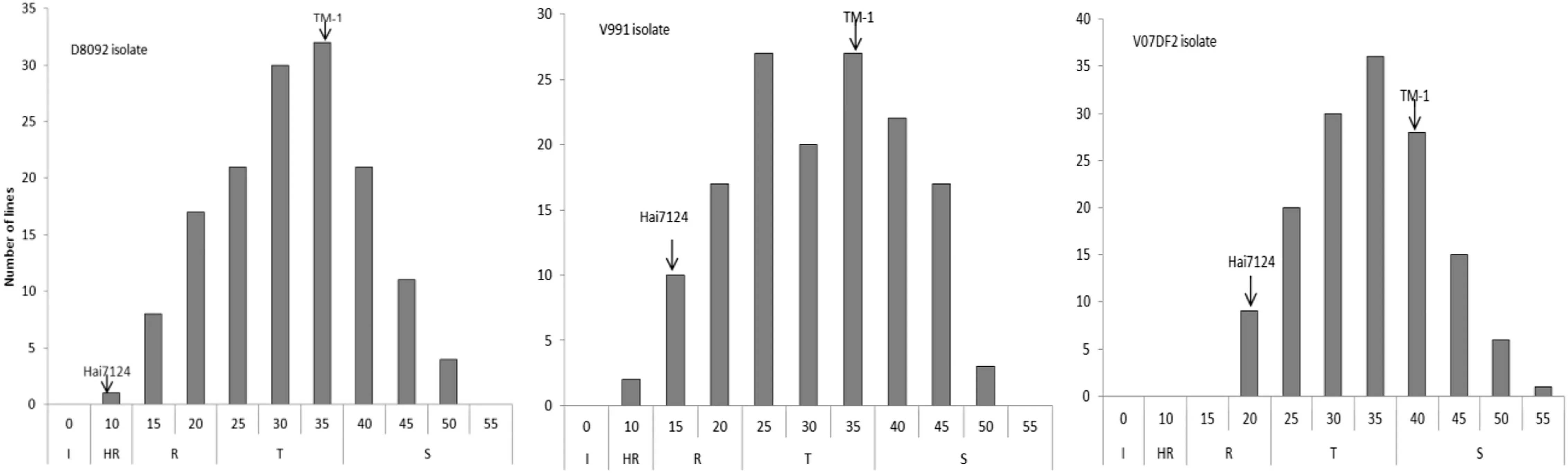
Fig.1-Distribution of relative disease indices (RDIs) of leaf traits caused by three V.dahliae isolates in greenhouse experiments at Nanjing Agricultural University (China).I: immunity to Verticillium wilt (RDI = 0); HR: high resistance to Verticillium wilt (RDI ≤ 10.0); R: resistance to Verticillium wilt (10.0 < RDI ≤ 20.0); T: tolerance to Verticillium wilt(20.0 < RDI ≤ 35.0); S: susceptibility to Verticillium wilt (RDI > 35.00).
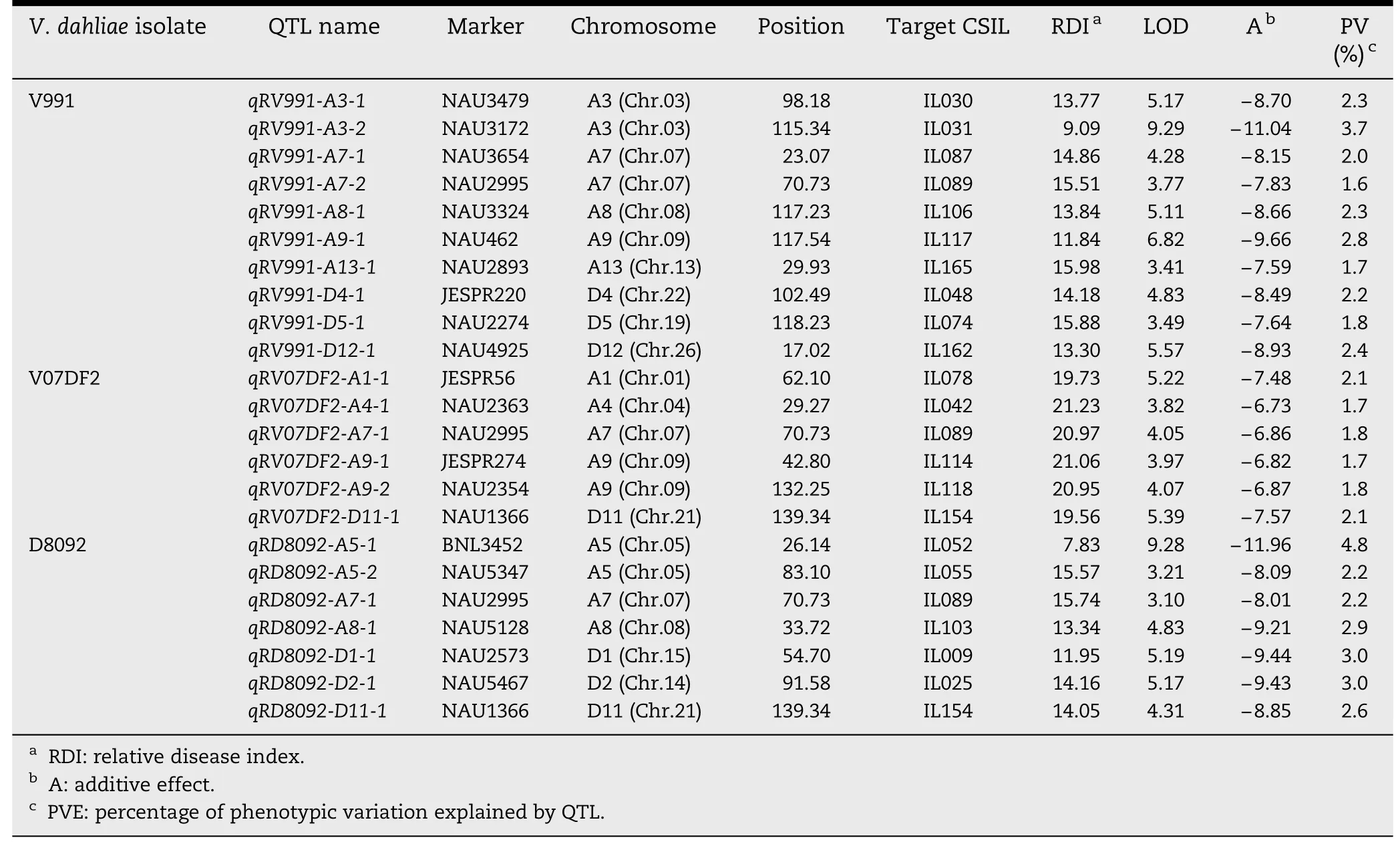
Table 2-QTL with resistance-increasing effect in response to three V.dahliae isolates in the CSIL population.
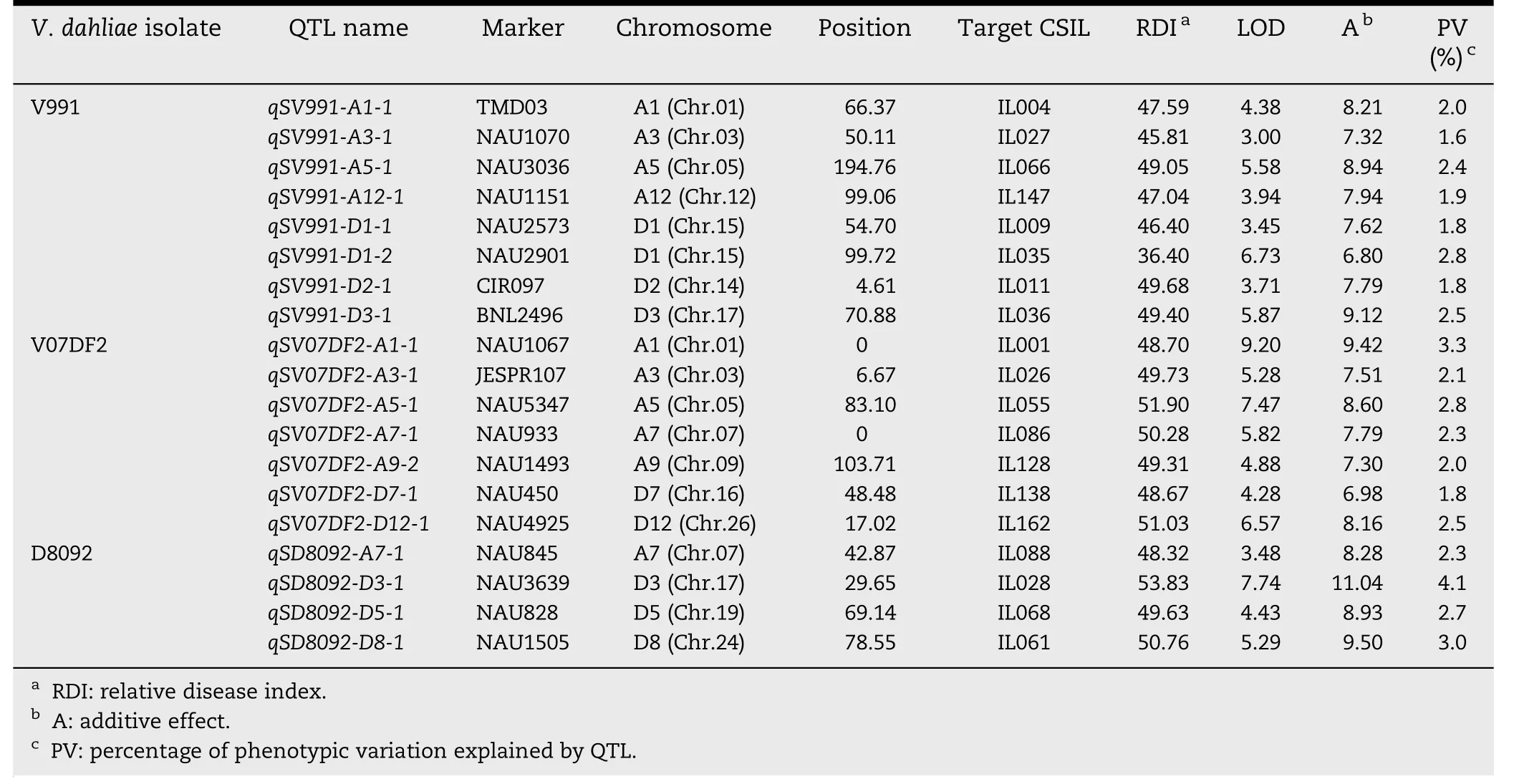
Table 3-QTL with resistance-decreasing effect in response to three V.dahliae isolates in the CSIL population.
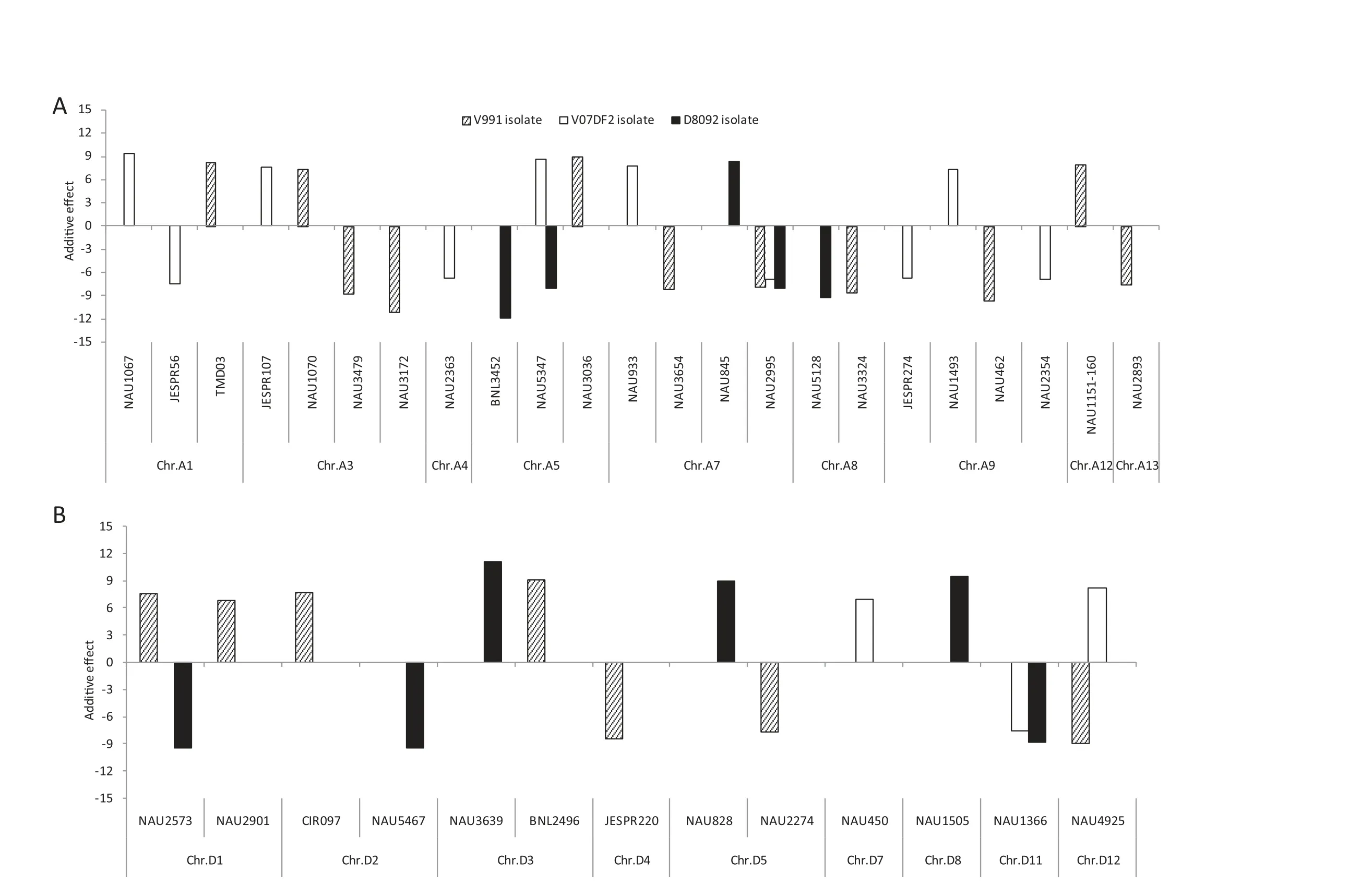
Fig.2-Additive effects and positions of QTL for resistance or susceptibility to three defoliating V.dahliae isolates that cause Verticillium wilt.A positive effect indicates that the allele in G.barbadense cv.Hai 7124 decreases resistance to Verticillium wilt,while a negative effect indicates that the allele in G.barbadense cv.Hai 7124 increases resistance.
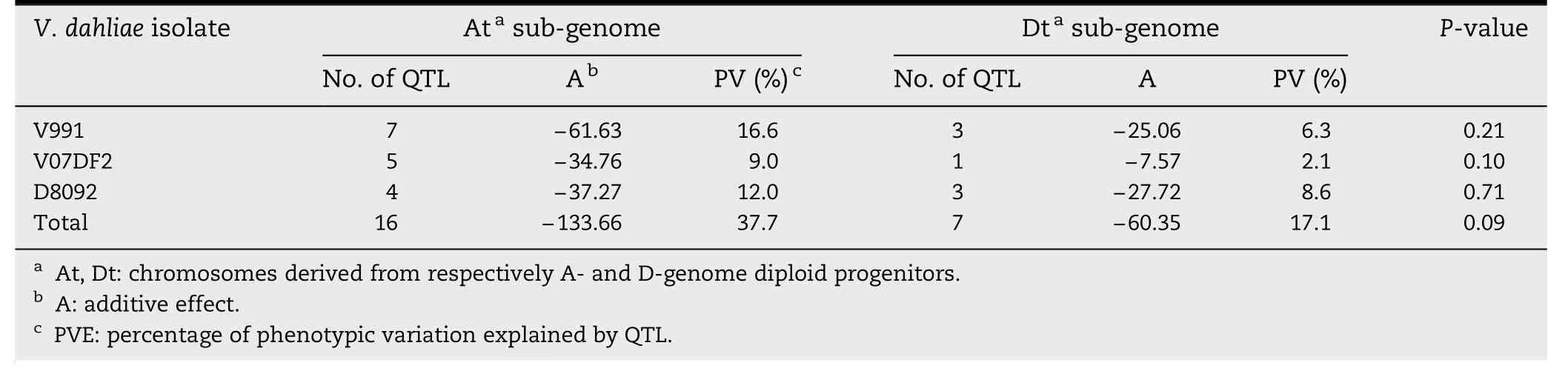
Table 4-Total additive effects,percentage of PV,and chi-square tests for the distribution of QTL with resistance-increasing effect in the At and Dt subgenomes.
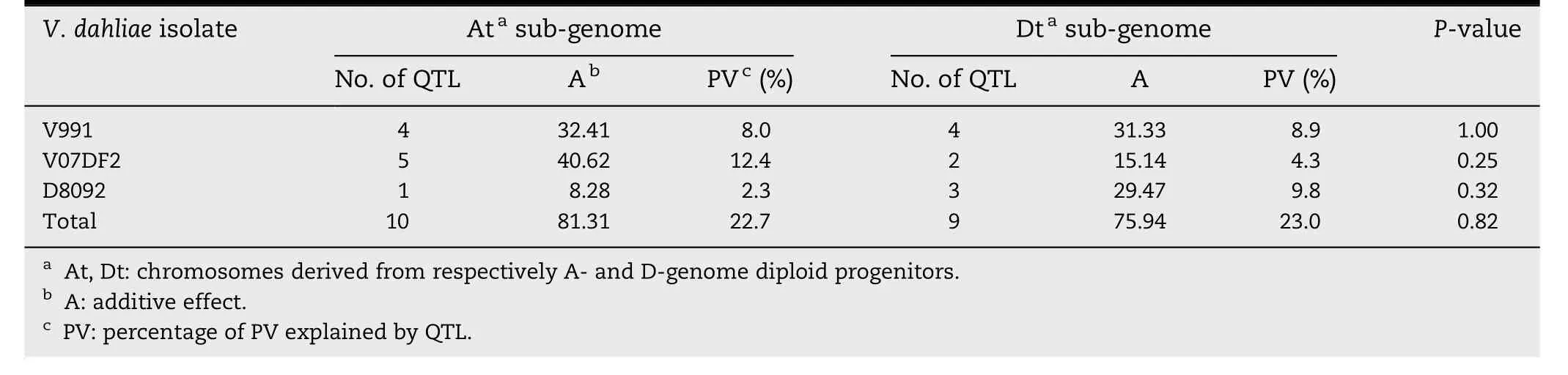
Table 5-Total additive effects,percentage of PV and chi-square tests for the distribution of QTL with resistance-decreasing effect in the At and Dt sub-genomes.
3.2.2.QTL associated with resistance to V.dahliae V07DF2
Six QTL for resistance to V.dahliae V07DF2 were detected in the greenhouse experiments in 2010.Based on the RDIs of the CSILs,these six QTL were distributed on five chromosomes: Chrs.A1,A4,A7,A9,and D11.Among six QTL,two QTL were located on Chr.A9.The additive effect of the increase in resistance to V.dahliae V07DF2 ranged from-7.57 to-6.43 for the resistance QTL and the percentage of PV ranged from 1.7 to 2.1%.In addition,seven susceptibility QTL were detected on Chrs.A1,A3,A5,A7,A9,D7,and D12,based on the RDIs of the CSIL population.The additive effect of the decrease in G.hirsutum cv.TM-1 resistance to the V.dahliae V07DF2 isolate ranged from 6.98 to 9.42 and the percentage of PV ranged from 1.8 to 3.3%.
3.2.3.QTL associated with resistance to V.dahliae D8092
Seven QTL for resistance to V.dahliae D8092 were detected in the greenhouse experiments in 2011.Based on RDIs of the CSILs,these QTL were found to be distributed on six chromosomes,Chrs.A5,A7,A8,D1,D2 and D11.Among the seven,two were located on Chr.A5.The additive effect of the increase in resistance to V.dahliae D8092 ranged from-11.96 to-8.01 for the resistance QTL and the percentage of PV ranged from 2.2 to-4.8%.Four susceptibility QTL were detected on Chrs.A7,D3,D5 and D8 based on the RDIs of the CSILs.The additive effect of the decrease in G.hirsutum cv.TM-1 resistance to V.dahliae D8092 ranged from 8.28 to 11.04 and the percentage of PV ranged from 2.3 to 4.1%.
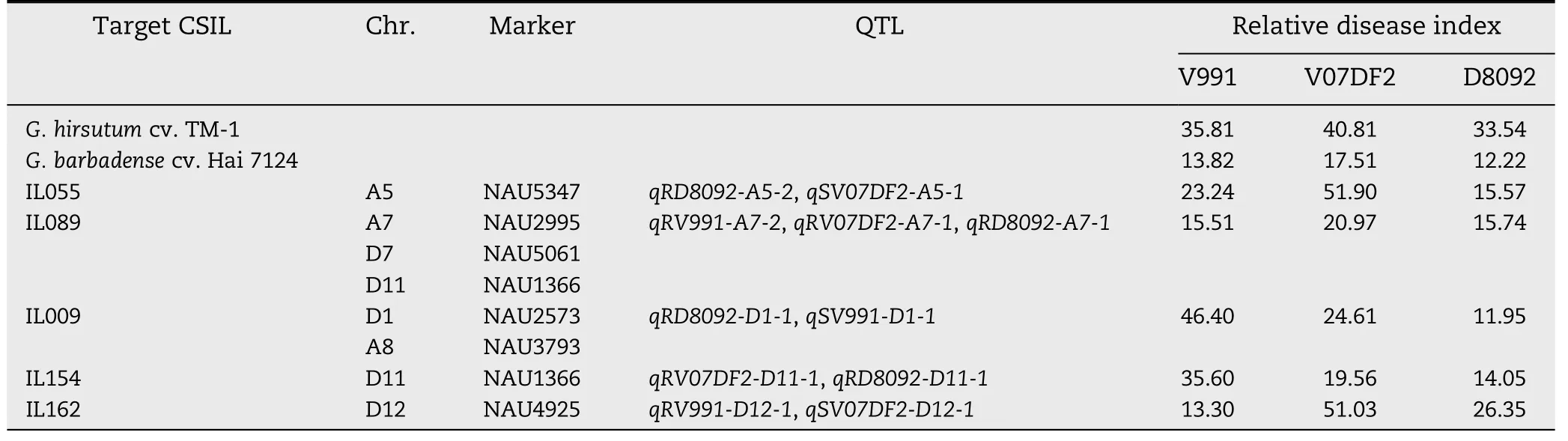
Table 6-Relative disease indices and markers of QTL detected in two or three cotton isolates.
3.2.4.Subgenomic distributions and effects of additive QTL
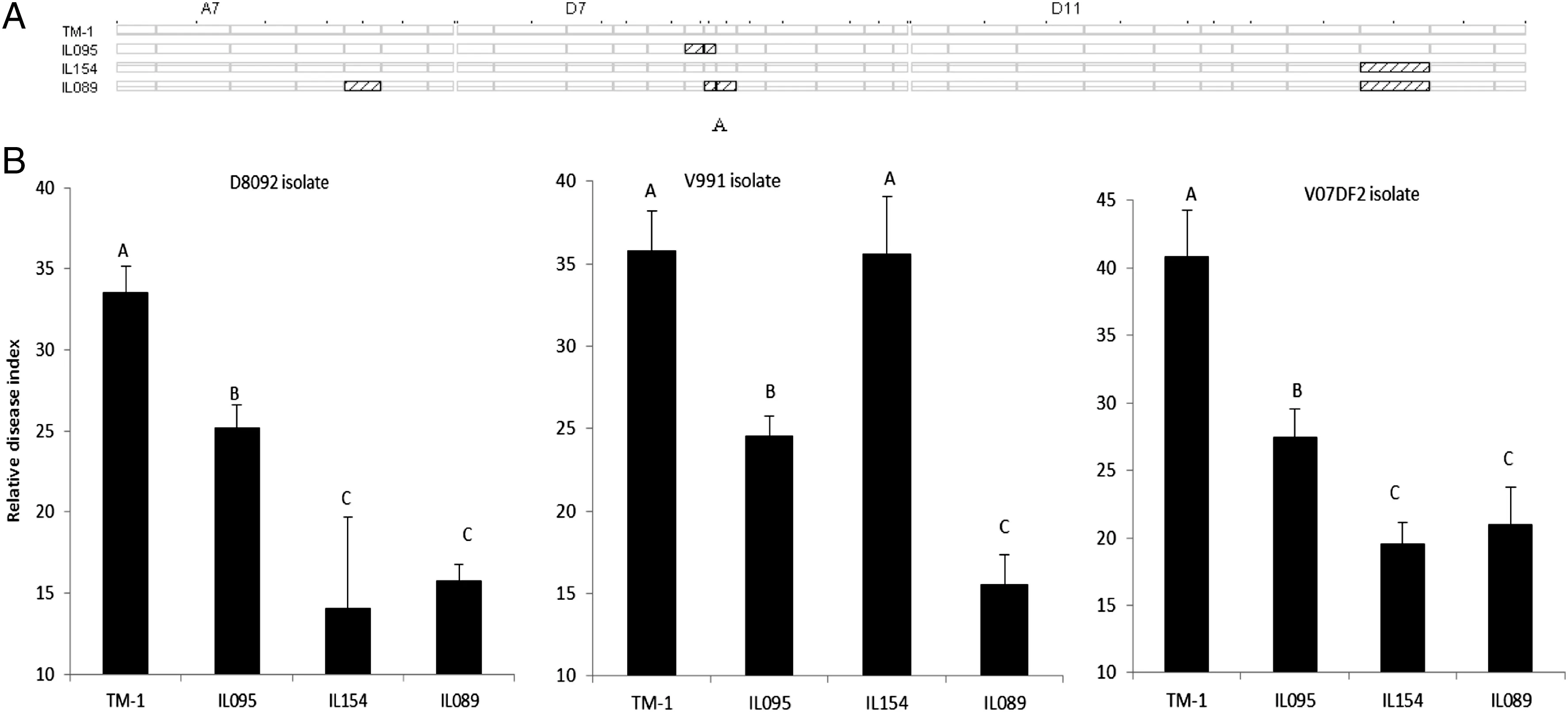
Fig.3-Graphical genotypes (A) and phenotypic performances (B) of G.hirsutumcv.TM-1 and three CSILs.Bars labeled with different letters are significantly different by multiple comparisons (Tukey's test).
There were seven QTL for resistance to V.dahliae V991 on the At subgenome,which was more than the three found on the Dt subgenome (Table 4).However,there was no significant difference between the numbers of resistance QTL on At and Dt subgenome chromosomes(P = 0.21)by chi-square test(Table 4).The total additive effect and PV of the V.dahliae V991 resistance QTL on the At subgenome chromosomes were-61.63 and 16.6%,respectively,and the total additive effect and PV of those on the Dt subgenome were-25.06 and 6.3%,respectively.The values for the other two V.dahliae isolates were similar to those obtained for the V991 isolate.These results indicate that the resistance effects of the QTL on the At subgenome are greater than those of the QTL on the Dt subgenome.
There were 10 QTL for susceptibility to all of the V.dahliae isolates in the At subgenome and nine in the Dt subgenome(Table 5).There was no significant difference in the numbers of susceptibility QTL located on At and Dt subgenome chromosomes(P = 0.82)(Table 5).The total additive effect and PV of the QTL for susceptibility to V.dahliae V991 on At subgenome chromosomes were 81.31% and 22.7%,respectively,and those of the susceptibility QTL on the Dt sub-genome was 75.94 and 23.0%,respectively.
3.3.Interaction between resistance QTL and V.dahliae isolates
The RDIs of five CSILs and G.hirsutum cv.TM-1 corresponding to 11 QTL are given in Table 6.Based on the RDIs,IL055 contained one introgressed segment on Chr.A5 and was resistant to V.dahliae D8092,tolerant to V.dahliae V991,but susceptible to V.dahliae V07DF2; IL162 contained one introgressed segment on Chr.D12 and was resistant to V991,tolerant to D8092,but susceptible to V07DF2; IL154 contained one introgressed segment on Chr.D11 and was resistant to V07DF2 and D8092 but susceptible to V991;IL009 contained two introgressed segments on Chrs.A8 and D1 and was resistant to D8092,tolerant to V07DF2,but susceptible to V991; and IL089 contained three introgressed segments on Chrs.A7,D7,and D11 and was resistant to D8092 and V991 and tolerant to V07DF2.Clearly,the CSILs showed variable resistance to each of the different V.dahliae isolates,suggesting that there might exist an additional effect between each resistance QTL and the different fungal strains.
The genotypes and resistance performances of three CSILs and G.hirsutum cv.TM-1 (recipient parent) are illustrated in Fig.3.IL095 and IL154 each contained one introgressed segment,located on Chrs.D7 and D11,respectively; whereas IL089 contained three introgressed segments located on Chrs.A7,D7 and D11,respectively (Fig.3-A).The two introgressed segments in IL089 on Chrs.D7 and D11 overlapped with the introgressed segments in IL095 and IL154.No resistance QTL was detected in IL095,but two QTL for resistance to V.dahliae D8092 and V07DF2 isolates were detected in IL154,and three QTL for resistance to all three V.dahliae isolates were detected in IL089.These three CSILs(IL095,IL154,and IL089)exhibited lower RDIs in response to the V.dahliae D8092 and V07DF2 isolates than G.hirsutum cv.TM-1(Fig.3-B).The RDIs of IL089 were between the values of IL095 and IL154,but the RDIs of IL809 did not differ significantly from those of IL154 to V.dahliae D8092 and V07DF2.Furthermore,IL095 and IL089 exhibited lower RDIs than G.hirsutum cv.TM-1,and IL154 exhibited the same RDI as G.hirsutum cv.TM-1 to V.dahliae V991.The RDI of IL089 was significantly lower than those of IL095 and IL154 to V.dahliae V991.These results support the presence of resistance QTL and further suggest the presence of additive effects of QTL for resistance to Verticillium wilt.
4.Discussion
4.1.Complex inheritance of resistance to V.dahliae in cotton
Genetic studies of Verticillium wilt resistance in cotton have reported different patterns of inheritance.Inheritance can be classified into two types according to the genetic basis of the resistance observed: major gene [9,20,28] and/or polygene[29–31].Owing to this genetic complexity,our understanding of disease resistance mechanisms remains limited.There are many difficulties encountered in the study of resistance to Verticillium wilt in cotton,including uncontrollable environmental influences on the development of the disease and minor background genetic effects.G.barbadense cv.Hai 7124 is used broadly in China as a resistant parent to develop cultivars with resistance to Verticillium wilt,but its mechanism of resistance to this pathogen is not well characterized.In previous greenhouse-based studies,resistance appeared to be due to qualitative inheritance,given that a 3:1 (resistant:susceptible) segregation was observed (provided that grades 0,1,and 2 were classified as resistant and grades 3 and 4 as susceptible) [4,9,20,28–31].In the present study,21 of the 23 resistance QTL conferred resistance to only one of the V.dahliae isolates assessed.However,fewer than 10% of the CSILs were resistant to Verticillium wilt in the greenhouse,and the RDIs of CSILs in the field were greater than observed in the greenhouse experiments.These results suggest that resistance to different V.dahliae isolates is controlled by distinct single genes and that interaction between resistance QTL or genes and fungal strains occurs.
Some progress has been achieved in mapping QTL for cotton resistance to Verticillium wilt[12,13,15,16].In the present study,a total of 42 QTL,including 23 resistant and 19 susceptible QTL,were identified and mapped on 18 chromosomes.Ten of the QTL were associated with resistance to V.dahliae V991,six to V.dahliae V07DF2,and seven to V.dahliae D8092.These QTL had high additive effects.We have detected 20 resistance QTL distributed on Chrs.A5,A7,A8,A9,D4,D5,and D11 using an F2and a BC1S2population derived from the cross between G.barbadense cv.Hai 7124 and G.hirsutum cv.Junmian 1 [4].In that study,15 resistance QTL were located on the same chromosomes using a CSIL population derived from the cross between G.barbadense cv.Hai 7124 and G.hirsutum cv.TM1,and many more resistance QTL identified were novel loci.Given that each of the CSILs used contained one and/or a few substituted segments from the donor G.hirsutum cv.TM-1,all the genetic variation between a CSIL and G.hirsutum cv.TM-1 is associated with the substituted segment(s).This circumstance minimizes background genetic effects and allows more reliable QTL detection and PV estimation.These results showed that CSIL populations are highly effective for studying resistance to Verticillium wilt.
In this study,four resistance QTL were found to be located on Chr.A7,with a further three on Chr.A9.Jiang et al.[13]mapped four QTL on Chr.D7 and four on Chr.D9 for V.dahliae BP2; five QTL on Chr.D7 and nine on Chr.D9 for V.dahliae VD8;four QTL on Chr.D7 and five on Chr.D9 for V.dahliae T9;and three QTL on Chr.D7 and seven on Chr.D7 for mixed pathogens in a F2:3population derived from the cross between G.hirsutum cv.60182 and G.hirsutum cv.Junmian 1.The QTL-mapping results revealed that QTL clusters with high additive effects were located on Chrs.A7 and A9.Bolek et al.[14]also detected three markers (CM12,STS1,and BNL3147-2) on Chr.A11 that conditioned resistance to Verticillium wilt in G.barbadense cv.Pima S-7.In the present study,one QTL for resistance to two defoliating V.dahliae isolates was found near the SSR marker BNL3147 on Chr.D11.As Chr.A11 and D11 area pair of homoeologous chromosomes,it is clear that these two homoeologous groups harbor resistance genes,and should be carefully considered in future Verticillium wilt-resistance breeding.
4.2.Pyramiding resistance QTL to breed cotton cultivars with broad-spectrum resistance
Verticillium wilt is a destructive disease with global consequences for cotton production.Breeding broad-spectrum cotton cultivars with resistance to this disease and others is considered to be one of the most effective means for reducing crop losses.Conventionally,breeding for disease resistance in cotton has involved selecting resistant individuals in the nursery or field from among plants suffering from serious disease.However,this approach is unsuitable for generating plants with resistance to Verticillium wilt [2].Furthermore,no significant breakthroughs in the breeding of resistance to Verticillium wilt have been achieved for a considerable time,owing largely to a lack of germplasm known to be immune or highly resistant to this fungal pathogen.Such problems can be caused by a lack of understanding of the pathogenetic differentiation of the pathogen,or the inheritance and mechanisms of resistance to Verticillium wilt,which has made breeding cotton cultivars resistant to this fungus very difficult.
The development and evaluation of CSILs is of great importance in molecular breeding,and such stocks have been employed successfully in rice,where many CSILs have been developed [32].Once favorable alleles in QTL/genes have been identified on introgressed segments,the CSILs become candidates for selection in subsequent molecular breeding strategies[26].In this present study,we found a broad-spectrum resistant CSIL,IL089,which carried three introgressed segments located on Chrs.A7,D7,and D11.The segment on Chr.D7 conferred tolerance to the three V.dahliae isolates used in this study.The segment on Chr.D11 was associated with resistance to the V.dahliae V07DF2 and D8092 isolates.When the two segments were combined in IL089,it was resistant to all three V.dahliae isolates.Combining different resistance QTL could allow breeding broad-spectrum resistant cultivars.For example,we could pyramid the following resistance QTL: qRV991-A3-2(resistant to V.dahliae V991),qRV07DF2-D11-1 (resistant to V.dahliae V07DF2) and qRD8092-A5-1 (resistant to V.dahliae D8092).These three high-resistance QTL could be combined to breed a cotton cultivar that exhibits broad-spectrum resistance to Verticillium wilt,using a modified backcrossing pyramiding breeding scheme with MAS.Such MAS breeding experiments are being conducted presently in our laboratory.
4.3.Effect of At/Dt sub-genomes on resistance QTL
Two cultivated tetraploid cotton species,G.hirsutum(AD)1and G.barbadense (AD)2,contain the A and D subgenomes.The effects of the two subgenomes on yield and fiber quality are important research objectives for the production of tetraploid cultivars.A meta-analysis revealed that cotton fiber QTL are enriched in the Dt subgenome [33],but a more recent study showed that the subgenomic distribution of fiber qualities is equally divided between the chromosomes of the two subgenomes[34].In the present study,the number of additive QTL detected in the At sub-genome was approximately equal to that found in the Dt sub-genome in the same CSIL population[18].This is the first report to consider the effect of the two subgenomes on resistance to Verticillium wilt.In the present study,we tried to analyze the effect of the two subgenomes on host resistance to Verticillium wilt.Eighteen QTL associated with resistance/susceptibility to one of the V.dahliae isolates assessed were detected and,of these,16 QTL were located in the At subgenome and seven in the Dt sub-genome.A chi-square test of QTL distribution on the At/Dt sub-genomes showed no significant difference in the distribution of the QTL between these subgenomes.Similar results were obtained for the other two V.dahliae isolates.These results suggest that the effects of the two subgenomes on the numbers of resistance and susceptibility QTL were insignificant.The total additive effect of resistance or susceptibility QTL on the At sub-genome was negative,but the total effect on the Dt subgenome was positive.This result is consistent with the total percentage of PV of resistance QTL in the At subgenome being greater than that in the Dt sub-genome,except for the total percentage of PV of QTL associated with resistance to V.dahliae D8092.These results show that the effect of the At subgenome on resistance to Verticillium wilt is greater than that of the Dt subgenome.
This study was supported by the National Natural Science Foundation of China(30730067 and 31171590),the Philosophy Doctoral Fund Program of Xinjiang Bingtuan Group(2010JC01),and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
[1] G.Sunilkumar,L.M.Campbell,L.Puckhaber,R.D.Stipanovic,K.S.Rathore,Engineering cotton seed for use in human nutrition by tissue-specific reduction of toxic gossypol,Proc.Natl.Acad.Sci.U.S.A.103 (2006) 18054–18059.
[2] Y.F.Cai,X.H.He,J.C.Mo,Q.Sun,J.P.Yang,J.G.Liu,Molecular research and genetic engineering of resistance to Verticillium wilt in cotton: a review,Afr.J.Biotechnol.8(2009)7363–7372.
[3] L.Gan,J.D.Lu,P.H.Wang,The relationship between glycoprotein toxin secreted from Verticillium dahliae of cotton and its pathogenicity,Sci.Agric.Sin.28(1995)58–65(in Chinese with English abstract).
[4] C.Yang,W.Z.Guo,G.Y.Li,F.Gao,S.S.Lin,T.Z.Zhang,QTLs mapping for Verticillium wilt resistance at seedling and maturity stages in Gossypium barbadense L.Plant Sci.174(2008)290–298.
[5] M.E.Göre,Ö.K.Caner,N.AltIn,M.H.AydIn,O.Erdoğan,F.Filizer,A.Büyükdöğerlioğlu,Evaluation of cotton cultivars for resistance to pathotypes of Verticillium dahliae,Crop.Prot.3(2009) 215–219.
[6] D.T.Bowman,Public cotton breeders –do we need them? J.Cotton Sci.3 (1999) 139–152.
[7] B.H.Zhang,F.Liu,C.B.Yao,K.B.Wang,Recent progress in cotton biotechnology and genetic engineering in China,Curr.Sci.79(2000) 37–44.
[8] G.L.Jian,C.Ma,C.L.Zheng,Y.F.Zou,Advances in cotton breeding for resistance to Fusarium and Verticillium wilt in the last fifty years in China,Agric.Sci.China 2(2003)280–288(in Chinese with English abstract).
[9] M.Mert,S.Kurt,O.Gencer,Y.Akiscan,K.Boyaci,F.M.Tok,Inheritance of resistance to Verticillium wilt (Verticillium dahliae) in cotton (Gossypium hirsutum L.),Plant Breed.124(2005) 102–104.
[10] J.F.Wendel,Genome evolution in polyploids,Plant Mol.Biol.42 (2000) 225–249.
[11] H.B.Zhang,Y.Li,B.H.Wang,P.W.Chee,Recent advances in cotton genomics,Int.J.Plant Genomics (2008),http://dx.doi.org/10.1155/2008/742304.
[12] W.P.Fang,S.M.Xu,Y.T.Sun,Z.J.Tang,J.D.Wang,The RAPD marker lined with Verticillium wilt resistance in cotton,J.Henan Agric.Sci.9(2001) 11–13(in Chinese with English abstract).
[13] F.Jiang,J.Zhao,L.Zhou,W.Z.Guo,T.Z.Zhang,Molecular mapping of Verticillium wilt resistance QTL clustered on chromosomes D7 and D9 in upland cotton,Sci.China C.Life Sci.52(2009) 872–884 (in Chinese with English abstract).
[14] Y.Bolek,K.M.El-Zik,A.E.Pepper,A.A.Bell,C.W.Magill,P.M.Thaxton,U.K.Reddy,Mapping of Verticillium wilt resistance genes in cotton,Plant Sci.168 (2005) 1581–1590.
[15] H.M.Wang,X.L.Zhang,D.H.He,Z.X.Lin,Y.C.Nie,Y.H.Li,W.Chen,DetectionofDNAmarkersassociatedwithresistanceto Verticilliumdahliae in cotton,ActaPhytopathol.Sin.35(2005)333–339(inChinesewithEnglishabstract).
[16] Y.Q.Gao,Y.C.Nie,X.L.Zhang,QTL mapping of genes resistant to Verticillium wilt in cotton,Cotton Sci.15(2003)73–78(in Chinese with English abstract).
[17] M.Ashikari,M.Matsuoka,Identification,isolation and pyramiding of quantitative trait loci for rice breeding,Trends Plant Sci.11(2006) 344–350.
[18] P.Wang,Y.J.Zhu,X.L.Song,Z.B.Cao,Y.Z.Ding,B.L.Liu,X.F.Zhu,S.Wang,W.Z.Guo,T.Z.Zhang,Genetic dissection of long staple fiber qualities in Gossypium barbadense using interspecific chromosome segment introgression lines,Theor.Appl.Genet.124 (2012) 1415–1428.
[19] R.J.Kohel,C.F.Lewis,T.R.Richmond,Texas marker-1:description of a genetic standard for Gossypium hirsutum L.Crop Sci.10(1970) 670–671.
[20] J.J.Pan,T.Z.Zhang,B.K.Kuai,X.P.Guo,M.Wang,Studies on the inheritance of resistance to Verticillium dahliae in cotton,J.Nanjing Agric.Univ.17 (1994) 8–18(in Chinese with English abstract).
[21] J.Zhang,W.Z.Guo,T.Z.Zhang,Molecular linkage map of allotetraploid cotton (Gossypium hirsutum L.×Gossypium barbadense L.) with a haploid population,Theor.Appl.Genet.105 (2002) 1166–1174.
[22] G.L.Jian,W.J.Sun,C.Ma,A new method for identifying the Verticillium wilt resistance: sprinkle the strain suspension on root in non bottom plastic film bottle,Acta Gossypii Sin.2(2001) 67–69(in Chinese with English abstract).
[23] Z.B.Wu,J.Li,C.D.Feng,J.F.Zhang,Studies on the identification techniques of cotton resistance to Verticillium wilt,Hubei Agric.Sci.5(1996)16–19(in Chinese with English abstract).
[24] S.Z.Li,P.Ma,H.C.Huang,X.H.Chen,Statistical basis for determining Verticillium wilt resistance of cotton cultivar/line according to relative disease index,Acta Gossypii Sin.2003(15) (2003) 344–347 (in Chinese with English abstract).
[25] J.Wang,X.Wan,J.Crossa,J.Crouch,J.Weng,H.Zhai,J.Wan,QTL mapping of grain length in rice (Oryza sativa L.) using chromosome segment substitution lines,Genet.Res.88(2006) 93–104.
[26] J.Wang,X.Wan,H.Li,W.Pfeiffer,J.Crouch,J.Wan,Application of identified QTL-marker associations in rice quality improvement through a design-breeding approach,Theor.Appl.Genet.115 (2007) 87–100.
[27] S.R.McCouch,Y.G.Cho,P.E.Yano,M.Blinstrub,H.Morishima,T.Kinoshita,Report on QTL nomenclature,Rice Genet.Newslett.14(1997) 11–13.
[28] J.R.Barrow,Heterozygosity in inheritance of Verticillium wilt tolerance in cotton,Phytopathology 60(1970) 301–303.
[29] L.M.Verhalen,L.A.Brinkerhoff,K.C.Fun,W.C.Morrison,A quantitative genetic study of Verticillium wilt resistance among selected lines of upland cotton,Crop Sci.11(1971)407–412.
[30] C.L.Roberts,G.Staten,Heritability of Verticillium wilt tolerance in crosses of American upland cotton,Crop Sci.12(1972)63–66.
[31] R.Zhen,X.F.Wang,Z.Y.Ma,G.Y.Zhang,X.Wang,A SSR marker linked with the gene of Verticillium wilt resistance in Gossypium barbadense,Cotton Sci.18(2006) 269–272(in Chinese with English abstract).
[32] J.M.Wan,Perspectives of molecular design breeding in crops,Acta Agron.Sin.32(2006) 455–462 (in Chinese with English abstract).
[33] J.K.Rong,F.A.Feltus,V.N.Waghmare,G.J.Pierce,P.W.Chee,X.Draye,Y.Saranga,R.J.Wright,T.A.Wilkins,O.L.May,C.W.Smith,J.R.Gannaway,J.F.Wendel,A.H.Paterson,Meta-analysis of polyploid cotton QTL shows unequal contributions of subgenomes to a complex network of genes and gene clusters implicated in lint fiber development,Genetics 176(2007)2577–2588.
[34] J.M.Lacape,D.Llewellyn,J.Jacobs,T.Arioli,D.Becker,S.Calhoun,Y.Al-Ghazi,S.M.Liu,O.Palaï,S.Georges,M.Giband,P.Augusto,V.Barroso,M.Claverie,G.Gawryziak,J.Jean,M.Vialle,C.Viot,Meta-analysis of cotton fiber quality QTLs across diverse environments in a Gossypium hirsutum × G.barbadense RIL population,BMC Plant Biol.10 (2010) 132.
- The Crop Journal的其它文章
- An expedited method for isolation of DNA for PCR from Magnaporthe oryzae stored on filter paper
- Morphological,cytological and molecular analyses of a synthetic hexaploid derived from an interspecific hybrid between Gossypium hirsutum and Gossypium anomalum
- The impacts of conservation agriculture on crop yield in China depend on specific practices,crops and cropping regions
- Exp2 polymorphisms associated with variation for fiber quality properties in cotton(Gossypium spp.)
- Isolation and characterization of a novel wall-associated kinase gene TaWAK5 in wheat(Triticum aestivum)
- Effect of subsoil tillage depth on nutrient accumulation,root distribution,and grain yield in spring maize

