Expression of an(E)-β-farnesene synthase gene from Asian peppermint in tobacco affected aphid infestation
Xiudo Yu,Yongjun Zhng,Youzhi M*,Zhoshi XuGenping WngLnqin Xi**
aInstitute of Crop Sciences,National Key Facility for Crop Gene Resources and Genetic Improvement,Chinese Academy of Agricultural Sciences, Beijing 100081,China
bSchool of Life Science and Technology,Nanyang Normal University,Nanyang,Henan 473061,China
cInstitute of Crop Protection,Chinese Academy of Agricultural Sciences,Beijing 100193,China
Expression of an(E)-β-farnesene synthase gene from Asian peppermint in tobacco affected aphid infestation
Xiudao Yua,b,Yongjun Zhangc,Youzhi Maa,*,Zhaoshi Xua,Genping Wanga,Lanqin Xiaa,**
aInstitute of Crop Sciences,National Key Facility for Crop Gene Resources and Genetic Improvement,Chinese Academy of Agricultural Sciences, Beijing 100081,China
bSchool of Life Science and Technology,Nanyang Normal University,Nanyang,Henan 473061,China
cInstitute of Crop Protection,Chinese Academy of Agricultural Sciences,Beijing 100193,China
A R T I C L E I N F O
Article history:
Received 7 March 2013
Received in revised form
15 May 2013
Accepted 28 May 2013
Available online 10 July 2013
EβF synthase gene
Aphids are major agricultural pests that cause significant yield losses in crop plants each year.(E)-β-farnesene(EβF)is the main or only component of an alarm pheromone involved in chemical communication within aphid species and particularly in the avoidance of predation.EβF also occurs in the essential oil of some plant species,and is catalyzed by EβF synthase.By using oligonucleotide primers designed from the known sequence of an EβF synthase gene from black peppermint(Mentha×piperita),two cDNA sequences,MaβFS1 and MaβFS2,were isolated from Asian peppermint(Mentha asiatica).Expression pattern analysis showed that the MaβFS1 gene exhibited higher expression in flowers than in roots,stems and leaves at the transcriptional level.Overexpression of MaβFS1 in tobacco plants resulted in emission of pure EβF ranging from 2.62 to 4.85 ng d−1g−1of fresh tissue.Tritrophic interactions involving peach aphids(Myzus persicae),and predatory lacewing(Chrysopa septempunctata)larvae demonstrated that transgenic tobacco expressing MaβFS1 had lower aphid infestation.This result suggested that the EβF synthase gene from Asian peppermint could be a good candidate for genetic engineering of agriculturally important crop plants. ©2013,Crop Science Society of China and Institute of Crop Science,CAAS.Production and hosting by Elsevier B.V.All rights reserved.
1.Introduction
Aphids are major agricultural pests which cause significant yield losses in crop plants each year by inflicting damage both through the direct effects of feeding and by vectoring harmful plant viruses[1,2].Annual worldwide crop losses due to aphids are estimated at hundreds of millions of dollars [3,4].Along with the application of nitrogen fertilizer and elevation of atmospheric CO2concentration,aphid infestation becomes more serious[5,6].For many crops,insecticides provide a simple strategy for aphid control.However,the application of chemicals is not desirable because of the development ofinsecticide resistance and pollution of the environment[7]. Transgenic crops engineered for enhanced resistance to aphids could be an efficient alternative strategy.Some plant lectins, including Galanthus nivalis agglutinin(GNA)and Pinellia ternate agglutinin(PTA),are toxic to aphids in transgenic plants[8,9]. However,GNA caused adverse effects in the food chain of predatory ladybirds and the parasitoid Aphidius ervi via aphids [10,11];for example,when aphids were fed on GNA transgenic wheat,the GNA ingested by aphids and transported into the honeydew negatively affected the survival of parasitoid A.ervi consuming the honeydew[11],resulting in concerns relating to biosafety issues for application of these lectin genes in agriculturally important crops for aphid control.Therefore,other safe and effective genes/genetic strategies for aphid control need to be found.
Aphids are attacked by a wide range of predators and parasitoids,which strongly influence the growth and persistence ofaphid colonies[1].When attacked by a predator,aphids secrete cornicle droplets containing an alarm pheromone.The sesquiterpene EβF is the predominant and sometimes only component ofmost aphid alarm pheromones[12–14].EβF is detected by their nearby conspecifics and triggers various escape behavioral reactions,such as dropping off the plant,or flying or walking away[15].Recently,EβF was also found to mediate and increase the production ofwinged offspring ofthe pea aphid(Acyrthosiphon pisum)[16],while alate aphids are usually more sensitive to EβF than the apterae ones and often leave host plants emitting EβF [17],thereby reducing the aphid numbers on host plants.As a consequence,many aphids do not survive during migration because of starvation or ground predators[18].Moreover,EβF may substantially enhance the effectiveness of pesticides and mycoinsecticides by increasing aphid mobility[19,20].EβF can also function as a kairomone(a chemical messenger emitted by organisms of one species but benefitting members of another species)in attracting aphid predators,including ladybirds[21,22], lacewings[21],hoverfly[23]and parasitoids[24],and thus recruit natural enemies for aphid control.
EβF occurs in the essential oils of plant species such as chamomile[25],Garland[26],Hemizygia petiolata[27],water pepper[28],and black peppermint[17].In field plot experiments,the numbers of pea aphid(A.pisum)were significantly reduced when sprayed with essential oil from H.petiolata,an oil that is rich in EβF(more than 70%EβF)[27].EβF is also a component of some plant volatiles.In natural environments, wild potato,Solanum berthaultii,releases high quantities of endogenous sesquiterpene EβF from specialized foliar trichomes, that are more repellent to the green peach aphid than the oil in commercial potato varieties which produce lower levels of EβF along withsome inhibitory compounds such as(E)-caryophyllene [29–31].EβF emission can be induced by herbivory in certain plants[32,33],and insect-induced EβF has been hypothesized to function either as a direct repellent to insects(i.e.,alarm pheromone function)or act as a kairomone for natural enemies of aphids[34].For example,caterpillar-damaged maize releasing EβF repels the corn leaf aphid Rhopalosiphum maidis[35],and oviposition-induced EβF from pine was shown to attract the parasitoid Chrysonotomyia ruforum[33,36].
The potential importance of EβF for aphid control in plants has prompted the cloning of genes related to its synthesis.EβF synthase genes that encode an enzyme converting farnesyl diphosphate(FPP)to EβF have been isolated and characterized in several plant species,including Douglas fir,Yuzu,sweet wormwood and black peppermint[37].In vitro analysis showed that EβF synthase from peppermint(Mentha×piperita L.)could convert FPP to EβF[17].Expression of the EβF synthase gene from black peppermint in Arabidopsis repelled aphids and attracted aphid parasitoids at a significant level[38].Moreover, EβF-emitting transgenic Arabidopsis allowed aphids to habituate to their own alarmpheromone;habituated aphids then showed no avoidance response to EβF,thereby increasing predator and parasitoid efficiency[34].Overexpression of sweet wormwood EβF synthase genes in tobacco also resulted in reduced aphid infestation[39].These results indicated that genetic engineering of plants to produce EβF for aphid control could be feasible.
However,not all of EβF synthase genes isolated so far are functional,such as MxpSS2 from black peppermint(GenBank accession number AJ786642).MxpSS2 encodes a protein with two amino acid differences from EβF synthase identified in a different black peppermint variety‘Black Mitcham'(GenBank accession number AF024615).One of the amino acid differences(leucine in MxpSS2 and serine in EβF synthase)at position 531 led to loss of EβF synthase activity in the MxpSS2 chemotype[17,40]possibly due to the L531 residue that lies in a J–K loop clamping down over the entrance to the active site[41].Therefore,it is necessary to study the effective and functional EβF synthase genes from a variety of plant varieties/species before their use in engineering other crop plants for aphid control.In the present study,two EβF synthase genes,designated as MaβFS1 and MaβFS2,were isolated from Asian peppermint.The tissue expression pattern of MaβFS1 was characterized.MaβFS1 transgenic tobacco plants were generated and molecularly characterized.EβF emission levels and aphid bioassays of transgenic tobacco plants were also investigated.
2.Materials and methods
2.1.Plant materials
Asian peppermint seedlings were purchased from Beijing Botanic Garden,Beijing,and planted in soil in a greenhouse at 20±5°C under 400 W HPS mercury vapor lamps.Roots,stems, leaves and flowers of the flowering Asian peppermint were excised,frozen in liquid nitrogen and stored at−80°C.Tobacco (Nicotiana tabacum L.,cv.W38)seedlings grown on standard MS medium were used for transformation.Commercial EβF was purchased from Tokyo Kasei Chemicals,Tokyo,Japan.
2.2.Isolation of EβF synthase genes from Asian peppermint by RT-PCR
Leaves of Asian peppermint plants were used to extract total RNA and genomic DNA(gDNA)using the RNAprep Pure Plant Kit and Plant Genomic DNA Kit(Tiangen Biotech,Beijing, China)according to the manufacturer's instructions.For RT-PCR,first strand cDNA synthesis was initiated with 2 μg of total RNA using 500 ng of random hexamers and M-MLV Reverse Transcriptase(TaKaRa,Dalian,China).PCR amplifications were done using the synthesized cDNA or gDNA as template.The specific primers were MaβFS F1 and MaβFS R1(listed in Table 1),where ATG and TTA are the start and stop codons of the published EβF synthase cDNA(GenBank accession no.AF024615).Reactions of 50 μL containing cDNA(50 ng) or g DNA(100 ng),dNTPs(0.2 mmol L−1of each),primers (0.2 μmol L−1of each),PrimeSTAR HS DNA Polymerase(1.25 U) and buffer were supplied with the polymerase(TaKaRa,Dalian). Reactions were conducted according to the following program: 98°C for 15 s;52°C for 10 s and 72°C for 2 min,40 cycles, followed by maintenance at 72°C for 10 min.The products obtained were separated by agarose gel electrophoresis(alongside DL2000 DNA marker or 250 bp DNA ladder marker to check the fragment size and approximate amounts)and then purified from the gel using a Tiangen Mini Purification Kit(Tiangen Biotech,Beijing).The purified PCR fragment was cloned into pEASY-Blunt vector(Tiangen Biotech,Beijing)and transformed into competent Escherichia coli DH5α cells.The cells were selected on LB agar plates containing kanamycin(100 mg L−1), 7 μL of 20%IPTG and 40 μL of 2%X-gal.

Table 1–Sequences of gene-specific primer pairs used in this study.
2.3.DNA sequencing and analysis
MaβFS plasmid DNA was isolated using a Tianpure Mini Plasmid Kit(Tiangen Biotech,Beijing)and inserts were sequenced using a Bigdye terminator chemistry kit(ABI,Perkin-Elmer)on an ABI 3130 XL DNA sequencer(ABI,Perkin-Elmer).DNA sequence data were assembled and analyzed using DNAMAN software,and putative amino acid sequences were analyzed in GenBank databases using the NCBI BLAST program.Schematic structures of MaβFS1 and MaβFS2 were drawn in a gene structure display server(GSDS,http://gsds.cbi.pku.edu.cn/).The theoretical isoelectric points(pI)and molecular weights(MW)of the proteins were computed using the Compute pI/MW Tool(http://www. expasy.org/tools/pi_tool.html).Alignment of the deduced protein sequences was performed using DNAMAN and CLUSTAL_X version 1.83.A joint unrooted phylogenetic tree was constructed by MEGA4 using the neighbor-joining method.
2.4.Tissue expression pattern analysis
TotalRNAofthe root,stem,leafandflower of Asian peppermint were extracted using the RNAprep Pure Plant Kit(Tiangen Biotech,Beijing),and a 2 μg aliquot of RNAper sample was used to synthesize first-strand cDNA.The expression levels of MaβFS were investigated using quantitative real time-PCR(qRT-PCR), which was performed with a Quant qRT-PCR Kit(Tiangen Biotech,Beijing)in an ABI PRISM 7000 sequence detection system(Applied Biosystems,Foster City,CA,USA),with reactions subjected to the following program:95°C for 1 min, 41 cycles of 95°C for 10 s,and 56°C for 30 s.To normalize the PCRs for the amount of added RNA the β-actin gene from peppermint(MaACT,GenBank accession no.AW255057)was selected as the endogenous control.For each sample,the MaβFS Ct value(meaning the number of cycles required for the fluorescence signal to cross the threshold)of each sample was normalized to the Ct value of β-actin.The relative value of gene expression was analyzed using the 2−ΔΔCtmethod[42].The relative expression levels of MaβFS in stems,leaves and flowers were presented relative to average root levels.The primer pairs, MaβFS F2 and MaβFS R2,and MaACT F and MaACT R,are listed in Table 1.
2.5.Tobacco transformation and regeneration
Compared with the commercial pBI121 vector,the modified pBI121 plasmid used here replaced the uidA gene(encoding GUS)of the original vector with a fragment possessing multiple cloning sites including Sma I and Spe I,but preserving the npt II gene encoding npt II gene driven by the NOS promoter and NOS terminator.The npt II gene confers resistance to aminoglycoside antibiotics,such as kanamycin.The full ORF sequence of the MaβFS1 gene with Sma I and Spe I was cloned into the Sma I and Spe I sites of the modified pBI121 to form the transformation vector MaβFS1-pBI121.The orientation and integrity of MaβFS1 in the construct were confirmed by sequencing.
The plasmids were then transferred into Agrobacterium tumefaciens strain AGL1.Transformation of tobacco cultivar W38 was performed by A.tumefaciens-mediated leaf disc transformation[43].Regenerated plantlets were identified by PCR(using primer pairs MaβFS F3 and MaβFS R3,Table 1),and positive lines (i.e.,tobacco lines successfully transformed with the target gene) were transferred to soilin pots,and grown in a greenhouse under 12:12 h light/dark at 25°C.T1and T2transgenic tobacco seeds from the pBI121 blank vector,and MaβFS1 transgenic lines were germinated on selective MS medium with 100 mg L−1kanamycin.The T2lines with acceptable RT-PCR results were transferred to soil in pots for further analysis with the transgenic line harboring the pBI121 blank vector as control.
2.6.Molecular characterization of transgenic tobacco plants
The presence and expression of the MaβFS1 gene in the transgenic tobacco plants were monitored by PCR and RT-PCR using the gene-specific primers MaβFS F3 and MaβFS R3.PCR and RT-PCR were performed in total volumes of 25 μL containing gDNA(100 ng),dNTPs(0.2 mmol L−1each),primers(0.2 μmol L−1each),Taq polymerase(1 U)and buffersupplied with the enzyme (TaKaRa,Dalian),and subjected to a program of initial denaturation at 94°C for 3 min,followed by 35 cycles of 45 s at 94°C, 45 s at 55°C,and 25 s at 72°C,and a final extension at 72°C for 10 min.The amplified product(330 bp)specific to the MaβFS1 gene was resolved on a 1.2%(W/V)agarose gel and visualized by ethidium bromide staining.The transcriptional expression levels of MaβFS1 were further analyzed by qRT-PCR,with the tobacco 18S rRNA gene(Nt18S,GenBank accession no.AJ236016)as a standard control(primer pairs Nt18S F and Nt18S R are listed in Table 1).
2.7.Air entrainment and GC–MS analysis
Transgenic and control plants were planted in soil in pots. Transgenic and control tobacco plants at flowering were subjected to volatile analysis,and volatiles from the T2transgenic lines with the pBI121 blank vector and MaβFS1 were respectively trapped by an Extract-Clean column(Grace Davison Discovery Sciences,Deerfield,IL,USA)with 50 mg Super Q(80/100 mesh;Alltech,Deerfield,IL,USA)as an adsorbent.The upper six leaves and flowers of each plant were enclosed in polythene bags(40 cm×60 cm).Air that passed through a charcoal-infused medium for purification and moistened to a relative humidity of 65%entered from the bottom of the bag.Volatiles emitted from the upper portion of plants enclosed within the bags were swept upward by the incoming laminar air flow.Air exited from the top of the bag through the trap column at a rate of 1 L/min by an automated flow controller.All collections were performed for periods ranging from 1 to 24 h(12 h light and 12 h darkness).
After a 24 h collection period,the traps were rinsed with 400 μL methylene chloride containing 1600 ng of n-octane as an internal standard.From each plant sample,1 μL was analyzed by HP6890/HP5973 GC–MS(Hewlett-Packard).Chromatographic resolution was done on an HP-5MS column (60 m×250 μm×0.25 μm;Hewlett-Packard)operated at 40°C for 1 min,and increased to 150°C at a rate of 4°C min−1,held at 100°C and 150°C for 1 min each;then increased to 280°C at a rate of10°C min−1and held at 280°Cfor 1 min.Hewlett-Packard Chemstation software was utilized for system control and data analysis.Quantification of all major components was based on comparisons with the internal standards.Individual components of the volatiles were identified by comparing the mass spectra and retention indices with those of the commercially available standards by the libraries of Wiley and the National Institute of Standards and Technology.
2.8.Aphid and predator bioassays
According to the results of GC–MS and RT-PCR,three MaβFS1 T2transgenic lines(Ma1,Ma4,Ma10)with higher EβF emissions were selected for aphid control assays with transgenic lines harboring the pBI121 blank vector as controls.For each assay, two independent experiments were performed and each was done in triplicate.All the bioassays were performed in a hexagon setup with a diameter of 1.5 m,and each of the Ma1, Ma4,and Ma10 transgenic plants and three control plants put in alternating order on the angle of the hexagon as described by Kappers et al.[44].This setup was totally enclosed by a white coarse-net cover in the greenhouse.Responses of aphids to MaβFS1 lines were tested by introduction of200 alate aphids into the chamber.The number of aphids on each plant was counted after 12 h.To assess the preliminary effect of aphid control by predator foraging and repellence,400 alate aphids and 10 lacewing larvae starved for 6 h were placed at the midpoint of the setup.Twelve hours later,the number of aphids on each plant was counted.Statistical analysis was performed using one-way analysis of variance and t-tests in Microsoft Excel[45].
3.Results
3.1.Cloning and sequence analysis of EβF synthase genes from Asian peppermint
Using gene-specific primers designed according to the published EβF synthase gene from black peppermint(GenBank accession number AF024615),EβF synthase cDNAs were isolated by RT-PCR.Sequencing of eight randomly selected clones identified two distinct c DNAs.One sequence,designated as MaβFS1(deposited in GenBank under accession number HQ337896)and 1653 bp in length with 5 nucleotide differences from AF024615,encoded a 550 amino acid protein with a theoretical pI of 5.27 and a 100%overall amino acid sequence identity with the published gene from black peppermint (GenBank accession number AF024615)(Fig.1).Another sequence designated as MaβFS2(deposited in GenBank under accession number HQ337897)was 1653 bp in length with 6 nucleotide differences from AF024615,encoded a 550 amino acid protein with a Val to Ala substitution at position 361 compared with the above published gene(Fig.1).Neither gene possessed a signal peptide at the N-terminal according to an iPSORT prediction; therefore,MaβFS1 and MaβFS2 were predicted to act in the cytoplasm,the supposed site for sesquiterpene biosynthesis[46].
Although there were remarkable sequence differences among EβF synthase genes fromvarious plant varieties/species, some regions,including the Asp-rich motifknown as DDXXD(at positions 301 to 305 in MaβFS1)and the NSE/DTE motif known as(N/D)DXX(S/T)XXX(E/D)(at positions 444 to 452 in MaβFS1), were highly conserved(Fig.1).Further phylogenetic reconstruction revealedthat MaβFS1 and MaβFS2 were more closely related to other terpene synthases from black peppermint or related species than to their counterparts from distant species(Fig.2).
PCR amplification of gDNA revealed that the whole length of the MaβFS1 genomic sequence was 2679 bp(deposited in GenBank under accession number HQ337898).It has seven exons of 114,256,376,219,139,246 and 303 bp,interspersed by six introns of approximately 102,68,368,124,287 and 77 bp, respectively(Fig.3-A).The length of the MaβFS2 genomic sequence was 2730 bp(deposited in GenBank under accession number HQ337899),with seven exons of 114,256,376,219,139, 246 and 303 bp interspersed by six introns of 102,76,409,124, 287 and 79 bp,respectively(Fig.3-B).
3.2.Tissue expression pattern of MaβFS1 in Asian peppermint
There was only one amino acid difference(Val to Ala at position 361)between MaβFS1 and MaβFS2,and it was not located in any putative functional domain.MaβFS1 was identical to the published EβF synthase gene from black peppermint(GenBank accession number AF024615)at the amino acid sequence level.As this gene had been reported to have activity in vitro[17]we chose MaβFS1 for further characterization.RNA was isolated from roots,stems,leaves and flowers of Asian peppermint at the flowering stage.To discriminate against amplification products from contaminating genomic DNA,specific primers(MaβFS F2 and MaβFS R2) were designed with the reverse primer spanning the fifth andsixth exons according to the MaβFS1 gene structure(Fig.3-A). qRT-PCR results indicated that MaβFS1 was not exclusively expressed in a certain tissue in Asian peppermint,but its expression level in the stem,leaf and flower was about 1.01, 1.31,and 1.78 times higher,respectively,than that in the root (Fig.4).This was consistent with EβF emission levels in Garland (Chrysanthemum coronarium)where expression was higher in reproductive organs than in other tissues[26].

Fig.1–Comparison of the deduced amino acid sequences of the plant-derived EβF synthases.1–4,EβF synthases from Douglas fir,sweet wormwood,Yuzu and black peppermint,respectively;5,MaβFS1;6,MaβFS2.Amino acids identical in all six genes are marked in black.Amino acids identical in five genes are marked in blue.The highly conserved DDXXD region is marked with boxІ,the NSE/DTE motif is marked with boxП,and the triangle indicates the Val to Ala substitution at position 361 of MaβFS2 compared with MaβFS1 and the published gene.
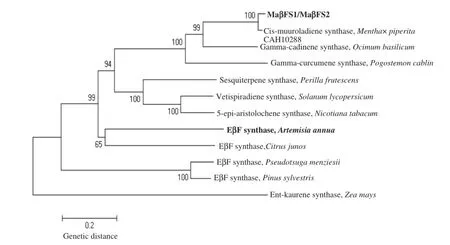
Fig.2–Phylogenetic analysis of MaβFS1,MaβFS2 and other representative sesquiterpene synthases.The joint unrooted tree was generated using MEGA4 by the neighbor-joining method.Bootstrap values from 10,000 replicates are indicated at each branch.The GenBank accession numbers of each appended protein are as follows:Cis-muuroladiene synthase (Mentha×piperita):CAH10288;Gamma-cadinene synthase(Ocimum basilicum):AAV63787;Gamma-curcumene synthase (Pogostemon cablin):AAS86319;Sesquiterpene synthase(Perilla frutescens):AAX16076;Vetispiradiene synthase(Solanum lycopersicum):AAG09949;5-epi-aristolochene synthase(Nicotiana tabacum):157837030;EβF synthase(Artemisia annua): AAX39387;EβF synthase(Citrus junos):AAK54279;EβF synthase(Pseudotsuga menziesii):AAX07265;EβF synthase(Pinus sylvestris):ADH29869;Ent-kaurene synthase(Zea mays):AAD34319.
3.3.Generation and molecular characterization of transgenic tobacco plants
To determine if transgenic plants containing MaβFS1 had enhanced ability to control aphids the pBI121 plasmids containing cDNAs of MaβFS1(Fig.5-A)were transferred into tobacco. Positive MaβFS1 transgenic tobacco plants in the T0–T2generations were selected by PCR(PCR results of the T2generation are shown in Fig.5-B)and RT-PCR analysis(data not shown);11 stably inherited MaβFS1 lines(designed Ma1 to Ma11)were obtained.According to the results of RT-PCR,three T2tobacco lines(Ma1,Ma4,Ma10)were chosen for further qRT-PCR analysis,which indicated that the expression levels of the transgenic lines were different(Fig.5-C).For example,the expression level in Ma4 was about 5.4 times higher than that of Ma1.
3.4.EβF emission from MaβFS1 transgenic lines
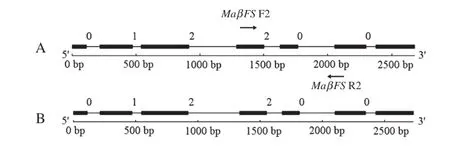
Fig.3–GSDS output of the MaβFS1 and MaβFS2 gene structure.MaβFS F2 and MaβFS R2 were the specific primers used for pattern analysis of tissue expression.Exons and introns are indicated by black rectangles and lines,respectively.0,1,and 2 represented the intron phases.A:GSDS output of the MaβFS1 gene structure.B:GSDS output of the MaβFS2 gene structure.
Volatiles from the control and three MaβFS1 transgenic lines (Ma1,Ma4,Ma10)were collected and analyzed by coupled GC–MS.MaβFS1 transgenic lines showed a unique peak compared with the control(Fig.6-A,B).The peak was identified as EβF with a retention time and mass spectrum identical to that of authentic EβF(Fig.6-C,D).EβF emission levels of the Ma1,Ma4,and Ma10 transgenic lines were 2.81,4.85 and 2.62 ng d−1g−1in fresh tissues,respectively.

Fig.4–qRT-PCR analysis of tissue-specific expression of MaβFS in Asian peppermint.The expression levels of MaβFS in stems,leaves and flowers are relative to average root levels. The average and SE ofthree technical replicates are presented.
3.5.Tritrophic interactions involving peach aphids,predatory lacewing larvae and MaβFS1 transgenic tobacco
To test the efficacy of the transgenic lines in control of aphids, two independent evaluations were conducted in a setup as indicated in Fig.7-A.In the repellence test,the numbers of aphids on transgenic and control tobacco plants were counted 12 h after the aphids were released.Compared with the control,aphids on transgenic lines Ma1,Ma4,and Ma10, were reduced by approximately 8.8%,10.4%and 7.7%,respectively,whereas about 25.5%of the aphids stayed on the net cover or died(Fig.7-B;Table 2).When 400 alate aphids and 10 lacewing larvae were simultaneously introduced into the setup,the numbers of aphids after 12 h were reduced by 19.2% in Ma1,29.5%in Ma4(P<0.05)and 16.7%in Ma10,compared with the control.Mostofthe surviving aphids were preyed upon by the lacewing larvae or stayed on the net cover(Fig.7-C; Table 3).Therefore,MaβFS1 transgenic tobacco plants showed a pleiotropic effect on aphid behavior,including repellence to aphids and attraction to aphid predators.Notably,in the presence of lacewing larvae,transgenic line Ma4 could recruit lacewing larvae that significantly affected aphid infestation.

Fig.5–Molecular analyses of tobacco plant lines transformed with MaβFS1.A:schematic of the MaβFS1 gene expression cassette in the pBI121 plasmid.Nos P:Nos promoter;Nos T:Nos terminator;CaMV 35S P:CaMV 35S promoter;B:PCR identification of MaβFS1 T2transgenic lines.M:DL2000 DNA marker;C+:MaβFS1-pBI121 plasmid;Ma1,Ma4 and Ma10:positive transgenic tobacco lines;C:qRT-PCR analysis of the expression of MaβFS1 in T2transgenic lines and vector control.Ma1,Ma4, and Ma10:positive transgenic tobacco lines.The expression levels of the target gene in Ma4 and Ma10 were relative to that in Ma1.The average and SE of three technical replicates are presented.
4.Discussion
4.1.Characteristics of the MaβFS1 and MaβFS2 genes isolated from Asian peppermint
In this study,we isolated MaβFS1 and MaβFS2 genes from Asian peppermint and showed that MaβFS1 was functional in tobacco.Although MaβFS1 was identical to the published EβF synthase gene from black peppermint(AF024615),it shared only 34.1%and 28.2%similarities at the amino acid sequence level with EβF synthases from Yuzu and Douglas fir,respectively,and only 34.0%similarity with that of the gene we isolated from sweet wormwood and characterized in vivo (Fig.1)[39].So far,the EβF synthase genes from Douglas fir, Yuzu,sweet wormwood and black peppermint have been isolated and characterized in vitro[37],and only the genes from black peppermint and sweet wormwood were successfully introduced into plants and proved to be functionalinvivo[38,39]. Furthermore,remarkable sequence differences were observed among the EβF synthase genes from various plant species/ varieties.However,some regions,including the Asp-rich motif known as DDXXD(at positions 301 to 305 in MaβFS1)and the NSE/DTE motif known as(N/D)DXX(S/T)XXX(E/D)(at positions 444 to 452 in MaβFS1),are highly conserved among the so far isolated plant-derived EβF synthase genes(Fig.1).These two conserved domains are supposed to be responsible for divalent metal ion–substrate binding during catalysis[47].This observation indicates that the steric effect and the physico-chemical nature of the amino acid rather than the sequence itself might have important roles underlying the EβF synthase catalysis mechanism.
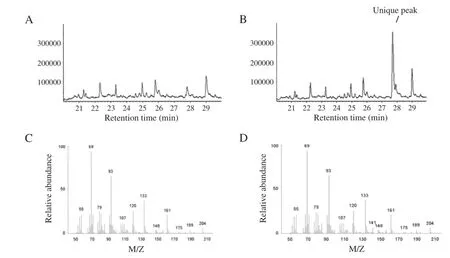
Fig.6–GC–MS profiles of volatiles emitted by MaβFS1 transgenic tobacco plants.A:volatiles from control plants;B:volatiles from MaβFS1 transgenic lines Ma4;C:mass spectrum of the unique peak;D:mass spectrum of commercially authentic EβF.
According to Trapp and Croteau[48]the terpene synthase genes can be classified into three classes by comparison of intron/exon patterns.Class I contains 12–14 introns,class II nine introns and class III six introns.The MaβFS1 and MaβFS2 genes described here had six introns and fall into class III; however,their counterpart from black peppermint isolated by Prosser et al.[40]had seven introns and did not belong to any of the above categories.It remains unclear how the variations of intron number affect the production of EβF or other terpenes.
Different terpene synthase genes have different tissue expression patterns.Of the 32 terpene synthase genes isolated in Arabidopsis,20 were expressed in flowers(six of them exclusively or almost exclusively so),11 were expressed in leaves,nine in stems and 12 in roots;eight genes were expressed in all of the selected organs[49].Based on the qRT-PCR analysis presented here(Fig.4),MaβFS1 expressed in allthe selected organs of Asian peppermint with the expression level in flowers being higher than in roots,stems and leaves.
4.2.The supply of FPP substrate might be a limiting factor in managing sesquiterpene synthesis by genetic manipulation

Fig.7–Tritrophic interactions involving peach aphids,lacewing larvae and transgenic tobacco plants expressing MaβFS1. Average numbers of aphids or lacewing larvae,SE were for three replicates.A:schematic of the setup used for testing.Dark plants represent the transgenic lines Ma1,Ma4 and Ma10;the gray plants are the control tobacco line.Alate aphids and lacewing larvae were released at the midpoint in each assay;B:repellence tests of aphids among MaβFS1 transgenic and control lines;C:evaluation of aphid control efficiency of MaβFS1 lines following release of 400 alate aphids and 10 lacewing larvae into the setup.Small letters indicate a significant differences at P=0.05.
To date,metabolic engineering of terpenoids in plants has met with some success,particularly monoterpenes.However,the low sesquiterpene production of transgenic plants overexpressing sesquiterpene synthase genes seems to be ageneral phenomenon,indicating that engineering sesquiterpene production in plants is a challenging task[37].For example,transgenic Arabidopsis plants overexpressing the FaNES1 gene also emitted the sesquiterpene nerolidol,but at a level 100 to 300-fold lower than that of linalool[50].Attempts to engineer synthesis of sesquiterpenes in tobacco have also been made with a fungal trichodiene synthase[51]and only small amounts of the expected sesquiterpenes were detected.When another sesquiterpene synthase,the amorpha-4,11-diene synthase gene from Artemisia annua,was transformed into tobacco,the production of amorpha-4,11-diene was 0.2 to 1.7 ng d−1g−1fresh weight[52].Overexpression of EβF synthase genes from sweet wormwood in tobacco emitted EβF at 1.55 to 4.65 ng g−1fresh tissue[39].Similarly,the EβF emission levels of MaβFS1 transgenic lines Ma1,Ma4 and Ma10 presented here were 2.81,4.85,and 2.62 ng d−1g−1fresh tissues.In these experiments, the strong and constitutive 35S promoter was used to direct the engineered sesquiterpene synthases targeting to the cytosol,the predicted location of FPP,the precursor for sesquiterpene synthesis.Therefore,the low emission level of EβF might be due to the limited supply of FPP substrate.

Table 2–Numbers of aphids on plants 12 h after release of 200 alate individuals.
4.3.Genetic engineering of plant-derived EβF synthase genes in other crop plants might be a valuable alternative strategy to recruit predators for aphid control
Exploring and characterizing different plant-derived EβF synthase genes can add value to the use of these genes in engineering other plants to produce EβF and hence to exploit the pheromonal properties of natural products for plant defense against aphids[37].In this study,transgenic tobacco continuously emitting EβF was generated.The phenotype might alter many other characteristics affecting aphid infestation,but here the GC–MS results of the transgenic tobacco plants showed that MaβFS1 transgenic tobacco lines had similar volatile profiles to the control(blank vector control), except for a unique peak identified as EβF(Fig.6-A,B).From this,we assumed that the effects of MaβFS1 transgenic lines on aphid and lacewing behavior were mainly due to the emission of EβF.However,as indicated in Fig.7-B,when both transgenic and the control plants were located in the same setup,no strong repellence of aphids was observed,and the reductions in aphid infestation levels on transgenic plants were statistically significant only when lacewing larvae were present (Fig.7-C),demonstrating that the reduction in aphid infestation was due to both repellence to aphids and attraction of lacewing predators.It was reported that the rate of EβF emission per day rather than the amountof EβF emitted regulated the proportion of wing offspring produced[16].In both the laboratory and field, applications of 1000 ng synthetic EβF three times a day were enough to be perceived by the aphids and to elicit alarm behavior although high volatility and a short atmospheric lifespan of EβF and airflow in the field very likely reduced the amount and concentration of EβF[16,53].In this context,the emission levels of EβF from the transgenic tobacco plants at flowering in this study were 2.62 to 4.85 ng d−1g−1of fresh tissue.Assuming that the fresh tissues of each transgenic tobacco plant weighed 1.000 g or more than that at flowering time,the EβF emission level of each transgenic plant would be enough to elicit aphid repellence(escape behavior).Therefore, we may presume that in addition to the limited amount of EβF released by the transgenic tobacco plants,the emission of other volatiles from both the control and transgenic plants might have inhibitory effects on the repellence of aphids.The full alarm response(repellence)of aphids in transgenic tobacco plants was not observed in this study.

Table 3–Numbers of aphids on plants following simultaneous release of 400 alate individuals and 10 lacewing larvae.
5.Conclusions
The research presented in this study isolated EβF synthase genes from Asian peppermint and elucidated the expression profile of MaβFS1 for the first time.It also suggested that an EβF synthase gene from Asian peppermint could be employed in genetic engineering of economically important crop plants for aphid control through continuous emission of EβF and recruitment of biological control agents.
Acknowledgments
This work was funded by the Research Initiative on Development of Transgenic Wheat Plants supported by the Chinese Ministry of Agriculture(2011ZX08002-001),the Natural Science Foundation of China(31171618),and the Chinese State Key Laboratory for Biology of Plant Diseases and Insects(SKLOF201307).
R E F E R E N C E S
[1]A.F.G.Dixon,Aphid Ecology:An Optimization Approach, Chapman and Hall,London,1998.
[2]F.L.Goggin,Plant–aphid interactions:molecular and ecological perspectives,Curr.Opin.Plant Biol.10(2007)399–408.
[3]R.L.Blackman,V.F.Eastop,Aphids on the World's Crops:An Identification and Information Guide,John Wiley,Chichester, 1984.476.
[4]W.P.Morrison,F.B.Peairs,Response model concept and economic impact,in:S.S.Quisenberry,F.B.Peairs(Eds.), Response Model for an Introduced Pest–The Russian WheatAphid,Entomological Society of America,Lanham,1998, pp.1–11.
[5]C.S.Awmack,R.Harrington,Elevated CO2affects the interactions between aphid pests and host plant flowering, Agric.For.Entomol.2(2000)57–61.
[6]M.A.Aqueel,S.R.Leather,Effect of nitrogen fertilizer on the growth and survival of Rhopalosiphum padi(L.)and Sitobion avenae(F.)(Homoptera:Aphididae)on different wheat cultivars,Crop Prot.30(2011)216–221.
[7]B.Sabater-Muñoz,F.Legeai,C.Rispe,J.Bonhomme,P. Dearden,C.Dossat,A.Duclert,J.P.Gauthier,D.Giblot Ducray, W.Hunter,P.Dang,S.Kambhampati,D.Martinez-Torres,T. Cortes,A.Moya,A.Nakabachi,C.Philippe,N. Prunier-Leterme,Y.Rahbé,J.C.Simon,D.Stern,P.Wincker,D. Tagu,Large-scale gene discovery in the pea aphid Acyrthosiphon pisum(Hemiptera),Genome Biol.7(2006)R21.
[8]V.A.Hilder,K.S.Powell,A.M.R.Gatehouse,J.A.Gatehouse,Y. Shi,W.D.O.Hamilton,A.Merryweather,C.A.Newell,J.C. Timans,W.J.Peumans,E.Van Damme,D.Boulter,Expression of snowdrop lectin in transgenic tobacco plants results in added protection against aphids,Transgenic Res.4(1995) 18–25.
[9]Y.Yu,Z.M.Wei,Increased oriental armyworm and aphid resistance in transgenic wheat stably expressing Bacillus thuringiensis(Bt)endotoxin and Pinellia ternate agglutinin (PTA),Plant Cell Tiss.Org.Cult.94(2008)33–44.
[10]A.N.E.Birch,I.E.Geoghegan,M.E.N.Majerus,J.W.McNicol, C.A.Hackett,A.M.R.Gatehouse,J.A.Gatehouse,Tritrophic interactions involving pest aphids,predatory 2-spot ladybirds and transgenic potatoes expressing snowdrop lectin for aphid resistance,Mol.Breed.5(1999)75–83.
[11]P.A.M.Hogervorst,F.L.Wäckers,J.Woodring,J.Romeis, Snowdrop lectin(Galanthus nivalis agglutinin)in aphid honeydew negatively affects survival of a honeydew-consuming parasitoid,Agric.For.Entomol.11 (2009)161–173.
[12]W.S.Bowers,L.R.Nault,R.E.Webb,S.R.Dutky,Aphid alarm pheromone:isolation,identification,synthesis,Science 177 (1972)1121–1122.
[13]J.A.Pickett,D.C.Griffiths,Composition of aphid alarm pheromones,J.Chem.Ecol.6(1980)349–360.
[14]F.Francis,S.Vandermoten,F.Verheggen,G.Lognay,E. Haubruge,Is the(E)-β-farnesene only volatile terpenoid in aphids?J.Appl.Entomol.129(2005)6–11.
[15]M.J.Lewis,I.M.Prosser,A.Mohib,L.M.Field,Cloning and characterisation of a prenyltransferase from the aphid Myzus persicae with potential involvement in alarm pheromone biosynthesis,Insect Mol.Biol.17(2008)437–443.
[16]G.Kunert,S.Otto,S.R.Rose,J.Gershenzon,W.W.Weisser, Alarm pheromone mediates production of winged dispersal morphs in aphids,Ecol.Lett.8(2005)596–603.
[17]J.Crock,M.Wildung,R.Croteau,Isolation and bacterial expression of a sesquiterpene synthase cDNA clone from peppermint(Mentha x piperita,L.)that produces the aphid alarm pheromone(E)-β-farnesene,Proc.Natl.Acad.Sci.U.S.A. 94(1997)12833–12838.
[18]P.Wohlers,Aphid avoidance of plants contaminated with alarm pheromone(E)-β-farnesene,Z.Angew.Ent.92(1981) 329–336.
[19]F.M.Elagamy,K.F.Haynes,Susceptibility of the pea aphid (Homoptera:Aphididae)to an insecticide and a predator in the presence of synthetic aphid alarm pheromone,J.Econ. Entomol.85(1992)794–798.
[20]E.Roditakis,I.D.Couzin,K.Balrow,N.R.Franks,A.K. Charnley,Improving secondary pick up of insect fungal pathogen conidia by manipulating host behaviour,Ann. Appl.Biol.137(2000)329–335.
[21]J.W.Zhu,A.A.Cossé,J.J.Obrycki,K.S.Boo,T.C.Baker, Olfactory reactions of the twelve-spotted lady beetle, Coleomegilla maculata and the green lacewing,Chrysoperla carnea to semiochemicals released from their prey and host plant:electroantennogram and behavioral responses, J.Chem.Ecol.25(1999)1163–1177.
[22]F.Francis,G.Lognay,E.Haubruge,Olfactory responses to aphid and host plant volatile releases:E-β-Farnesene an effective kairomone for the predator Adalia bipunctata,J. Chem.Ecol.30(2004)741–755.
[23]N.Harmel,R.Almohamad,M.L.Fauconnier,P.Du Jardin,F. Verheggen,M.Marlier,E.Haubruge,F.Francis,Role of terpenes from aphid-infested potato on searching and oviposition behavior of Episyrphus balteatus,Insect Sci.14 (2007)57–63.
[24]S.P.Foster,I.Denholm,R.Thompson,G.M.Poppy,W.Powell, Reduced response of insecticide-resistant aphids and attraction of parasitoids to aphid alarm pheromone;a potential fitness trade-off,Bull.Entomol.Res.95(2005)37–46.
[25]E.Máday,É.Szöke,Z.Muskath,E.Lemberkovičs,A study of the production of essential oils in chamomile hairy root cultures, Eur.J.Drug Metab.Pharmacokinet.24(1999)303–308.
[26]G.Flamini,P.L.Cioni,I.Morelli,Differences in the fragrances of pollen,leaves,and floral parts of Garland(Chrysanthemum coronarium)and composition of the essential oils from flowerheads and leaves,J.Agric.Food Chem.51(2003)2267–2271.
[27]T.J.A.Bruce,M.A.Birkett,J.Blande,A.M.Hooper,J.L.Martin,B. Khambay,I.Prosser,L.E.Smart,L.J.Wadhams,Response of economically important aphids to components of Hemizygia petiolata essential oil,Pest Manag.Sci.61(2005)1115–1121.
[28]M.Miyazawa,N.Tamura,Components of the essential oil from sprouts of Polygonum hydropiper L.(‘Benitade'),Flavour Fragr.J.22(2007)188–190.
[29]R.W.Gibson,J.A.Pickett,Wild potato repels aphids by release of aphid alarm pheromone,Nature 302(1983)608–609.
[30]G.W.Dawson,D.C.Griffiths,J.A.Pickett,M.C.Smith,C.M. Woodcock,Natural inhibition of the aphid alarm pheromone, Entomol.Exp.Appl.36(1984)197–199.
[31]D.A.Avé,P.Gregory,W.M.Tingey,Aphid repellent sesquiterpenes in glandular trichomes of Solanum berthaultii and S.tuberosum,Entomol.Exp.Appl.44(1987)131–138.
[32]T.C.J.Turlings,M.Bernasconi,R.Bertossa,F.Bigler,G.Caloz, S.Dorn,The induction of volatile emissions in maize by three herbivore species with different feeding habits:possible consequences for their natural enemies,Biol.Control 11 (1998)122–129.
[33]R.Mumm,K.Schrank,R.Wegener,S.Schulz,M.Hilker,Chemical analysis of volatiles emitted by Pinus sylvestris after induction by insect oviposition,J.Chem.Ecol.29(2003)1235–1252.
[34]M.De Vos,W.Y.Cheng,H.E.Summers,R.A.Ragusob,G. Jandera,Alarm pheromone habituation in Myzus persicae has fitness consequences and causes extensive gene expression changes,Proc.Natl.Acad.Sci.U.S.A.107(2010)14673–14678.
[35]M.L.Bernasconi,T.C.J.Turlings,L.Ambrosetti,P.Bassetti,S. Dorn,Herbivore-induced emissions of maize volatiles repel the corn leaf aphid,Rhopalosiphum maidis,Entomol.Exp.Appl. 87(1998)133–142.
[36]R.Mumm,M.Hilker,The significance of background odour for an egg parasitoid to detect plants with host eggs,Chem. Senses 30(2005)1–7.
[37]X.D.Yu,J.A.Pickett,Y.Z.Ma,T.Bruce,J.Napier,J.A.Pickett, H.D.Jones,L.Q.Xia,Metabolic engineering of plant-derived (E)-β-farnesene synthase genes for a novel type of aphid-resistant GM crop plants,J.Integr.Plant Biol.54(2012) 282–299.
[38]M.H.Beale,M.A.Birkett,T.J.A.Bruce,K.Chamberlain,L.M. Field,A.K.Huttly,J.L.Martin,R.Parker,A.L.Phillips,J.A. Pickett,I.M.Prosser,P.R.Shewry,L.E.Smart,L.J.Wadhams, C.M.Woodcock,Y.Zhang,Aphid alarm pheromone produced by transgenic plants affects aphid and parasitoid behavior, Proc.Natl.Acad.Sci.U.S.A.103(2006)10509–10513.
[39]X.D.Yu,H.D.Jones,Y.Z.Ma,G.P.Wang,Z.S.Xu,B.M.Zhang,Y.J. Zhang,G.W.Ren,J.A.Pickett,L.Q.Xia,(E)-β-Farnesene synthase genes affect aphid(Myzus persicae)infestation in tobacco (Nicotiana tabacum),Funct.Integr.Genomics 12(2012)207–213.
[40]I.M.Prosser,R.J.Adams,M.H.Beale,N.D.Hawkins,A.L. Phillips,J.A.Pickett,L.M.Field,Cloning and functional characterisation of a cis-muuroladiene synthase from black peppermint(Mentha×piperita)and direct evidence for a chemotype unable to synthesise farnesene,Phytochemistry 67(2006)1564–1571.
[41]C.M.Starks,K.W.Back,J.Chappell,J.P.Noel,Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase,Science 277(1997)1815–1820.
[42]K.J.Livak,T.D.Schmittgen,Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCtmethod,Methods 25(2001)402–408.
[43]R.B.Horsch,R.T.Fraley,S.G.Rogers,P.R.Sanders,A.Lloyd,N. Hoffmann,Inheritance of functional foreign genes in plants, Science 223(1984)496–498.
[44]I.F.Kappers,A.Aharoni,T.W.J.M.van Herpen,L.L.P. Luckerhoff,M.Dicke,H.J.Bouwmeester,Genetic engineering of terpenoid metabolism attracts bodyguards to Arabidopsis, Science 309(2005)2070–2072.
[45]K.N.Berk,P.Carey,Data Analysis with Microsoft Excel, Duxbury,Pacific Grove,2004.579.
[46]J.Chappell,Biochemistry and molecular biology of the isoprenoid biosynthetic pathway in plants,Annu.Rev.Plant. Physiol.Plant.Mol.Biol.46(1995)521–547.
[47]J.P.Noel,N.Dellas,J.A.Faraldos,M.Zhao,B.A.Hess Jr.,L. Smentek,R.M.Coates,P.E.O'Maille,Structural elucidation of cisoid and transoid cyclization pathways of a sesquiterpene synthase using 2-fluorofarnesyl diphosphates,ACS Chem. Biol.5(2010)377–392.
[48]S.C.Trapp,R.B.Croteau,Genomic organization of plant terpene synthases and molecular evolutionary implications, Genetics 158(2001)811–832.
[49]F.Chen,D.Tholl,J.C.D'Auria,A.Farooq,E.Pichersky,J. Gershenzon,Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers,Plant Cell 15(2003) 481–494.
[50]A.Aharoni,A.P.Giri,S.Deuerlein,F.Griepink,W.J.de-Kogel, F.W.A.Verstappen,H.A.Verhoeven,M.A.Jongsma,W. Schwab,H.J.Bouwmeester,Terpenoid metabolism in wildtype and transgenic Arabidopsis plants,Plant Cell 15 (2003)2866–2884.
[51]T.M.Hohn,J.B.Ohlrogge,Expression of a fungal sesquiterpene cyclase gene in transgenic tobacco,Plant Physiol.97(1991)460–462.
[52]T.E.Wallaart,H.J.Bouwmeester,J.Hille,L.Poppinga,N.C.A. Maijers,Amorpha-4,11-diene synthase:cloning and functional expression of a key enzyme in the biosynthetic pathway of the novel antimalarial drug artemisinin,Planta 212(2001)460–465.
[53]E.Hatano,G.Kunert,W.W.Weisser,Aphid wing induction and ecological costs of alarm pheromone emission under field conditions,PLoS One 5(2010)e11188.
*Corresponding author.Tel.:+86 10 82109718.
**Corresponding author.Tel.:+86 10 82105804.
E-mail addresses:mayouzhi@caas.cn(Y.Ma),xialanqin@caas.cn(L.Xia).
Peer review under the responsibility of Crop Science Society of China and Institute of Crop Science,CAAS.
Mentha asiatica
Myzus persicae
Nicotiana tabacum
- The Crop Journal的其它文章
- Zea mays(L.)P1 locus for cob glume color identified as a post-domestication selection target with an effect on temperate maize genomes
- Anatomical and chemical characteristics associated with lodging resistance in wheat
- Difference between resistant and susceptible maize to systematic colonization as revealed by DsRed-labeled Fusarium verticillioides
- Identification and fine mapping of two blast resistance genes in rice cultivar 93-11
- Zea mays(L.) P1 locus for cob glume color identified as a post-domestication selection target with an effect ontemperate maize genomes
- Identification of unconditional and conditional QTL for oil,protein and starch content in maize

