Molecular approaches unravel the mechanism of acid soil tolerance in plants
Mio Bin,Meixue Zhou,Dongf Sun,Chengdo Li*
aTasmanian Institute of Agriculture,University of Tasmania,P.O.Box 46,Kings Meadows,Tas 7249,Australia
bCollege of Plant Science and Technology,Huazhong Agricultural University,Wuhan,Hubei 430070,China
cDepartment of Agriculture&Food WA,3 Baron-Hay Court,South Perth,WA 6155,Australia
Molecular approaches unravel the mechanism of acid soil tolerance in plants
Miao Biana,b,c,Meixue Zhoua,Dongfa Sunb,Chengdao Lic,*
aTasmanian Institute of Agriculture,University of Tasmania,P.O.Box 46,Kings Meadows,Tas 7249,Australia
bCollege of Plant Science and Technology,Huazhong Agricultural University,Wuhan,Hubei 430070,China
cDepartment of Agriculture&Food WA,3 Baron-Hay Court,South Perth,WA 6155,Australia
A R T I C L E I N F O
Article history:
Received 13 June 2013
Received in revised form 16 July 2013
Accepted 12 August 2013
Available online 27 August 2013
Soil acidity
Acid soil is a worldwide problem to plant production.Acid toxicity is mainly caused by a lack of essential nutrients in the soil and excessive toxic metals in the plant root zone.Of the toxic metals,aluminum(Al)is the most prevalent and most toxic.Plant species have evolved to variable levels of tolerance to aluminum enabling breeding of high Al-tolerant cultivars. Physiological and molecular approaches have revealed some mechanisms of Al toxicity in higher plants.Mechanisms of plant tolerance to Al stress include:1)exclusion of Al from the root tips,and 2)absorbance,but tolerance of Al in root cells.Organic acid exudation to chelate Al is a feature shared by many higher plants.The future challenge for Al tolerance studies is the identification of novel tolerance mechanisms and the combination of different mechanisms to achieve higher tolerance.Molecular approaches have led to significant progress in explaining mechanisms and detection of genes responsible for Al tolerance. Gene-specific molecular markers offer better options for marker-assisted selection in breeding programs than linked marker strategies.This paper mainly focuses on recent progress in the use of molecular approaches in Al tolerance research.
Crown Copyright©2013 Production and hosting by Elsevier B.V.on behalf of Crop Science Society of China and Institute of Crop Science,CAAS.All rights reserved.
Contents
1.Introduction..........................................................92
2.Acid soil and its toxic effects.................................................92
2.1.Acid soil and its distributions.............................................92
2.2.Causes of soil acidity.................................................92
2.3.pH level and acid soil toxicity.............................................94
2.4.Effects of aluminum toxicity on plant growth....................................94
2.5.Solutions to overcome acid soil toxicity:breeding for tolerance to soil acidity..................95
3.Mechanisms of alleviating Al toxicity in plants........................................96
3.1.External mechanisms.................................................96
3.2.Internal mechanisms.................................................96
4.Molecular approaches to reveal mechanisms of Al tolerance................................96
4.1.Molecular marker development and their application in studies of Al tolerance and marker-assisted selection(MAS)............................................96
4.2.QTL mapping and inheritance of Al tolerance in plants..............................97
4.3.Association mapping.................................................97
4.4.Identification of functional genes for Al tolerance.................................97
4.5.Heterologous expression studies...........................................98
4.6.Application of molecular markers and QTL mapping in marker-assisted selection...............98
4.7.Transgenic approaches................................................98
4.8.Transcriptional approaches.............................................99
5.Conclusions..........................................................99
Acknowledgments.........................................................100
.............................................................100
1.Introduction
Acid soils are widespread and limit plant production all over the world.They cover 30%–40%of arable land and more than 70%of potential arable land[1].Constraints to production in acid soils are caused by a combination of lack of essential nutrients, reduced water uptake and mineral toxicity.The initial visual symptom on plant growth is reduced root length[2].Although approaches such as adding lime,magnesium or calcium to the soil can ameliorate adverse effects on plant growth,they are both costly and ecologically unsound.Breeding tolerantcultivars is the most efficient way to cope with soil acidity.Plants vary significantly in acid soiltolerance.Variation in acid soiltolerance makes it possible to breed tolerant cultivars.The success of breeding programs relies on an understanding of the physiology, genetics and gene regulatory information of acid soil tolerance. Decades of study have revealed that the tolerance is due to both internal and external mechanisms.The external mechanism, organic acid exudation,is common in higher plants.Various genes and QTL in different species are responsible for different tolerance mechanisms.Molecular markers have been developed to assist gene cloning and to provide useful resources for marker-assisted selection for breeding tolerant cultivars.This paper reviews recent progress in molecular approaches to improve Al tolerance in plants.
2.Acid soil and its toxic effects
Soil pH has significant adverse effects on the availability of plant nutrients[3],solubility of toxic heavy metals[4],soil microorganism activity[5],breakdown of root cells[6],and cation exchange capacity in soils[7].The toxic effects can be classified as morphological and physiological.Both lead to poor plant development and consequently yield reduction [8].
2.1.Acid soil and its distributions
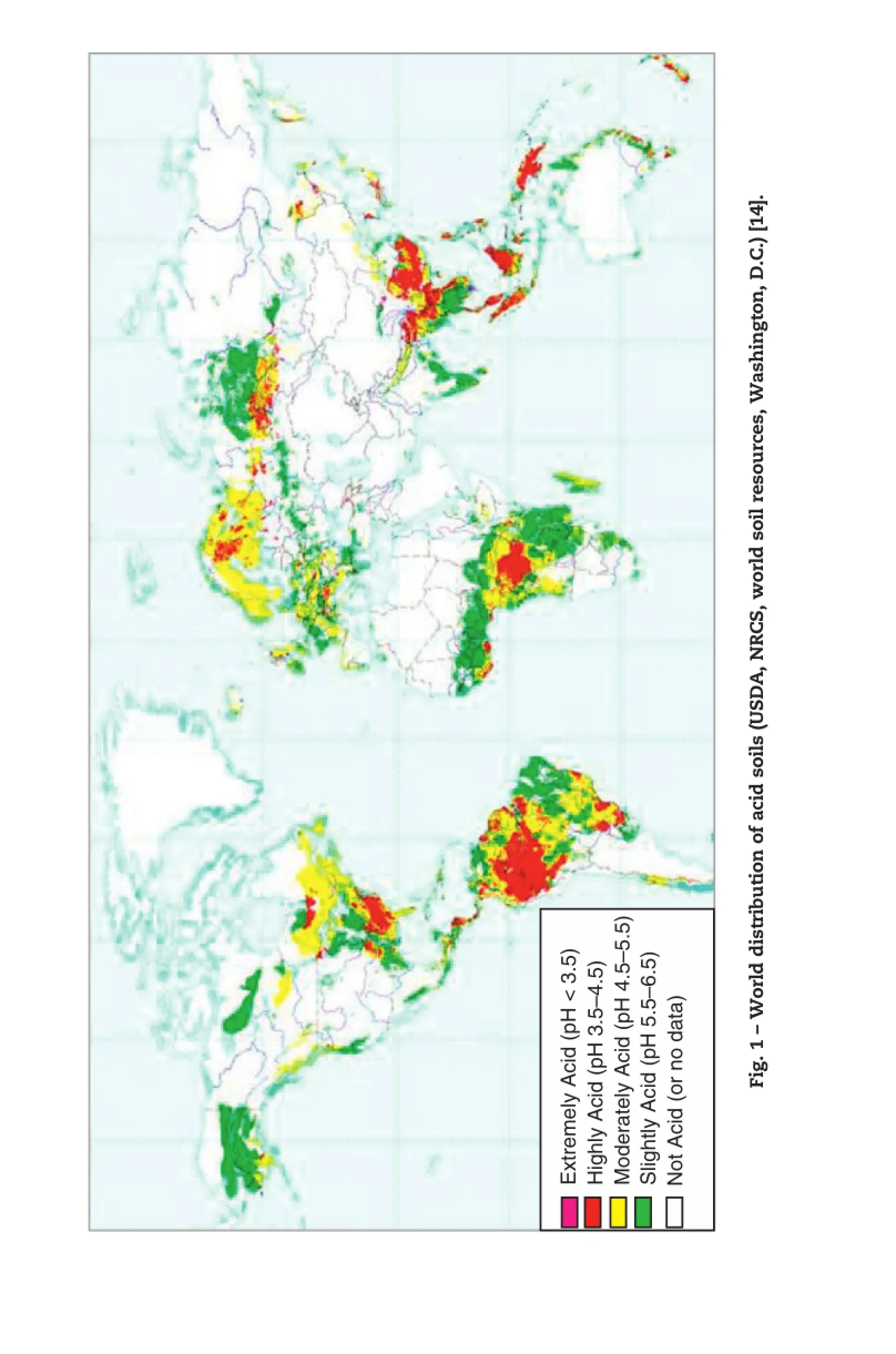
Acid soil is a worldwide problem(Fig.1)mainly located in two belts:viz.,the northern belt in the cold humid temperate zone covering North America,South Asia and Russia;and the southern belt in humid high rainfall tropical areas including South Africa,South America,Australia and parts of New Zealand[1].There are 3950 million ha of arable land affected by soil acidity.It affects about 38%of farmland in Southeast Asia,31%in Latin America,20%in East Asia,56% in Sub-Saharan Africa,and parts of North America[9,10]. In the Americas,1616 million ha is affected,mostly in South America.In Australia and New Zealand,239 million ha of agricultural land is acidic[11].In China and India, 212 million ha or 12%of agricultural land is classified as acidic.
Acid soils not only cause plant production losses,but also affect plant distribution.For example,barley–the fourth most important cereal in the world–with its diverse origin and high importance in agriculture[12],is well known for its wide tolerance to abiotic stress,such as drought,alkaline conditions,cold and heat[13].Due to its high stress tolerance, barley is distributed all over the world.Its growing areas extend from subtropical to temperate zones including North America,Europe,Northwestern Africa,Eastern Asia,Oceania and the Andeans countries of South America(Fig.2).However, as can be seen in Figs.1 and 2,the intensive barley production areas are mainly non-acid soilregions of Europe,North America and Australia.
2.2.Causes of soil acidity
In addition to natural soil acidity,many agricultural and industrial activities lead to increased soil acidity,including acid rainfall[16],fertilizer use,especially acid-forming nitrogen fertilizers[17],and organic matter decay[18].H+ions in acid rain interact with soil cations and displace them from original binding sites;cation exchange capacity reduces and H+concentrations in soil water increase, resulting in leaching[19].When crops are harvested and removed from fields,some basic materials for balancing soil acidity are also lost,thus leading to increased soil acidity.Guo et al.[17]reported that intensive farming and overuse of N fertilizer contribute to soil acidification in China.
2.3.pH level and acid soil toxicity
Acid soil toxicity is caused by a combination of heavy metal toxicity,lack of essential nutrients and acidity per se[20].Large amounts of H+ions have adverse effects on the availability of soil nutrients;availability decreases with falls in soil pH[2,21]. Low pH also increases the solubility of heavy metal elements, such as iron(Fe),copper(Cu),manganese(Mn),zinc(Zn)and aluminum(Al)(Fig.3).Only small amounts of these heavy metals are needed by plants and excessive amounts of soluble ions make them toxic to plant growth[22].
Aluminum,the third most common element in the earth's crust,is one of the most toxic[23].Above a soil pH of 6.0, aluminum forms non-soluble chemical components,with only a small proportion in soluble form in the rhizosphere (Fig.3).When soil pH decreases,Al becomes soluble and causes deleterious effects[24].
A high concentration of H+ions in acid soil is also toxic to higher plants,a feature that has been underestimated for several decades[26].Acidity toxicity and Al toxicity cannot be separated since Al is only soluble in acid solution.Excessive H+ions compete with other mineral elements such as phosphorus (P),magnesium(Mg),calcium(Ca),and Fe for plant absorption and disrupt transportation and uptake of other nutrients, resulting in reduced plant growth[27].Kinraide[26]reported that H+toxicity was dominant at low Al concentration.After screening different collections of the grasses Holcus lanatus L. and Betula pendula Roth under different levels of pHand Al,Kidd and Proctor[2]found that collections from acid organic soils were H+tolerant,whereas those from acidic mineral soils were Al3+tolerant but not necessarily H+tolerant.The authors emphasized that pH toxicity was an important limiting factor in very acid soils.
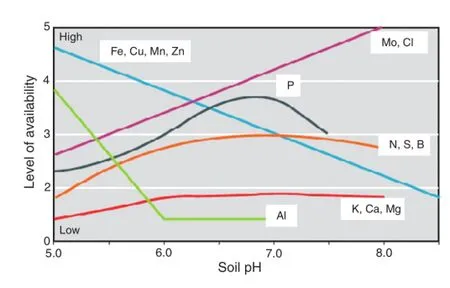
Fig.3–Relationships between availability of elements and soil pH[25].
2.4.Effects of aluminum toxicity on plant growth
Aluminumions(Al3+)cause severe damage to plants.The effects of Al toxicity can be classified as morphological and physiological.Morphological effects refer to symptoms on different plant parts,whereas physiological effects refer to the strong binding effect of soluble Al3+in acid soils where it can interact with multiple sites of the cell,including the cell wall,cell membrane and cell cytosol with consequent toxic effects[28].
The first and most significant morphological symptom of Al toxicity is inhibition and reduction of root growth.It can be detected within several minutes after Al addition[29]. Aluminum limits the ability of roots to scavenge for nutrients and restricts the depth of penetration,resulting in a poorly developed root system,nutrient deficiencies and eventually reduced grain yields[30].Hecht-Buchholz and Foy[31]found typical symptoms of Al toxicity on newly-emerging lateral roots of barley.Root tips were stunted and inhibited in barley varieties differing in tolerance,but the onset of symptoms in the tolerant genotype was several days later than in the sensitive genotype.Tamas et al.[32]observed that Al treatment induced root growth inhibition and loss ofcellviability in barleyroot cells during germination.In white clover,the number of root hairs decreased when the root was treated with Alsolution. An increased Al3+concentration caused root hairs to disappear and stunted root growth[33].
Compared with roots,symptoms of Al toxicity are not so easily identifiable on leaves[20].One of the symptoms is nutrient deficiency,probably a result of low nutrient transport from damaged roots[28].Phosphorus deficiency is manifested by overall stunting,small,dark green leaves,late maturity, purpling of stems,leaves and leaf veins,and yellowing and death of leaf tips[20].Calcium deficiency in the presence of Al can be observed as curling or rolling of young leaves and collapse of growing points or petioles[34].Thus Al inhibition of leaf development may be a response to Al-induced stress in roots[35].Thornton et al.[36]found that leaf size and expansion rates of honey locust seedlings were significantly lower than those in the controls.The size and thickness of leaf blades also decreased,as did the size of leaf cells in seedlings of red pepper when exposed to Al stress[37].
Physiological symptoms include severe inhibition of DNA synthesis[38],blockage of cell division[3],disjunction of cell walls,disruption of plasma membrane integrity,inhibition of signal transduction pathways,and changes in cytoskeleton structure[32].Liu et al.[39]reported that aluminum chloride induced mitotic irregularities and extrusion of nuclear material into the cytoplasm in root tip cells of garlic.Ikeda and Tadano[40]observed alterations of root tip cells in barley treated with Al.These alterations included thickened cell walls, accumulation of small vesicles around the Golgi apparatus and swollen endoplasmic reticulum in cells of the peripheral cap. The activities of different enzymes during seed imbibition and early growth of barley seedlings were also affected by Al3+. Antioxidative enzymes such as peroxidase,superoxide and dismutase had elevated activities in the presence of Al3+. Hydrolytic enzymes including phosphatases,glucosidase and esterase were strongly inhibited at high Al3+solutions[41]. Zhang et al.[42]reported that Al treatment altered lipid composition on cell membranes.In the tolerant wheat cultivar PT741,phosphatidylcholine levels increased dramatically and sterol lipids decreased,but no such changes occurred in the sensitive cultivar Katepwa.
2.5.Solutions to overcome acid soil toxicity:breeding for tolerance to soil acidity
Toxicity of acid soils is mainly caused by low pH,thus agronomic practices to overcome this problem are primarily based on increasing soil pH.Application of lime has been the most common practice for many years.It was reported that the use of lime in Western Australia increased by 57,143 tons per year from 2004 to 2010(http://www.nrm.gov.au/funding/ agriculture/innovation/pubs/soil-acidification.docx).The addition of lime increases root cell growth,lowers absorption of Al and enhances the protective ability of the cell[43,44]. However,this practice has disadvantages[55,56],including Zn and Mn deficiency[45].
Magnesium has been reported to be more efficient than lime in alleviating Al toxicity since the addition of Mg can enhance the efflux of organic acids[46].However,when Mg is present in excess,it becomes toxic[47].Other substances, such as boron(B)and silicon(Si),also help to alleviate Al toxicity[48,49].These strategies were reported to be dependent on species or even genotypes.Nevertheless,of all practices,improving plant tolerance to acid soilthrough breeding is still the best solution to cope with Al toxicity.Traditional breeding methods,such as backcrossing,intercrossing,single seed descent and topcrossing can be used in breeding cereals for acid soil tolerance.With advances in molecular techniques,such as marker-assisted selection(MAS),breeding for acid soil tolerance becomes more effective.However,the effectiveness ofusing MAS relies on the closeness of markers linked to the tolerance genes.
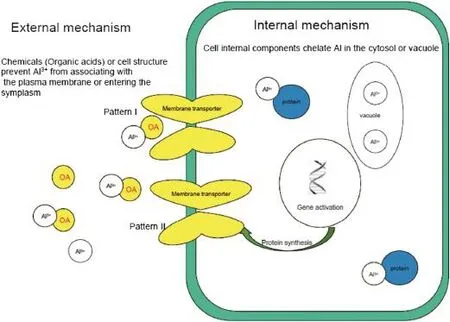
Fig.4–Internal and external mechanisms employed by higher plants in coping with aluminum toxicity.
3.Mechanisms ofalleviating Altoxicity in plants
Plant species differ significantly in Al tolerance.Various studies suggested that Al tolerance follows the order of pea (Pisum sativum L.)<two-rowed barley(Hordeum vulgare L.)<oat (Avena sativa L.)<rye(Secale cereale L.)<rice(Oryza sativa L.)[50]; rye>oat>millet(Pennisetum americanum L.)>bread wheat (Triticum aestivum L.)>barley>durum wheat(Triticum turgidum L.) [51,52].Al tolerance also differs among genotypes within species [53,54].Different mechanisms are employed by higher plants to adapt to acidic environments,which can be classified as external or internal depending on different means of Al binding [29].
3.1.External mechanisms
External mechanisms refer to external structures of the root, such as cell wall,cell membrane or chemical exudates including organic acids[55],phenolic compounds[56]and phosphates[57]that can prevent Al from entering and accumulating in cells(Fig.4).Of various chemicals secreted by cells,organic acids are the most studied[58].For example, in wheat,tolerance is related to citrate[59]and malate exudation[60].Citrate exudation is a major tolerance mechanism for Cassia tora L.[61],snap bean(Phaseolus vulgaris L.) [62],barley[63],and soybean(Glycine max L.)[64].Oxalate exudation was reported in buckwheat(Fagopyrum esculentum M.) [65]and taro(Colocasia esculenta[L.]Schott)[66].These organic acids chelate Al and form non-toxic Al organic acid complexes to prevent Al from interacting with root apices [67].The effects of their amelioration on plant growth under Al stress were demonstrated by exogenous addition of organic acids[68].Different organic acids have different abilities to chelate Al:oxalic acid>citric acid>malic acid>succinic acid, depending on the carboxyl number.Exudation of organic acids can occur immediately upon Al treatment of wheat[69] and tobacco(Nicotiana tabacum)[70].A delay between Al treatment and organic acid extrusion was observed in soybean [64]and triticale(Triticosecale Wittmack)[71].This process of Al-stimulated exudation of organic acids is independent of organic acid and protein synthesis,as well as cell metabolism (Fig.4).
Other external mechanisms such as cell wall composition and cell membrane effect were also reported.Cell-wall pectin content was much lower in Al-resistant buckwheat cultivars than Al-sensitive cultivars.When treated with Al,an Al-sensitive cultivar tended to have more low-methyl-ester pectins and less high-methyl-ester pectins[54].Yang et al.[72] observed that in most cell walls Al accumulated in the hemicellulose 1 fraction and absorption decreased when the hemicellulose 1 was removed in Arabidopsis.The contents of cell wall polysaccharides,which can bind more Al in cell walls,were much higher in Al-tolerant cultivars than Al-sensitive ones[73].The activity of H+-ATPase on plasma membranes was also reported to be correlated with Al-induced root growth inhibition[74].
3.2.Internal mechanisms
Internal mechanisms refer to cell internal components or structures that chelate Al to form non-toxic components. These include the chelating of Al in the cytosol,compartmentalization in the vacuole,Al-binding proteins and Al-tolerant isoenzymes[29].Little is known about the internal mechanism that alleviates Al toxicity since it is very complicated and there are numerous chemicals and targets responding to Al toxicity[75].For example,Watanabe and Osaki[76]reported that the melastoma could accumulate high concentrations of Al in leaves.When Al was translocated from roots to leaves,it formed different chemicals including Al-citrate and Al-oxalate complexes.Flavonoid-type phenolics can possibly detoxify Al inside plant cells.Kidd et al.[77]found that phenolics including catechol and quercetin were released in maize treated with Al and Si,and the release was dependent on Al concentration. However,due to a lack of efficient methodologies,our understanding ofinternalmechanisms of Altolerance in plants is still fragmentary.
4.Molecular approaches to reveal mechanisms of Al tolerance
4.1.Molecular marker development and their application in studies of Al tolerance and marker-assisted selection(MAS)
Genetic markers are useful tools to reveal Al tolerance mechanisms in higher plants following their detection by inheritance studies and identification of relevant genes or loci.During the last two decades,molecular markers based on DNA sequence variations were widely used to study Al tolerance.By detecting molecular markers,the gene or trait could be easily identified and traced[78].Based on the techniques used,molecular markers could be classified as PCR-based or hybridization-based[79].DArT(Diversity Arrays Technology)and RFLP(restriction fragment length polymorphism)are hybridization-based markers,whereas AFLP (amplified fragment length polymorphism),RAPD(randomly amplified of polymorphic DNA),SSR(simple sequence repeat) and SNP(single nucleotide polymorphism)are based on polymerase chain reaction(PCR)techniques.PCR-based markers are preferred and widely used as they are highly efficient,use less DNA,are less labor intensive and amenable to automation and avoidance of autoradiography[80].The use of molecular markers in Al-tolerance studies includes Al-tolerance gene/loci identification and molecular mapping as well as MAS.
One RFLP marker bcd1230,co-segregating with a major gene for Al tolerance,on wheat chromosome 4DL,explained 85%of the phenotypic variation in Al tolerance[81].Using an F2population derived from barley varieties Dayton and Harlan,three RFLP markers,Xbcd1117,Xwg464 and Xcdo1395, were closely linked to Alp on chromosome 4H[82].The authors pointed out that Al tolerance in barley was controlled by a single gene that could be an ortholog of AltBHon wheat chromosome 4D.Five AFLP markers,AMAL1,AMAL2,AMAL3, AMAL4 and AMAL5,were closely linked to,and flanked Alt3 on the long arm of chromosome 4R[83].After screening 35Al-tolerant wheat landrace accessions using ten AFLP primer combinations,Stodart et al.[84]found that these accessions had diverse genetic background and were therefore valuable germplasms for Al tolerance breeding.RAPD marker OPS14705 was linked to the Alt3 locus in rye.A SCAR marker ScOPS14705derived from a RAPD marker,was further shown to be linked to Alt3 locus[85].Ma et al.[86]reported SSR markers Xwmc331 and Xgdm125 flanking the ALMT locus and they indicated that these markers could be used for MAS in breeding Al-tolerant wheat cultivars.In barley,several SSR markers,Bmag353, HVM68 and Bmac310,were closely linked with an Al tolerance gene[87,88].Wang et al.[89]identified a candidate gene HvMATE(Multidrug and Toxin Efflux)for Al tolerance by fine mapping and the gene was closely linked with markers ABG715,Bmag353,GBM1071,GWM165 and HvGABP.
DArT is a hybridization-based molecular marker system.It has been used in barley[90],wheat[91],rye[92]and triticale [93].It is particularly noted for its high-throughput,quickness, high reproducibility and low cost[94].Hundreds to thousands of polymorphisms can be detected very quickly[95].The use of DArT markers to perform whole-genome mapping in some Brazilian wheat cultivars validated the citrate efflux mechanism for Al tolerance[59].DArT markers combined with SSR and STS markers also validated the candidate Al tolerance gene HvMATE on chromosome 4H in barley[89].
4.2.QTL mapping and inheritance of Al tolerance in plants
Genetic mapping refers to the mapping of gene/loci to specific chromosome locations using linked genetic markers[96].Some cereal crops,such as wheat[97],barley,sorghum(Sorghum bicolor L.)and oat were reported to have simple genetic mechanisms of Al tolerance,whereas rice and maize(Zea mays L.)have more complicated inheritance with numerous genes/loci involved.
Generally,a single dominant gene is responsible for Al tolerance in wheat[98];however,there are exceptions in some cultivars[99].Using different populations,genes/loci for Al tolerance were mapped on different wheat chromosomes. Single loci for Al tolerance were identified on chromosomes 4DL,4D,4BL or 3BL,which had phenotypic contributions as high as 85%(locus on 4DL),50%(4D),50%(4BL)and 49%(3BL) [59,81,86,100].In addition,genes/loci on chromosomes 6AL, 7AS,2DL,5AS,3DL and 7D had roles in Al tolerance in wheat [101,102].Complex inheritance of Al tolerance was found in wheat.Zhou et al.[103]identified a secondary QTL for Al resistance on chromosome 3BL in Atlas 66,which was effective only when the epistatic gene on 4DL was absent.Cai et al.[104] mapped three QTL responsible for Al tolerance on wheat chromosomes 4DL,3BL and 2A,which collectively explained 80%of the phenotypic variation.
In sorghum,Al tolerance was simply inherited[105]. Magalhaes et al.[106]reported a major locus AltSBon chromosome 3 for Al tolerance using comparative mapping.In rye,Al tolerance was reported to be controlled by several loci;at least fourindependentloci,Alt1 on6RS[107],Alt2 on 3RS[101],Alt3 on 4RL[83]and Alt4 on 7RS[108],were validated by QTL analysis. The genes on 3R,6RS and 4R were validated using wheat addition and substitution lines with rye chromosomes[101]. Gallego and Benito[109]reported that Al tolerance in rye was controlled by dominant loci Alt1 and Alt3;the latter on chromosome 4RL was validated using recombinant inbred lines[83].Alt4 on chromosome 7RS was identified in three different F2populations[108].
In Arabidopsis,Al tolerance seems to be multi-genetically controlled.Two major QTL accounting for approximately 40% of the phenotypic variance in Al tolerance were identified using recombinant inbred lines derived from the sensitive ecotype Landsberg erecta and tolerant ecotype Columbia[110]. Another two QTL explaining 43%of phenotype variation were detected on chromosomes 1 and 4 in a different cross[111]. The QTL on chromosome 1 was common to both crosses.
In rice and maize,Al tolerance seemed to be quantitatively inherited and QTL analysis showed that multiple loci/genes may control the trait.Nguyen et al.[112]detected 10 QTL for Al tolerance in rice using a double haploid population.They also identified three QTL using recombinant inbred lines derived from a cross between one cultivar and one wild species[113]. In maize,five QTL were identified on chromosomes 2,6 and 8, accounting for 60%of the phenotype variation[114].Two QTL responding to Al tolerance in maize were mapped on the short arms of chromosomes 6 and 10 in a different study[115].
Considerable effort was made in searching for genes involved in Al tolerance in barley;one gene along with additional minor gene effects were detected[52,116].Major QTL,Alp[117],Pht[118], Alt[119]and Alp3[120]on chromosome 4H,were reported,but it is unknown whether these QTL/genes are the same or allelic[52]. Minor QTLforaluminumtolerance were identified on 2H,3Hand 4H in the Oregon Wolfe Barley(OWB)mapping population [100,121].The reason that different QTL were detected in the different populations may be the heterogeneity between different parents[122].More information is required to validate all QTL for Al tolerance in cereals.
4.3.Association mapping
Association mapping is based on associations between molecular markers and traits that can be attributed to the strength of linkage disequilibrium in large populations without crossing[123].It differs from bi-parental QTL mapping that evaluates only two alleles.Association mapping can evaluate numerous alleles simultaneously and is useful for studying the inheritance of complex traits controlled by multiple QTL[124].Using association mapping,six genes in different metabolic pathways were significantly associated with response to Al stress in maize[125].In triticale,several molecular markers had strong associations with phenotypic data from232 advanced breeding lines and the marker wPt-3564 on chromosome 3R was validated by various approaches[126].
4.4.Identification of functional genes for Al tolerance
Using multiple molecular approaches,several genes responding to Al tolerance in plants were identified.These genes mainly belong to the MATE(multidrug and toxic compound extrusion)and ALMT(aluminum-activated malate transporters) families.MATE genes encode transporters excreting a broad range of metabolites and xenobiotics in eukaryotes and prokaryotes[127]and ALMT family members encode vacuolar malate channels[128].
In wheat,Al tolerance is mainly controlled by two genes. TaALMT1 which encodes a malate transporter on chromosome 4D is constitutively expressed on root apices[129]. TaMATE1 reportedly responds to Al stress based on citrate efflux[59].Two genes were reportedly responsible for organic acid extrusion in barley;HvMATE encodes a citrate transporter responsible for Al tolerance[130];and HvALMT,on chromosome 2H,is responsible for malate acid extrusion.Although transgenic plants showed increased Al tolerance,the gene was more likely responsible for anion homeostasis in the cytosol and osmotic adjustment in barley[131].Al tolerance in sorghum is controlled by SbMATE which is the major Al-tolerant locus AltSBon chromosome 3[132].Two genes were reportedly responsible for Al tolerance in Arabidopsis; AtALMT1 encodes a malate transporter responsible for malate efflux on chromosome 1[10]and AtMATE encodes an Al-activated citrate transporter[133].These two genes function independently and both are regulated by the C2H2-type zinc finger transcription factor STOP1[133]which is also reportedly related with low pH tolerance[134].In rye,ScALMT1,which is mainly expressed in the root apex and up-regulated by Al, co-segregates with the Alt4 locus on chromosome 7RS[135]. Another candidate gene ScAACT1 on chromosome 7RS was mapped 25 cM from ScALMT1[136].In maize,ZmMATE1 and ZmMATE2 co-segregated with two major Al-tolerant QTL[114]. ZmMATE1 was induced by Al and related with Al tolerance, whereas ZmMATE2 did not respond to Al[137].
Other reports reveal further genes that do not relate to organic acid extrusion and do not belong to the MATE or ALMT families.For example,the cell-wall-associated receptor kinase gene WAK1 was reportedly involved in Al stress in Arabidopsis [138].In rice,two genes,STAR1 and STAR2,encoding a bacterial-type ATP binding cassette(ABC)transporter,are essential for detoxifying Al[139].
Although some genes have been identified in plants, knowledge of the functional regulation of these genes is still fragmentary.Recent studies showed that gene sequence variation led to different gene expression.For example,allelic variation within the wheat Al-tolerance gene TaALMT1 was demonstrated.There were repeats in the upstream region and the number of repeats was positively correlated with gene expression and Al tolerance[140].In barley,a 1 kb insertion in the upstream region of HvAACT1 enhanced gene expression and altered the location of expression to root tips in some Asian barley cultivars[141].In maize,the copy number of ZmMATE1 was the basis of the phenotypic variation in Al tolerance[142].
4.5.Heterologous expression studies
Heterologous expression is a particularly useful approach for validation of gene function in Al-tolerance studies.Different types of material such as Escherichia coli,yeast,Xenopus oocytes,onion and tobacco cells have been used for heterologous expression study of Al tolerance.For example,TaALMT1 in wheat[129],HvAACT1[130]in barley,ZmMATE1 and ZmMATE2 in maize[137]were heterologously expressed in Xenopus oocytes to validate transport activity in Al tolerance.Huang et al.[139] found that rice genes STAR1 and STAR2 interacted with each other;these two genes were heterologously expressed in onion epidermal cells,rice protoplasts and yeast.The products of the two genes formed a complex with efflux transport activity specific for UDP-glucose,of which exogenous addition protected rootgrowth under Al stress.Protein activity of Al-tolerance genes BnALMT1 and BnALMT2 in Brassica was tested in tobacco cells and Xenopus oocytes and showed that they conferred malate efflux, and transgenic tobacco cells had enhanced tolerance to Al toxicity[143].
4.6.Application of molecular markers and QTL mapping in marker-assisted selection
The rapid development of molecular markers and QTL mapping of Al tolerance permits MAS for Al tolerance in breeding programs.Traditional breeding has benefited from conventional selection based on phenotyping;however, phenotypic selection is reportedly difficult,inefficient and laborious due to its dependence on specific environments [144].MAS is based on associations between molecular markers and superior alleles of genetic traits of interest. After QTL are validated,tightly-linked markers can be used to detect,transfer and accumulate desirable genome regions into superior genotypes,a process that is much faster than phenotypic selection.The major advantages of MAS compared to conventional phenotypic selection are cost-effectiveness,simplicity of selection,time-saving and screening precision[145].
Different types of markers have been developed to trace interesting genes or loci.As discussed in a previous section, molecular markers including RFLP,AFLP,RAPD,SSR,DArT and SNP have been developed and used in Al-tolerance studies.These have proved efficient in MAS in breeding programs.With increasing numbers of genes for Al tolerance being identified and sequenced in plants,PCR-based genespecific markers developed from gene sequencing are preferred in MAS for their easy identification,high polymorphism and good reproducibility[146].In wheat,Raman et al.[158] developed SSR markers,ALMT1-SSR3a and ALMT1-SSR3b and a CAPS marker from the repetitive InDels and substitution region of the TaALMT1 gene.These PCR-based markers co-segregating with the tolerance locus should be efficient tools for MAS[147]. In barley,one gene-specific marker,HvMATE-21indel,was developed from the tolerance gene HvMATE.The marker increased the explained phenotypic variation compared with the other SSR markers.It can also be used for selecting the tolerance gene from multiple tolerance sources[148].With additional and different types of molecular markers being developed for Al tolerance,breeding programs could be accelerated by using these markers in MAS[78].
4.7.Transgenic approaches
Transgenic methods are very efficient for validating gene function in Al-tolerance studies.The first report on a transgenic approach to increasing Al tolerance in plants was in 1997 when De La Fuente et al.[149]reported that an overexpressed citrate synthase gene enhanced citrate efflux and led to improved root Al tolerance in transgenic tobacco. Nodule enhanced malate dehydrogenase and phosphoenolpyruvate carboxylase expression in alfalfa caused increased organic acid exudation in transgenic alfalfa[150].ALMT1 is asingle major gene for Al tolerance in wheat.Delhaize et al. [151]reported that wheat malate transporter gene ALMT1 significantly improved Al tolerance in transgenic barley. Transgenic plants showed robust root growth and unaffected root apices under certain levels of Al stress.Similar results were also reported by Pereira et al.[152]who transformed TaALMT1 into wheat line ET8 using particle bombardment. T-2 lines showed increased gene expression,malate efflux and Al3+resistance.HvALMT,a barley malate transporter gene,on chromosome 2H is mainly expressed in stomatal guard cells and expanding root cells[153].When this gene was overexpressed in transgenic barley plants there was enhanced exudation of organic compounds and improved Al resistance.The efflux was validated to be independent of Al3+[131].
4.8.Transcriptional approaches
Transcriptional approaches,such as transcriptional profiling, RT-PCR,RNAi,Northern blotting,and RNA sequencing[154] facilitated the identification of pathway-related genes and verification of gene function in Al tolerance.Northern analysis of ALS3,which was reported to encode an ABC transporter-like protein related to Al tolerance in Arabidopsis, revealed that gene expression occurred in all organs and expression increased in roots treated with Al[155].Chandran et al.[156]reported over 3000 genes by transcription profiling in an Al-sensitive Medicago truncatula cultivar under Al treatment. These genes were involved in cell wall modification,cell metabolism,protein synthesis and processing,and abiotic and biotic stress responses.RNA-induced silencing also proved that two ofthese genes,pectin acetylesterase and annexin,increased sensitivity to Al.Using a suppression subtractive hybridization technique,Chen et al.[157]identified 229 functional ESTs in the roots of Al-sensitive alfalfa cultivar YM1 after treatment with 5 μmol L−1Al stress.Of them,137 were known Al-response genes,while the other 92 were novel genes potentially related to Al tolerance.The author also noticed that some novel genes related to metabolism and energy were up-regulated and RT-PCR validated the same result.
5.Conclusions
Al is one of the most abundant metals in the earth's crust and prevails in acid soils all over the world.Due to the increasing world population,there is an urgent need to ameliorate Al toxicity to increase plant production on acid soils.Although several approaches for adding exogenous chemicals have proved effective,breeding for tolerance seems to be the most promising.
Over recent decades,molecular approaches have contributed greatly in unraveling genetic mechanisms.Although plants vary significantly in Al tolerance,it seems that they share common tolerance mechanisms.Many researchers have shown that an external mechanism,especially organic acid exudation,plays a major role in detoxifying Al.These genes in wheat either belong to the MATE family encoding a citrate transporter or to the ALMT family which encodes a malate transporter on membranes.Multiple types of markers including SSR,RFLP and SNP were developed to trace the interesting genes.These markers provide not only efficient tools for genetic studies but also important resources for molecular marker-assisted selection.Marker-assisted selection has shifted from linked markers to gene-specific molecular markers for direct tracing of genes of interest.Gene-specific markers developed from wheat Al tolerance gene TaALMT1 and barley Al tolerance gene HvAACT1 co-segregate with the respective tolerance genes and thus should be efficient in MAS [148,158].As shown in Fig.5,the gene-specific marker HvMATE-21indel can be used to differentiate tolerant andsensitive barley cultivars.Genetic behavior of the tolerance of some plant species has been clarified with some genes responding for Al tolerance being identified.In some genotypes of barley[141],wheat[140],and maize[142],gene expression was reportedly affected by variation in gene sequence.However,regulatory networks affecting gene expression remainpoorly understood.The future challenge for studying Al tolerance is the identification of new tolerance mechanisms.For example,it was reported that citrate exudation is the main mechanismand HvAACT1 is the responsible gene for Al tolerance in barley. However,as shown in Fig.6,the gene-specific marker based on the 1 kb InDel does not differentiate tolerant cultivars from sensitive ones[148].The function of the other gene,HvALMT1, for malate acid exudation in barley is still unclear.
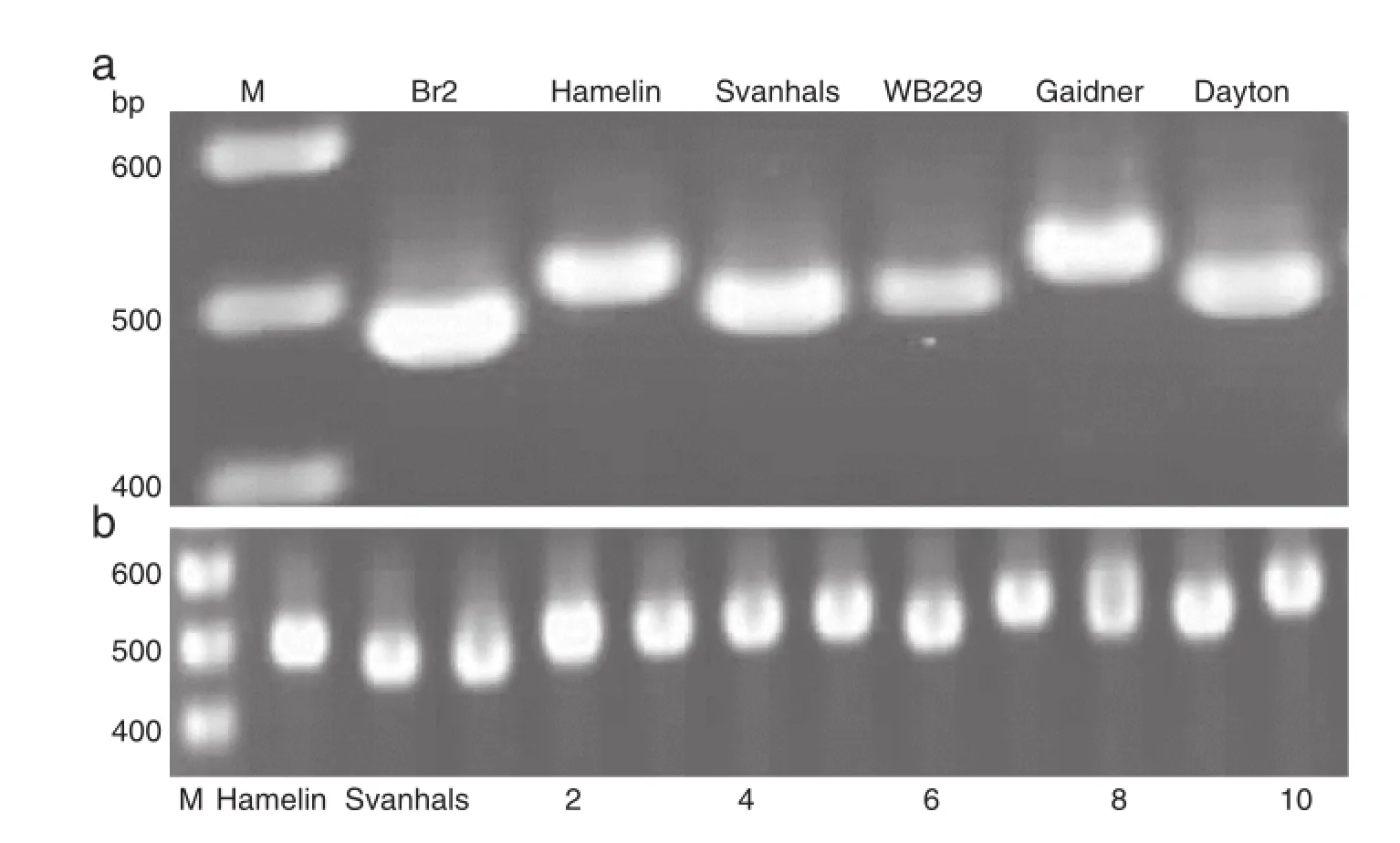
Fig.5–HvMATE-21indel amplicons in six barley cultivars and a DH population separated on 2%agarose gel.a:HvMATE-21indel showing different amplicons in six cultivars:the cultivars with higher bands are sensitive,while those with lower bands are tolerant;b:Polymorphism was confirmed in the Hamelin/Svanhals population;lanes 1–10,Hamelin/Svanhals DH lines.The marker can differentiate different cultivars and the polymorphism was validated in one DH population[148].
Due to recent advances in marker development,a stronger impact of marker-assisted selection in breeding is expected. Although MAS is used successfully for Al tolerance,current markers are still some distance from the Al-tolerance genes. Closer markers or gene-specific markers will make selection more efficient.Combinations of different tolerance mechanisms may achieve better tolerance,thus the discovery of new genes remains a priority for improved Al tolerance in crop plants.
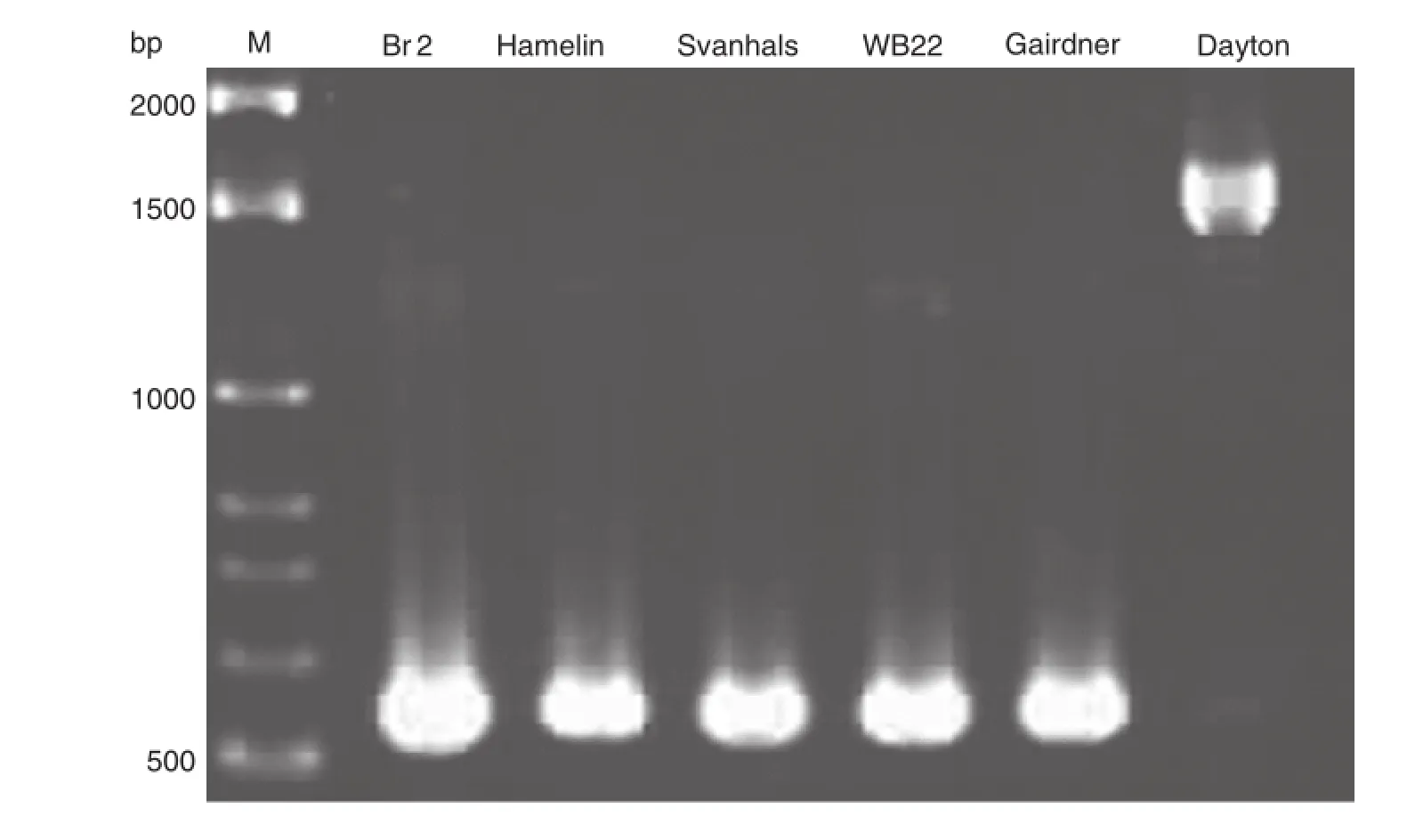
Fig.6–Amplicons of INT0rr+INT2f primer covering a 1 kb insertion in six barley cultivars.The insertion is only found in the sensitive cultivar Dayton;the other two sensitive cultivars Hamelin and WB229 had lower bands as per the three tolerant cultivars Br2,Svanhals and Gairdner[148].
Acknowledgments
This study was supported by the Australian Grains Research and Development Corporation.
R E F E R E N C E S
[1]H.R.Von Uexkuell,E.Mutert,Global extent,development and economic impact of acid soils,Plant Soil 171(1995) 1–15.
[2]P.S.Kidd,J.Proctor,Why plants grow poorly on very acid soils:are ecologists missing the obvious?J.Exp.Bot.52 (2001)791–799.
[3]C.Merino-Gergichevich,M.Alberdi,A.G.Ivanov,M. Reyes-Diaz,Al3+–Ca2+interaction in plants growing in acid soils:Al-phytotoxicity response to calcareous amendments, J.Soil Sci.Plant Nutr.10(2010)217–243.
[4]M.C.Chuan,G.Y.Shu,J.C.Liu,Solubility of heavy metals in a contaminated soil:effects of redox potential and pH,Water Air Soil Pollut.90(1996)543–556.
[5]J.Rousk,P.C.Brookes,E.Baath,Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization,Appl.Environ. Microbiol.75(2009)1589–1596.
[6]H.Koyama,T.Toda,T.Hara,Brief exposure to low-pH stress causes irreversible damage to the growing root in Arabidopsis thaliana:pectin–Ca interaction may play an important role in proton rhizotoxicity,J.Exp.Bot.52(2001) 361–368.
[7]G.P.Gillman,Effects of pH and ionic-strength on the cation-exchange iapacity of soils with variable charge,Aust. J.Soil Res.19(1981)93–96.
[8]D.A.Samac,M.Tesfaye,Plant improvement for tolerance to aluminum in acid soils:a review,Plant Cell Tissue Organ Cult.75(2003)189–207.
[9]S.Wood,K.Sebastian,S.J.Scherr,Pilot Analysis of Global Ecosystems:Agroecosystems,WRI and IFPRI,Washington, 2000.
[10]O.A.Hoekenga,L.G.Maron,M.A.Pineros,G.M.A.Cancado, J.Shaff,Y.Kobayashi,P.R.Ryan,B.Dong,E.Delhaize,T. Sasaki,H.Matsumoto,Y.Yamamoto,H.Koyama,L.V. Kochian,AtALMT1,which encodes a malate transporter,is identified as one of several genes critical for aluminum tolerance in Arabidopsis,Proc.Natl.Acad.Sci.U.S.A.103 (2006)9738–9743.
[11]H.R.Uexküll,E.Mutert,Global extent,development and economic impact of acid soils,Plant Soil 171(1995)1–15.
[12]D.Fei,E.Nevo,D.Z.Wu,J.Comadran,M.X.Zhou,L.Qiu,Z.H. Chen,A.Beiles,G.X.Chen,G.Zhang,Tibet is one of the centers of domestication of cultivated barley,Proc.Natl. Acad.Sci.U.S.A.109(2012)16969–16973.
[13]B.P.Forster,R.P.Ellis,W.T.B.Thomas,A.C.Newton, R.Tuberosa,D.This,R.A.El-Enein,M.H.Bahri, M.Ben Salem,The development and application of molecular markers for abiotic stress tolerance in barley, J.Exp.Bot.51(2000)19–27.
[14]Soil Survey Staff.Natural Resources Conservation Service, United States Department of Agriculture,Available online at http://websoilsurvey.nrcs.usda.gov/.
[15]B.Leff,N.Ramankutty,J.A.Foley,Geographic distribution of major crops across the world,Global Biogeochem.Cycles 18 (2004)GB1009.
[16]E.C.Krug,C.R.Frink,Acid rain on acid soil–a new perspective,Science 221(1983)520–525.
[17]J.H.Guo,X.J.Liu,Y.Zhang,J.L.Shen,W.X.Han,W.F.Zhang, P.Christie,K.W.T.Goulding,P.M.Vitousek,F.S.Zhang, Significant acidification in major Chinese croplands, Science 327(2010)1008–1010.
[18]J.F.Ma,P.R.Ryan,Understanding how plants cope with acid soils,Funct.Plant Biol.37(2010)III–VI.
[19]J.E.Zhang,Y.Ouyang,D.J.Ling,Impacts of simulated acid rain on cation leaching from the Latosol in South China, Chemosphere 67(2007)2131–2137.
[20]C.D.Foy,R.L.Chaney,M.C.White,The physiology of metal toxicity in plants,Annu.Rev.Plant Physiol.29(1978) 511–566.
[21]M.Kasai,M.Sasaki,Y.Yamamoto,H.Matsumoto, Aluminum stress increases K+efflux and activities of ATP-and PPj-Dependent H+pumps of tonoplast-enriched membrane vesicles from barley roots,Plant Cell Physiol.33 (1992)1035–1039.
[22]R.Ginocchio,L.M.de la Fuente,P.Sanchez,E.Bustamante, Y.Silva,P.Urrestarazu,P.H.Rodriguez,Soil acidification as a confounding factor on metal phytotoxicity in soils spiked with copper-rich mine wastes,Environ.Toxicol.Chem.28 (2009)2069–2081.
[23]P.R.Ryan,E.Delhaize,The convergent evolution of aluminium resistance in plants exploits a convenient currency,Funct.Plant Biol.37(2010)275–284.
[24]E.Delhaize,P.R.Ryan,Aluminum toxicity and tolerance in plants,Plant Physiol.107(1995)315–321.
[25]W.J.Goedert,E.Lobato,S.Lourenco,Nutrient use efficiency in Brazilian acid soils:nutrient management and plant efficiency,in:A.C.Moniz,et al.,(Eds.),Plant−Soil Interactions at Low pH,Brazilian Soil Science Society,1997, pp.97–104.
[26]T.B.Kinraide,Aluminum enhancement of plant-growth in acid rooting media:a case of reciprocal alleviation of toxicity by 2 toxic cations,Physiol.Plant.88(1993)619–625.
[27]C.Poschenrieder,M.Llugany,J.Barcelo,Short-term effects of pH and aluminum on mineral-nutrition in maize varieties differing in proton and aluminum tolerance,J.Plant Nutr.18 (1995)1495–1507.
[28]G.R.Rout,S.Samantaray,P.Das,Aluminium toxicity in plants:a review,Agronomie 21(2001)3–21.
[29]L.V.Kochian,O.A.Hoekenga,M.A.Pineros,How do crop plants tolerate acid soils?–Mechanisms of aluminum tolerance and phosphorous efficiency,Annu.Rev.Plant Biol. 55(2004)459–493.
[30]C.D.Foy,Plant adaptation to acid aluminum-toxic soils, Commun.Soil Sci.Plant Anal.19(1988)959–988.
[31]C.Hecht-Buchholz,C.Foy,Effect of aluminium toxicity on root morphology of barley,Plant Soil 63(1981)93–95.
[32]L.Tamas,M.Simonovicova,J.Huttova,I.Mistrik,Changes in the composition of cell wall proteins in barley roots during germination and growth in aluminium presence,Plant Soil Environ.49(2003)327–331.
[33]D.A.Care,The effect of aluminium concentration on root hairs in white clover(Trifolium repens L.),Plant Soil 171(1995) 159–162.
[34]A.Roy,A.Sharma,G.Talukder,Some aspects of aluminum toxicity in plants,Bot.Rev.54(1988)145–178.
[35]P.B.Larsen,L.V.Kochian,S.H.Howell,Al inhibits both shoot development and root growth in als3,an Al-sensitive Arabidopsis mutant,Plant Physiol.114(1997) 1207–1214.
[36]F.C.Thornton,M.Schaedle,D.J.Raynal,Effects of aluminum on growth,development,and nutrient composition of honey locust(Gleditsia triacanthos L.)seedlings,Tree Physiol. 2(1986)307–316.
[37]A.Konarska,Effects of aluminum on growth and structure of red pepper(Capsicum annuum L.)leaves,Acta Physiol. Plant.32(2010)145–151.
[38]S.U.Wallace,I.C.Anderson,Aluminum toxicity and DNA synthesis in wheat roots,Agron.J.76(1984)5–8.
[39]D.H.Liu,W.S.Jiang,D.S.Li,Effects of aluminum ion on root-growth,cell-division,and nucleoli of garlic(Allium sativum L.),Environ.Pollut.82(1993)295–299.
[40]H.Ikeda,T.Tadano,Ultrastructural changes of the root tip cells in barley induced by comparatively low concentration of aluminum,Soil Sci.Plant Nutr.39(1993)109–117.
[41]M.Simonovicova,L.Tamas,J.Huttova,B.Siroka,I.Mistrik, Activity of some enzymes in barley caryopses during imbibition in aluminium presence,Plant Soil Environ.50 (2004)189–195.
[42]G.Zhang,J.J.Slaski,D.J.Archambault,G.J.Taylor, Alternation of plasma membrane lipids in aluminum-resistant and aluminum-sensitive wheat genotypes in response to aluminum stress,Physiol. Plant.99(1997)302–308.
[43]T.R.Guo,Y.Chen,Y.H.Zhang,Y.F.Jin,Alleviation of Al toxicity in barley by addition of calcium,Agric.Sci.China 5 (2006)828–833.
[44]T.B.Kinraide,Three mechanisms for the calcium alleviation of mineral toxicities,Plant Physiol.118(1998)513–520.
[45]I.M.Rao,R.S.Zeigler,R.Vera,S.Sarkarung,Selection and breeding for acid-soil tolerance in crops,Bioscience 43(1993) 454–465.
[46]J.Bose,O.Babourina,Z.Rengel,Role of magnesium in alleviation of aluminium toxicity in plants,J.Exp.Bot.62 (2011)2251–2264.
[47]S.Venkatesan,S.Jayaganesh,Characterisation of magnesium toxicity,its influence on amino acid synthesis pathway and biochemical parameters of tea,Res.J. Phytochem.4(2010)67–77.
[48]K.E.Hammond,D.E.Evans,M.J.Hodson,Aluminium/silicon interactions in barley(Hordeum vulgare L.)seedlings,Plant Soil 173(1995)89–95.
[49]M.Yu,R.Shen,H.Xiao,M.Xu,H.Wang,H.Wang,Q.Zeng, J.Bian,Boron alleviates aluminum toxicity in pea(Pisum sativum),Plant Soil 314(2009)87–98.
[50]J.J.Ślaski,Differences in the metabolic responses of root tips of wheat and rye to aluminium stress,Plant Soil 167(1994) 165–171.
[51]L.Bona,R.J.Wright,V.C.Baligar,J.Matuz,Screening wheat and other small grains for acid soil tolerance,Landsc.Urban Plan.27(1993)175–178.
[52]J.P.Wang,H.Raman,G.P.Zhang,N.Mendham,M.X.Zhou, Aluminium tolerance in barley(Hordeum vulgare L.): physiological mechanisms,genetics and screening methods,J.Zhejiang Univ.Sci.B 7(2006)769–787.
[53]F.Klotz,W.J.Horst,Genotypic differences in aluminum tolerance of soybean(Glycine max L.)as affected by ammonium and nitrate-nitrogen nutrition,J.Plant Physiol. 132(1988)702–707.
[54]J.L.Yang,X.F.Zhu,C.Zheng,Y.J.Zhang,S.J.Zheng, Genotypic differences in Al resistance and the role of cell-wall pectin in Al exclusion from the root apex in Fagopyrum tataricum,Ann.Bot.107(2011)371–378.
[55]J.F.Ma,P.R.Ryan,E.Delhaize,Aluminium tolerance in plants and the complexing role of organic acids,Trends Plant Sci.6(2001)273–278.
[56]R.Tolra,J.Barcelo,C.Poschenrieder,Constitutive and aluminium-induced patterns of phenolic compounds in two maize varieties differing in aluminium tolerance,J.Inorg. Biochem.103(2009)1486–1490.
[57]D.M.Pellet,L.A.Papernik,L.V.Kochian,Multiple aluminum-resistance mechanisms in wheat–roles of root apical phosphate and malate exudation,Plant Physiol.112 (1996)591–597.
[58]J.F.Ma,Role of organic acids in detoxification of aluminum in higher plants,Plant Cell Physiol.41(2000)383–390.
[59]P.R.Ryan,H.Raman,S.Gupta,W.J.Horst,E.Delhaize,A second mechanism for aluminum resistance in wheat relies on the constitutive efflux of citrate from roots,Plant Physiol. 149(2009)340–351.
[60]E.Delhaize,P.R.Ryan,P.J.Randall,Aluminum tolerance in wheat(Triticum aestivum L.):II.Aluminum-stimulated excretion of malic acid from root apices,Plant Physiol.103 (1993)695–702.
[61]J.F.Ma,S.J.Zheng,H.Matsumoto,Specific secretion of citric acid induced by Al stress in Cassia tora L,Plant Cell Physiol. 38(1997)1019–1025.
[62]S.C.Miyasaka,J.G.Buta,R.K.Howell,C.D.Foy,Mechanism of aluminum tolerance in snapbeans–root exudation of citric-acid,Plant Physiol.96(1991)737–743.
[63]J.F.Ma,S.Nagao,K.Sato,H.Ito,J.Furukawa,K.Takeda, Molecular mapping of a gene responsible for Al-activated secretion of citrate in barley,J.Exp.Bot.55(2004)1335–1341.
[64]Z.M.Yang,H.Nian,M.Sivaguru,S.Tanakamaru,H. Matsumoto,Characterization of aluminium-induced citrate secretion in aluminium-tolerant soybean(Glycine max) plants,Physiol.Plant.113(2001)64–71.
[65]M.J.Feng,S.Hiradate,H.Matsumoto,High aluminum resistance in buckwheat:II.Oxalic acid detoxifies aluminum internally,Plant Physiol.117(1998)753–759.
[66]Z.Ma,S.C.Miyasaka,Oxalate exudation by taro in response to Al,Plant Physiol.118(1998)861–865.
[67]H.Matsumoto,Cell biology of aluminum toxicity and tolerance in higher plants,Int.Rev.Cytol. (2000)1–46(Academic Press).
[68]I.D.Prijambada,E.Proklamasiningsih,Effect of organic acids amendment on the growth and yield of soybean(Glycine max)in Ultisol,Int.J.Agric.Biol.12(2010)566–570.
[69]P.R.Ryan,E.Delhaize,P.J.Randall,Characterization of Al-stimulated efflux of malate from the apices of Al-tolerant wheat roots,Planta 196(1995)103–110.
[70]E.Delhaize,D.M.Hebb,P.R.Ryan,Expression of a Pseudomonas aeruginosa citrate synthase gene in tobacco is not associated with either enhanced citrate accumulation or efflux,Plant Physiol.125(2001)2059–2067.
[71]J.F.Ma,S.Taketa,Z.M.Yang,Aluminum tolerance genes on the short arm of chromosome 3R are linked to organic acid release in triticale,Plant Physiol.122(2000) 687–694.
[72]J.L.Yang,X.F.Zhu,Y.X.Peng,C.Zheng,G.X.Li,Y.Liu,Y.Z. Shi,S.J.Zheng,Cell wall hemicellulose contributes significantly to aluminum adsorption and root growth in Arabidopsis,Plant Physiol.155(2011)1885–1892.
[73]J.L.Yang,Y.Y.Li,Y.J.Zhang,S.S.Zhang,Y.R.Wu,P.Wu,S.J. Zheng,Cell wall polysaccharides are specifically involved in the exclusion of aluminum from the rice root apex,Plant Physiol.146(2008)602–611.
[74]Y.S.Kim,W.Park,H.Nian,T.Sasaki,B.Ezaki,Y.S.Jang,G.C. Chung,H.J.Bae,S.J.Ahn,Aluminum tolerance associated with enhancement of plasma membrane H plus-ATPase in the root apex of soybean,Soil Sci.Plant Nutr.56(2010) 140–149.
[75]M.A.Rounds,P.B.Larsen,Aluminum-dependent root-growth inhibition in Arabidopsis results from AtATR-regulated cell-cycle arrest,Curr.Biol.18(2008) 1495–1500.
[76]T.Watanabe,M.Osaki,Influence of aluminum and phosphorus on growth and xylem sap composition in Melastoma malabathricum L,Plant Soil 237(2001)63–70.
[77]P.S.Kidd,M.Llugany,C.Poschenrieder,B.Gunse,J.Barcelo, The role of root exudates in aluminium resistance and silicon-induced amelioration of aluminium toxicity in three varieties of maize(Zea mays L.),J.Exp.Bot.52(2001) 1339–1352.
[78]C.Inostroza-Blancheteau,B.Soto,C.Ibanez,P.Ulloa,F. Aquea,P.Arce-Johnson,M.Reyes-Diaz,Mapping aluminum tolerance loci in cereals:a tool available for crop breeding, Electron.J.Biotechnol.13(2010)4.
[79]P.Kumar,V.K.Gupta,A.K.Misra,D.R.Modi,B.K.Pandey, Potential of molecular markers in plant biotechnology,Plant Omics J.2(2009)141–162.
[80]J.Nagaraju,K.D.Reddy,G.M.Nagaraja,B.N.Sethuraman, Comparison of multilocus RFLPs and PCR-based marker systems for genetic analysis of the silkworm,Bombyx mori, Heredity 86(2001)588–597.
[81]C.R.Riede,J.A.Anderson,Linkage of RFLP markers to an aluminum tolerance gene in wheat,Crop.Sci.36(1996) 905–909.
[82]Y.Tang,M.E.Sorrells,L.V.Kochian,D.F.Garvin, Identification of RFLP markers linked to the barley aluminum tolerance gene Alp,Crop.Sci.40(2000)778–782.
[83]Miftahudin,G.J.Scoles,J.P.Gustafson,AFLP markers tightly linked to the aluminum-tolerance gene Alt3 in rye(Secale cereale L.),Theor.Appl.Genet.104(2002)626–631.
[84]B.J.Stodart,H.Raman,N.Coombes,M.Mackay,Evaluating landraces of bread wheat Triticum aestivum L.for tolerance to aluminium under low pH conditions,Genet.Resour.Crop Evol.54(2007)759–766.
[85]C.Benito,J.Silva-Navas,G.Fontecha,M.V. Hernandez-Riquer,M.Eguren,N.Salvador,F.J.Gallego,From the rye Alt3 and Alt4 aluminum tolerance loci to orthologous genes in other cereals,Plant Soil 327(2010) 107–120.
[86]H.X.Ma,G.H.Bai,B.F.Carver,L.L.Zhou,Molecular mapping of a quantitative trait locus for aluminum tolerance in wheat cultivar Atlas 66,Theor.Appl.Genet. 112(2005)51–57.
[87]H.Raman,A.Karakousis,J.S.Moroni,R.Raman,B.J.Read, D.F.Garvin,L.V.Kochian,M.E.Sorrells,Development and allele diversity of microsatellite markers linked to the aluminium tolerance gene Alp in barley,Aust.J.Agric.Res. 54(2003)1315–1321.
[88]J.Wang,H.Raman,B.Read,M.Zhou,N.Mendham,S. Venkatanagappa,Validation of an Alt locus for aluminium tolerance scored with eriochrome cyanine R staining method in barley cultivar Honen(Hordeum vulgare L.),Aust.J. Agric.Res.57(2006)113–118.
[89]J.Wang,H.Raman,M.Zhou,P.Ryan,E.Delhaize,D.Hebb,N. Coombes,N.Mendham,High-resolution mapping of the Alp locus and identification of a candidate gene HvMATE controlling aluminium tolerance in barley(Hordeum vulgare L.),Theor.Appl.Genet.115(2007)265–276.
[90]P.Wenzl,H.Li,J.Carling,M.Zhou,H.Raman,E.Paul,P. Hearnden,C.Maier,L.Xia,V.Caig,J.Ovesna,M.Cakir,D. Poulsen,J.Wang,R.Raman,K.P.Smith,G.J.Muehlbauer,K.J. Chalmers,A.Kleinhofs,E.Huttner,A.Kilian,A high-density consensus map of barley linking DArT markers to SSR,RFLP and STS loci and agricultural traits,BMC Genomics 7(2006) 206.
[91]P.Mantovani,M.Maccaferri,M.C.Sanguineti,R.Tuberosa,I. Catizone,P.Wenzl,B.Thomson,J.Carling,E.Huttner,E.DeAmbrogio,A.Kilian,An integrated DArT-SSR linkage map of durum wheat,Mol.Breed.22(2008)629–648.
[92]H.Bolibok-Bragoszewska,K.Heller-Uszynska,P.Wenzl,G. Uszynski,A.Kilian,M.Rakoczy-Trojanowska,DArT markers for the rye genome-genetic diversity and mapping,BMC Genomics 10(2009)578.
[93]K.V.Alheit,J.C.Reif,H.P.Maurer,V.Hahn,E.A.Weissmann, T.Miedaner,T.Wurschum,Detection of segregation distortion loci in triticale(×Triticosecale Wittmack)based on a high-density DArT marker consensus genetic linkage map,BMC Genomics 12(2011).
[94]K.Semagn,A.Bjornstad,H.Skinnes,A.G.Maroy,Y. Tarkegne,M.William,Distribution of DArT,AFLP,and SSR markers in a genetic linkage map of a doubled-haploid hexaploid wheat population,Genome 49(2006)545–555.
[95]C.P.Sansaloni,C.D.Petroli,J.Carling,C.J.Hudson,D.A. Steane,A.A.Myburg,D.Grattapaglia,R.E.Vaillancourt,A. Kilian,A high-density diversity arrays technology(DArT) microarray for genome-wide genotyping in Eucalyptus, Plant Methods 6(2010)16.
[96]K.Semagn,A.Bjornstad,M.N.Ndjiondjop,Principles, requirements and prospects of genetic mapping in plants, Afr.J.Biotechnol.5(2006)2569–2587.
[97]D.J.Somers,J.P.Gustafson,The expression of aluminum stress induced polypeptides in a population segregating for aluminum tolerance in wheat(Triticum aestivum L),Genome 38(1995)1213–1220.
[98]E.Delhaize,S.Craig,C.D.Beaton,R.J.Bennet,V.C.Jagadish, P.J.Randall,Aluminum tolerance in wheat(Triticum aestivum L.):I.Uptake and distribution of aluminum in root apices, Plant Physiol.103(1993)685–693.
[99]Y.Tang,D.F.Garvin,L.V.Kochian,M.E.Sorrells,B.F.Carver, Physiological genetics of aluminum tolerance in the wheat cultivar Atlas 66,Crop.Sci.42(2002)1541–1546.
[100]S.Navakode,A.Weidner,R.K.Varshney,U.Lohwasser, U.Scholz,M.S.Roeder,A.Boerner,A genetic analysis of aluminium tolerance in cereals,Agric.Conspec.Sci.75 (2010)191–196.
[101]A.Aniol,J.P.Gustafson,Chromosome location of genes controlling aluminum tolerance in wheat,rye and triticale, Can.J.Genet.Cytol.26(1984)701–705.
[102]A.Aniol,Genetics of tolerance to aluminum in wheat Triticum aestivum L.Thell,Plant Soil 123(1990)223–228.
[103]L.L.Zhou,G.H.Bai,H.X.Ma,B.F.Carver,Quantitative trait loci for aluminum resistance in wheat,Mol.Breed.19(2007) 153–161.
[104]S.Cai,G.H.Bai,D.Zhang,Quantitative trait loci for aluminum resistance in Chinese wheat landrace FSW, Theor.Appl.Genet.117(2008)49–56.
[105]N.Bassam,M.Dambroth,B.C.Loughman,P.R.Furlani,C.R. Bastos,Genetic control of aluminium tolerance in sorghum, Genetic Aspects of Plant Mineral Nutrition,Springer, Netherlands,1990.215–219.
[106]J.V.Magalhaes,D.F.Garvin,Y.Wang,M.E.Sorrells,P.E.Klein, R.E.Schaffert,L.Li,L.V.Kochian,Comparative mapping of a major aluminum tolerance gene in sorghum and other species in the Poaceae,Genetics 167(2004)1905–1914.
[107]F.J.Gallego,B.Calles,C.Benito,Molecular markers linked to the aluminium tolerance gene Alt1 in rye(Secale cereale L.), Theor.Appl.Genet.97(1998)1104–1109.
[108]M.Matos,M.V.Camacho,V.Perez-Flores,B.Pernaute,O. Pinto-Carnide,C.Benito,A new aluminum tolerance gene located on rye chromosome arm 7RS,Theor.Appl.Genet. 111(2005)360–369.
[109]F.J.Gallego,C.Benito,Genetic control of aluminium tolerance in rye(Secale cereale L.),Theor.Appl.Genet.95 (1997)393–399.
[110]O.A.Hoekenga,T.J.Vision,J.E.Shaff,A.J.Monforte,G.P.Lee, S.H.Howell,L.V.Kochian,Identification and characterization of aluminum tolerance loci in Arabidopsis (Landsberg erecta×Columbia)by quantitative trait locus mapping.A physiologically simple but genetically complex trait,Plant Physiol.132(2003)936–948.
[111]Y.Kobayashi,H.Koyama,QTL analysis of Al tolerance in recombinant inbred lines of Arabidopsis thaliana,Plant Cell Physiol.43(2002)1526–1533.
[112]V.T.Nguyen,B.D.Nguyen,S.Sarkarung,C.Martinez,A.H. Paterson,H.T.Nguyen,Mapping of genes controlling aluminum tolerance in rice:comparison of different genetic backgrounds,Mol.Genet.Genomics 267(2002)772–780.
[113]B.D.Nguyen,D.S.Brar,B.C.Bui,T.V.Nguyen,L.N.Pham,H.T. Nguyen,Identification and mapping of the QTL for aluminum tolerance introgressed from the new source, Oryza rufipogon Griff.,into indica rice(Oryza sativa L.),Theor. Appl.Genet.106(2003)583–593.
[114]F.Ninamango-Cárdenas,C.Teixeira Guimarães,P.Martins, S.Netto Parentoni,N.Portilho Carneiro,M.Lopes,J.Moro, E.Paiva,Mapping QTLs for aluminum tolerance in maize, Euphytica 130(2003)223–232.
[115]S.T.Sibov,M.Gaspar,M.J.Silva,L.M.M.Ottoboni,P.Arruda, A.P.Souza,Two genes control aluminum tolerance in maize:genetic and molecular mapping analyses,Genome 42(1999)475–482.
[116]C.L.Echart,J.Fernandes Barbosa-Neto,D.F.Garvin,S. Cavalli-Molina,Aluminum tolerance in barley: methods for screening and genetic analysis,Euphytica 126 (2002)309–313.
[117]D.A.Reid,Genetic control of reaction to aluminum in winter barley,International Barley Genetics Symposium, Washington State University Press,1971,pp.409–413.
[118]O.StøLen,S.Andersen,Inheritance of tolerance to low soil pH in barley,Hereditas 88(1978)101–105.
[119]H.Raman,J.S.Moroni,K.Sato,B.J.Read,B.J.Scott, Identification of AFLP and microsatellite markers linked with an aluminium tolerance gene in barley(Hordeum vulgare L.),Theor.Appl.Genet.105(2002)458–464.
[120]H.Raman,S.Moroni,R.Raman,A.Karakousis,B.Read,K. Sato,B.J.Scott,A genomic region associated with aluminium tolerance in barley,Proceedings of the 10th Australian Barley Technical Symposium,Canberra,2001, (http://wwwregionalorgau/au/abts/2001/t3/raman.htm).
[121]S.Navakode,A.Weidner,R.Varshney,U.Lohwasser,U. Scholz,A.Börner,A QTL analysis of aluminium tolerance in barley,using gene-based markers,Cereal Res.Commun.37 (2009)531–540.
[122]J.B.Holland,Genetic architecture of complex traits in plants, Curr.Opin.Plant Biol.10(2007)156–161.
[123]C.Zhu,M.Gore,E.S.Buckler,J.Yu,Status and prospects of association mapping in plants,Plant Genome 1(2008) 5–20.
[124]E.S.Buckler IV,J.M.Thornsberry,Plant molecular diversity and applications to genomics,Curr.Opin.Plant Biol.5(2002) 107–111.
[125]A.M.Krill,M.Kirst,L.V.Kochian,E.S.Buckler,O.A.Hoekenga, Association and linkage analysis of aluminum tolerance genes in maize,PLoS One 5(2010)e9958.
[126]A.Niedziela,P.T.Bednarek,H.Cichy,G.Budzianowski,A. Kilian,A.Aniol,Aluminum tolerance association mapping in triticale,BMC Genomics 13(2012)67.
[127]L.A.Weston,P.R.Ryan,M.Watt,Mechanisms for cellular transport and release of allelochemicals from plant roots into the rhizosphere,J.Exp.Bot.63(2012)3445–3454.
[128]P.Kovermann,S.Meyer,S.Hoertensteiner,C.Picco, J.Scholz-Starke,S.Ravera,Y.Lee,E.Martinoia,The Arabidopsis vacuolar malate channel is a member of the ALMT family,Plant J.52(2007)1169–1180.
[129]T.Sasaki,Y.Yamamoto,B.Ezaki,M.Katsuhara,S.J.Ahn,P.R. Ryan,E.Delhaize,H.Matsumoto,A wheat gene encoding analuminum-activated malate transporter,Plant J.37(2004) 645–653.
[130]J.Furukawa,N.Yamaji,H.Wang,N.Mitani,Y.Murata,K. Sato,M.Katsuhara,K.Takeda,J.F.Ma,An aluminum-activated citrate transporter in barley,Plant Cell Physiol.48(2007)1081–1091.
[131]B.D.Gruber,E.Delhaize,A.E.Richardson,U.Roessner,R.A. James,S.M.Howitt,P.R.Ryan,Characterisation of HvALMT1 function in transgenic barley plants,Funct.Plant Biol.38 (2011)163–175.
[132]J.V.Magalhaes,J.Liu,C.T.Guimaraes,U.G.Lana,V.M.Alves, Y.H.Wang,R.E.Schaffert,O.A.Hoekenga,M.A.Pineros,J.E. Shaff,P.E.Klein,N.P.Carneiro,C.M.Coelho,H.N.Trick,L.V. Kochian,A gene in the multidrug and toxic compound extrusion(MATE)family confers aluminum tolerance in sorghum,Nat.Genet.39(2007)1156–1161.
[133]J.Liu,J.V.Magalhaes,J.Shaff,L.V.Kochian, Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance,Plant J.57(2009) 389–399.
[134]S.Luchi,H.Koyama,A.Iuchi,Y.Kobayashi,S.Kitabayashi, Y.Kobayashi,T.Ikka,T.Hirayama,K.Shinozaki,M. Kobayashi,Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance,Proc.Natl.Acad.Sci.U.S.A.104(2007) 9900–9905.
[135]G.Fontecha,J.Silva-Navas,C.Benito,M.A.Mestres,F.J. Espino,M.V.Hernandez-Riquer,F.J.Gallego,Candidate gene identification of an aluminum-activated organic acid transporter gene at the Alt4 locus for aluminum tolerance in rye(Secale cereale L.),Theor.Appl.Genet.114(2007)249–260.
[136]J.Silva-Navas,C.Benito,B.Téllez-Robledo,D.Abd El-Moneim,F.Gallego,The ScAACT1 gene at the Qalt5 locus as a candidate for increased aluminum tolerance in rye (Secale cereale L.),Mol.Breed.(2011)1–12.
[137]L.G.Maron,M.A.Pineros,C.T.Guimaraes,J.V.Magalhaes, J.K.Pleiman,C.Mao,J.Shaff,S.N.J.Belicuas,L.V.Kochian, Two functionally distinct members of the MATE(multi-drug and toxic compound extrusion)family of transporters potentially underlie two major aluminum tolerance QTLs in maize,Plant J.61(2010)728–740.
[138]M.Sivaguru,B.Ezaki,Z.H.He,H.Tong,H.Osawa,F.Baluska, D.Volkmann,H.Matsumoto,Aluminum-induced gene expression and protein localization of a cell wall-associated receptor kinase in Arabidopsis,Plant Physiol.132(2003) 2256–2266.
[139]C.F.Huang,N.Yamaji,N.Mitani,M.Yano,Y.Nagamura, J.F.Ma,A bacterial-type ABC transporter is involved in aluminum tolerance in rice,Plant Cell 21(2009)655–667.
[140]T.Sasaki,P.R.Ryan,E.Delhaize,D.M.Hebb, Y.Ogihara,K.Kawaura,K.Noda,T.Kojima,A.Toyoda,H. Matsumoto,Y.Yamamoto,Sequence upstream of the wheat (Triticum aestivum L.)ALMT1 gene and its relationship to aluminum resistance,Plant Cell Physiol.47(2006) 1343–1354.
[141]M.Fujii,K.Yokosho,N.Yamaji,D.Saisho,M.Yamane,H. Takahashi,K.Sato,M.Nakazono,J.F.Ma,Acquisition of aluminium tolerance by modification of a single gene in barley,Nat.Commun.3(2012)713.
[142]L.G.Maron,C.T.Guimaraes,M.Kirst,P.S.Albert,J.A.Birchler, P.J.Bradbury,E.S.Buckler,A.E.Coluccio,T.V.Danilova,D. Kudrna,J.V.Magalhaes,M.A.Pineros,M.C.Schatz,R.A. Wing,L.V.Kochian,Aluminum tolerance in maize is associated with higher MATE1 gene copy number,Proc. Natl.Acad.Sci.U.S.A.110(2013)5241–5246.
[143]A.Ligaba,M.Katsuhara,P.R.Ryan,M.Shibasaka,H. Matsumoto,The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells,Plant Physiol.142(2006)1294–1303.
[144]B.C.Y.Collard,D.J.Mackill,Marker-assisted selection:an approach for precision plant breeding in the twenty-first century,Philos.Trans.R.Soc.B Biol.Sci.363(2008)557–572.
[145]S.P.Moose,R.H.Mumm,Molecular plant breeding as the foundation for 21st century crop improvement,Plant Physiol.147(2008)969–977.
[146]P.Poczai,I.Varga,M.Laos,A.Cseh,N.Bell,J.P.T.Valkonen,J. Hyvonen,Advances in plant gene-targeted and functional markers:a review,Plant Methods 9(2013)6.
[147]H.Raman,P.R.Ryan,R.Raman,B.J.Stodart,K.Zhang,P. Martin,R.Wood,T.Sasaki,Y.Yamamoto,M.Mackay,D.M. Hebb,E.Delhaize,Analysis of TaALMT1 traces the transmission of aluminum resistance in cultivated common wheat(Triticum aestivum L.),Theor.Appl.Genet.116(2008) 343–354.
[148]M.Bian,I.Waters,S.Broughton,X.-Q.Zhang,M.Zhou,R. Lance,D.Sun,C.Li,Development of gene-specific markers for acid soil/aluminium tolerance in barley(Hordeum vulgare L.),Mol.Breed.(2013)1–10.
[149]J.M.De La Fuente,V.Ramirez-Rodriguez,J.L.Cabrera-Ponce, L.Herrera-Estrella,Aluminum tolerance in transgenic plants by alteration of citrate synthesis,Science 276(1997) 1566–1568.
[150]M.Tesfaye,S.J.Temple,D.L.Allan,C.P.Vance,D.A.Samac, Overexpression of malate dehydrogenase in transgenic alfalfa enhances organic acid synthesis and confers tolerance to aluminum,Plant Physiol.127(2001)1836–1844.
[151]E.Delhaize,P.R.Ryan,D.M.Hebb,Y.Yamamoto,T.Sasaki,H. Matsumoto,Engineering high-level aluminum tolerance in barley with the ALMT1 gene,Proc.Natl.Acad.Sci.U.S.A.101 (2004)15249–15254.
[152]J.F.Pereira,G.Zhou,E.Delhaize,T.Richardson,M.Zhou,P.R. Ryan,Engineering greater aluminium resistance in wheat by over-expressing TaALMT1,Ann.Bot.106(2010)205–214.
[153]B.D.Gruber,P.R.Ryan,A.E.Richardson,S.D.Tyerman,S. Ramesh,D.M.Hebb,S.M.Howitt,E.Delhaize,HvALMT1 from barley is involved in the transport of organic anions,J.Exp. Bot.61(2010)1455–1467.
[154]Z.Wang,M.Gerstein,M.Snyder,RNA-Seq:a revolutionary tool for transcriptomics,Nat.Rev.Genet.10(2009)57–63.
[155]P.B.Larsen,M.J.B.Geisler,C.A.Jones,K.M.Williams,J.D. Cancel,ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis,Plant J.41(2005)353–363.
[156]D.Chandran,N.Sharopova,S.Ivashuta,J.S.Gantt,K.A. VandenBosch,D.A.Samac,Transcriptome profiling identified novel genes associated with aluminum toxicity, resistance and tolerance in Medicago truncatula,Planta 228 (2008)151–166.
[157]Q.Chen,X.D.Zhang,S.S.Wang,Q.F.Wang,G.Q.Wang,H.J. Nian,K.Z.Li,Y.X.Yu,L.M.Chen,Transcriptional and physiological changes of alfalfa in response to aluminium stress,J.Agric.Sci.149(2011)737–751.
[158]H.Raman,R.Raman,R.Wood,P.Martin,Repetitive inDel markers within the ALMT1 gene conditioning aluminium tolerance in wheat(Triticum aestivum L.),Mol.Breed.18 (2006)171–183.
*Corresponding author.
E-mail address:chengdao.li@agric.wa.gov.au(C.Li).
Peer review under responsibility of Crop Science Society of China and Institute of Crop Science,CAAS
Aluminum
Marker-assisted selection
Mechanism
- The Crop Journal的其它文章
- QTL mapping of starch granule size in common wheat using recombinant inbred lines derived from a PH82-2/Neixiang 188 cross
- Identification and functional analysis of miRNAs in developing kernels of a viviparous mutant in maize
- Enhanced tolerance to drought in transgenic rice plants overexpressing C4photosynthesis enzymes
- Enhanced tolerance to drought in transgenic rice plants overexpressing C4 photosynthesis enzymes
- HrcQ is necessary for Xanthomonas oryzae pv.oryzae HR-induction in non-host tobacco and pathogenicity in host rice
- Isolation and characterization of an isoamylase gene from rye

