Identification and fine mapping of two blast resistance genes in rice cultivar 93-11
Cilin Lei,Kun Ho,Yilong Yng,Jin M,Shui Wng,Jiulin Wng,Zhijun Cheng,Shsh Zho,Xin Zhng,Xiuping Guo,Chunming Wng,Jinmin Wn,,*
aInstitute of Crop Science,Chinese Academy of Agricultural Sciences,The National Key Facility for Crop Gene Resources and Genetic Improvement,Beijing 100081,China
bKey Laboratory of Crop Genetics and Germplasm Enhancement,Jiangsu Provincial Center of Plant Gene Engineering,Nanjing Agricultural University,Nanjing,Jiangsu 210095,China
1.Introduction
Rice blast,caused by the fungus Magnaporthe oryzae,is an important disease in most rice production regions of the world because of its devastating effects on yield.In this pathosystem,pathotype-or race-specific resistance follows the gene-for-gene relationship[1].M.oryzae is highly variable,and loss of resistance in varieties is quite common [2,3],especially when resistance is based on a single resistance (R) gene [4–6].Nevertheless,the utilization of R genes is still considered to be the most effective and economical method to control the disease.A major strategy to develop more durable resistance is to combine multiple R genes that confer overlapping resistance spectra to multiple isolates/races of M.oryzae in a single variety [7].In this regard,continued identification of new R genes in genetic resource materials is essential.
Genetic studies on blast resistance began as early as the 1960s and were intensified with the availability of genome sequences of the two subspecies of cultivated rice,Oryza sativa (ssp.japonica cultivar(cv.)Nipponbare and spp.indica cv.93-11)and abundant genetic markers[8–10].To date,more than 70 R genes and some quantitative trait loci(QTL)have been identified and mapped on rice chromosomes[11–13].These R genes are largely clustered on chromosomes 6,11 and 12,and involve different specificities[11–15].Development of DNA markers closely linked to the R genes not only sets the stage for marker-assisted selection(MAS)in rice breeding programs,but also facilitates map-based cloning.Some blast R genes have been finely mapped [11,12,15–21],and among them Pib[22],Pita[23],Pi9[24],Piz-t and Pi2[25],Pid2[26],Pi36[27],Pi37[28],Pikm[29],Pi5[30],Pid3/Pi25[31,32],Pit[33],Pish[34],pi21 [35],Pb1 [36],Pia/PiCO39 [37,38],Pi-kh/Pi54 [39],Pik [40],Pik-p[41]and Pi1[21]have been isolated.Markers tightly linked to the R genes,and more recently,markers derived from cloned R genes should greatly facilitate pyramiding of the R genes into cultivars by MAS; for example,markers developed from Pita[42,43]and Pib[18].
The sequenced indica cv.93-11 is a widely grown blast resistant cultivar and hybrid rice restorer in China[9,44–47].It is resistant to M.oryzae races ZA49,ZE3 and ZG1 from Jiangsu,China[44],and to 80%of 45 M.oryzae isolates(22 from japonica and 23 from indica)from other provinces in China[48].Although blast R gene Pi41 was previously reported in cv.93-11 [47],additional R genes must also be present [26,31,49–53].The objectives of the present study were to evaluate blast resistance in cv.93-11 using a wide range of Chinese M.oryzae isolates,and to identify and map R genes additional to Pi41.
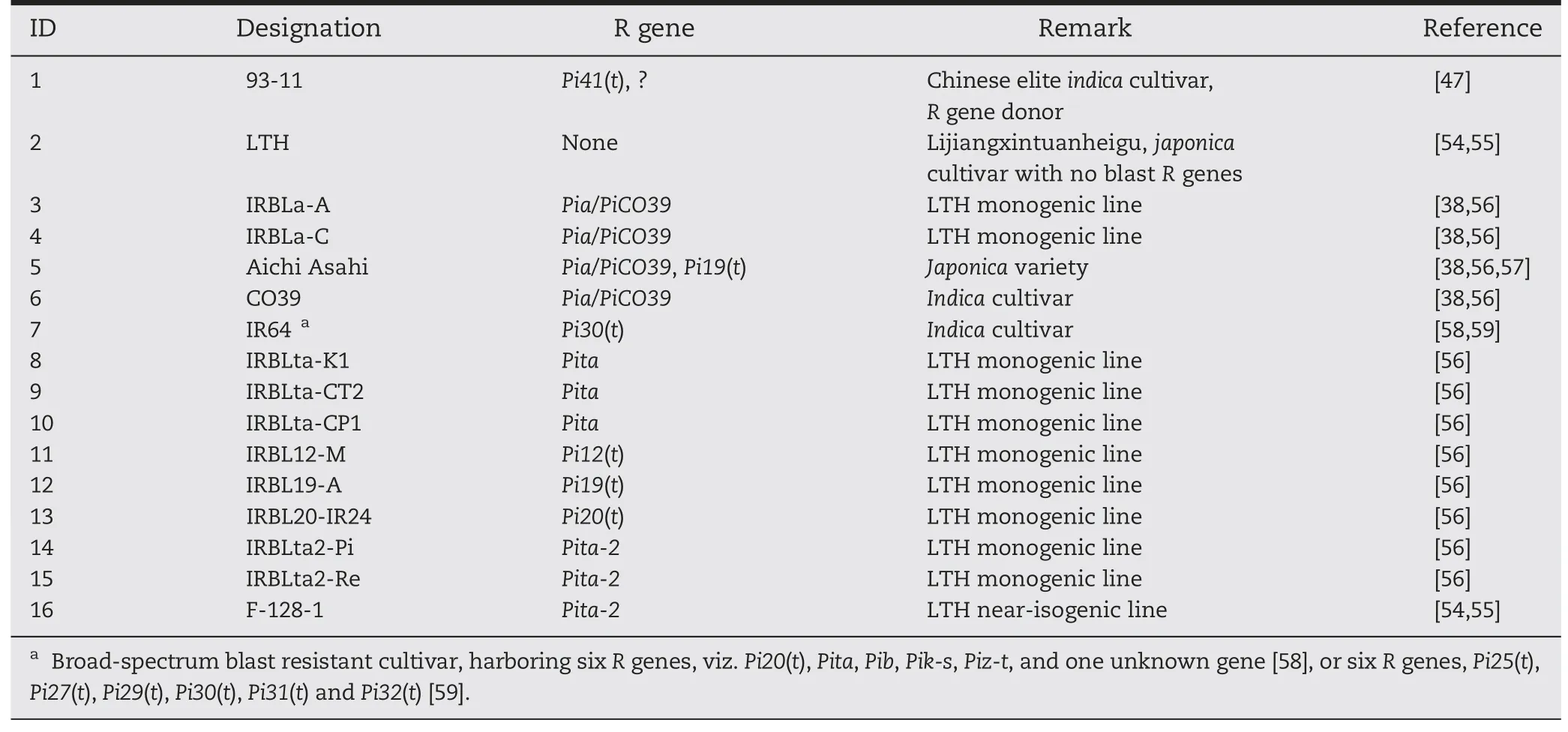
Table 1-Rice genotypes evaluated for blast resistance.
2.Materials and methods
2.1.Rice genotypes and culture
Five rice cultivars,one near-isogenic line (NIL) and ten monogenic lines(MLs)were used in this study(Table 1).Indica cv.93-11(resistant,male parent) and japonica cv.Lijiangxintuanheigu(LTH,susceptible,female parent)were evaluated for reaction to M.oryzae isolates,and crossed to develop F2and F3populations for genetic analysis and gene mapping.Cultivars CO39,Aichi Asahi and IR64 together with 11 NILs/MLs,each carrying a single R gene(Table 1),were used as reference lines to differentiate the genes mapped in 93-11 from previously reported R genes.93-11,LTH,Aichi Asahi,IR64 and F-128-1 are maintained at the Institute of Crop Science,Chinese Academy of Agricultural Sciences(CAAS),Beijing,China.CO39 and the other 10 MLs were kindly provided by Dr.Yoshimichi Fukuta,International Rice Research Institute(IRRI),Los Baños,The Philippines.
Seedlings were grown in 60 cm × 30 cm × 5 cm plastic seedling trays in a greenhouse.Pre-soaked seeds of test materials were sown in rows together with the parents.For genetic analysis and gene mapping,300 seeds of the F2population and 15 seeds of each parent were sown per tray.For differentiating R genes,15 seeds of each of the two parents and 11 reference lines(Table 1)were sown in each tray with two replications.
2.2.M.oryzae isolates and conidial inoculation
A total of 495 M.oryzae isolates from different rice growing regions were used to evaluate the reaction of cv.93-11.Among them,five isolates – three indica-derived isolates,001-99-1(pathotype 241.6,from Jiangsu of China),RB17 (pathotype 423.2,from Hunan of China),and GZ26(pathotype 541.0,from Guizhou of China),and two japonica-derived isolates,99-26-2(pathotype 437.1,from Beijing of China) and P-2b (pathotype 303.0,from Japan) – provided clear resistant or susceptible reactions on the two parents and 11 reference lines (Table 1)and were chosen for further studies.
All isolates are stored at the Institute of Crop Science,CAAS,Beijing,China.Inoculum preparation and seedling inoculation followed the procedure described by Chen et al.[60].Disease reactions were evaluated 6–7 days after inoculation on a numerical scale ranging from 0–3(resistant)to 4–5(susceptible)as described by Mackill and Bonman[4].Observed reactions were verified in the field following injection-inoculation as described by Lei et al.[61].

Table 2-Primer sequences of InDel markers used for mapping gene Pi60(t).
2.3.Development of markers and construction of a linkage map
Genomic DNA was extracted from seedling leaves using the CTAB method [62].Two sets of DNA bulks,each containing a resistant pool and a susceptible pool,were prepared following the methods described by Yang et al.[47].The first set (set 1,for Pi60(t)) was assembled with equimolar amounts of DNA from 15 isolate 001-99-1-resistant or 15 susceptible F2individuals,and the second set (set 2,for Pi61(t)) from 15 isolate 99-26-2-resistant or susceptible F2individuals.
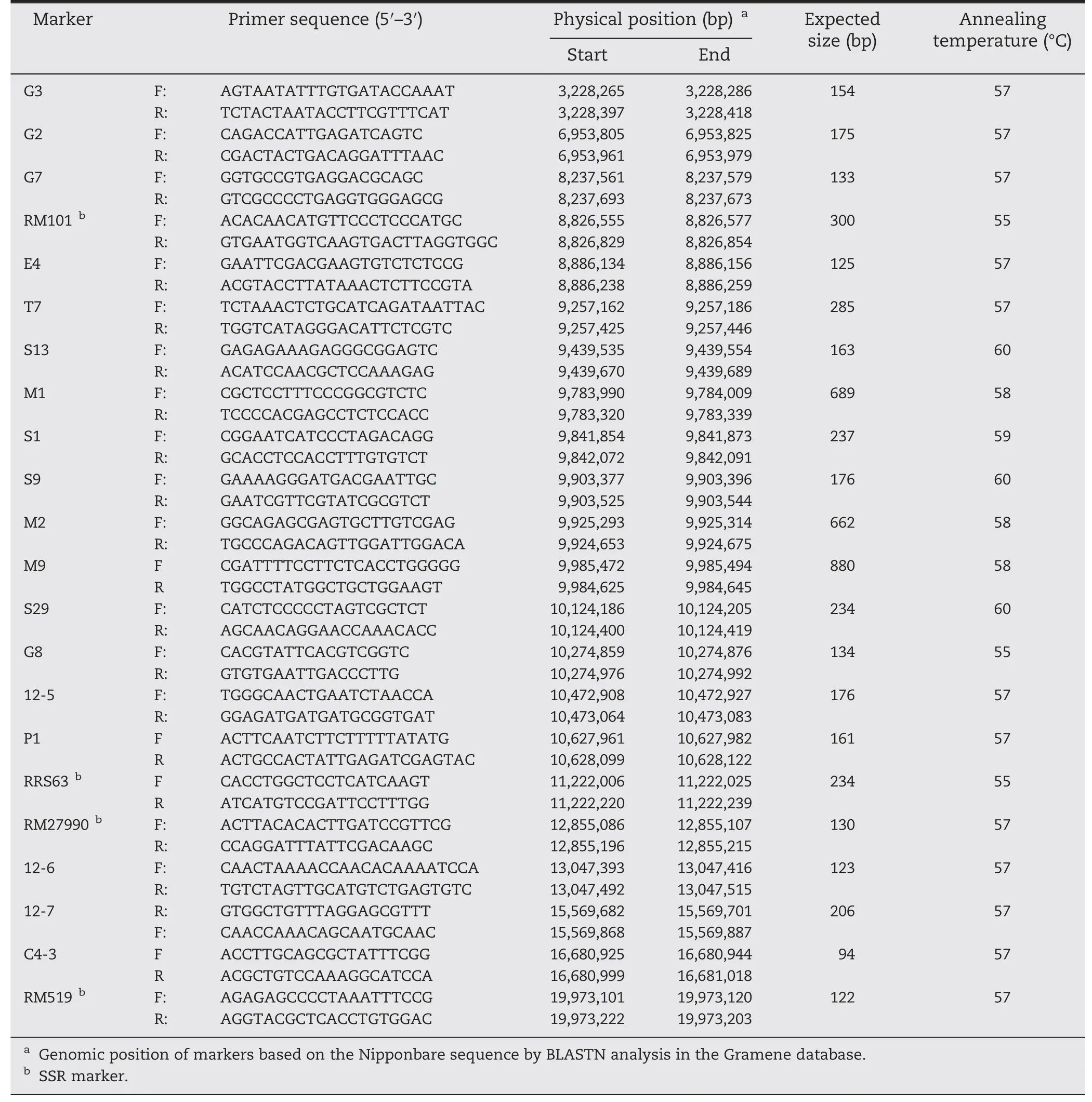
Table 3-Primer sequences of InDel and SSR markers used for mapping gene Pi61(t).
Two hundred and ninety simple sequence repeats(SSR)and 212 insertion–deletion(InDel)markers distributed evenly across the genome were screened for polymorphisms between the respective parents.Polymorphic markers were then subjected to bulked segregant analysis (BSA) combined with recessive class analysis (RCA) [63,64].Candidate markers linked with resistant phenotypes were further confirmed using the F2individuals comprising the resistant and susceptible pools.To finely map the R genes,two populations were developed.The first consisted of 1629 F2individuals that were extremely susceptible to isolate 001-99-1 and 725 that were extremely resistant.The second consisted of 1911 F2individuals extremely susceptible to isolate 99-26-2.Additional sets of SSR and InDel markers in the target R gene regions identified by the initial linkage analysis were used for alignment within the critical region of the genomic sequences of 93-11 and Nipponbare using the software Premier 3(http://www.premierbiosoft.com/).
作者应自留底稿,采用与否一律不退稿。向本刊投稿者,视为自愿遵守以上约定。来稿请用电子邮件发送到本刊信箱(E-mail:linyekeji@vip.tom.com或Linyekeji@163.net)。本刊地址:350012 福州市新店上赤桥35号;联系电话:0591-87911427;网址:http:∥fjlk.chinajournal.net.cn;http∥fjlykj.periodicals.net.cn。
The PCR amplifications were performed in 25 μL volumes containing 50 ng template,0.2 μmol L-1of each primer,1.5 mmol L-1MgCl2,0.02 μmol L-1dNTP and 1 U Taq polymerase.The PCR cycling profile consisted of initial denaturation at 94 °C for 5 min,followed by 35 cycles of 94 °C for 60 s,55–58 °C for 30 s,and 72 °C for 60 s,with a final extension at 72 °C for 7 min.PCR products were separated on 8% non-denaturing polyacrylamide gels and visualized using the silver staining method described by Sanguinetti et al.[65].InDel and SSR primers linked to the R genes in cv.93-11 are listed in Tables 2 and 3.Genetic distances between adjacent loci were estimated as Nr/2NT(Nrbeing the number of recombinants,and NTthe overall population size)[47,66].
2.4.Construction of physical maps and identification of candidate R genes
The physical map of the target locus was constructed based on Nipponbare contigs (http://www.gramene.org/).The 93-11 contigs were also anchored to this framework using the linked markers.Candidate genes within the target region were predicted and annotated using the Gramene database (http://www.gramene.org/).Each candidate NBS-LRR (nucleotide binding site-leucine rich repeat)gene in cv.93-11 was amplified using specific primers (Table 4)designed from the Nipponbare sequence in the Gramene database,and then sequenced by Beijing Biomed Co.Ltd.,Beijing.DNA and protein sequences were predicted using the softberry program (http://linux1.softberry.com/),and then aligned with Nipponbare homologues using the Gramene and EBI needle programs (http://www.ebi.ac.uk/).
3.Results
3.1.Blast resistance spectrum of cv.93-11
A total of 495 M.oryzae isolates were evaluated (Table 5).Cultivar 93-11 was resistant to 86.5% (186/215) of japonicaderived isolates collected from northern China(Beijing,Tianjin,Liaoning,Jilin,Hebei,Heilongjiang and Ningxia)and Japan,and 67.1%(188/280)of indica-derived isolates collected from southern China(Jiangsu,Yunnan,Guangdong,Zhejiang,Sichuan,Hunan,Fujian,Guizhou and Guangxi),an average of 75.6%(374/495).
In a more local context,93-11 was resistant to 92%–100%of 150 isolates from Beijing,Tianjin,Liaoning,Jilin,Hebei,Jiangsu of China and Japan(Table 5).This indicated that 93-11 could be used as a resistance resource in most japonica growing regions and in some indica regions,such as Jiangsu.
3.2.Genetic analysis of blast resistance in cv.93-11
The F2population derived from the cross LTH × 93-11 segregated 3R:1S when challenged with the indica-derived isolate 001-99-1 and japonica-derived isolate 99-26-2(Table 6),suggesting that the resistance of 93-11 to each of the two isolates was controlled by one dominant R gene.
To determine whether the same genes were involved,153 001-99-1-susceptible F2individuals were planted and injectioninoculated with isolate 99-26-2.A 3R:1S segregation ratio was observed,indicating that the genes were different and genetically independent.We tentatively named them Pi60(t) and Pi61(t),effective against isolates 001-99-1 and 99-26-2,respectively.

Table 4-Primers used for PCR amplification of entire genomic fragments of related candidate NBS-LRR genes in cv.93-11.
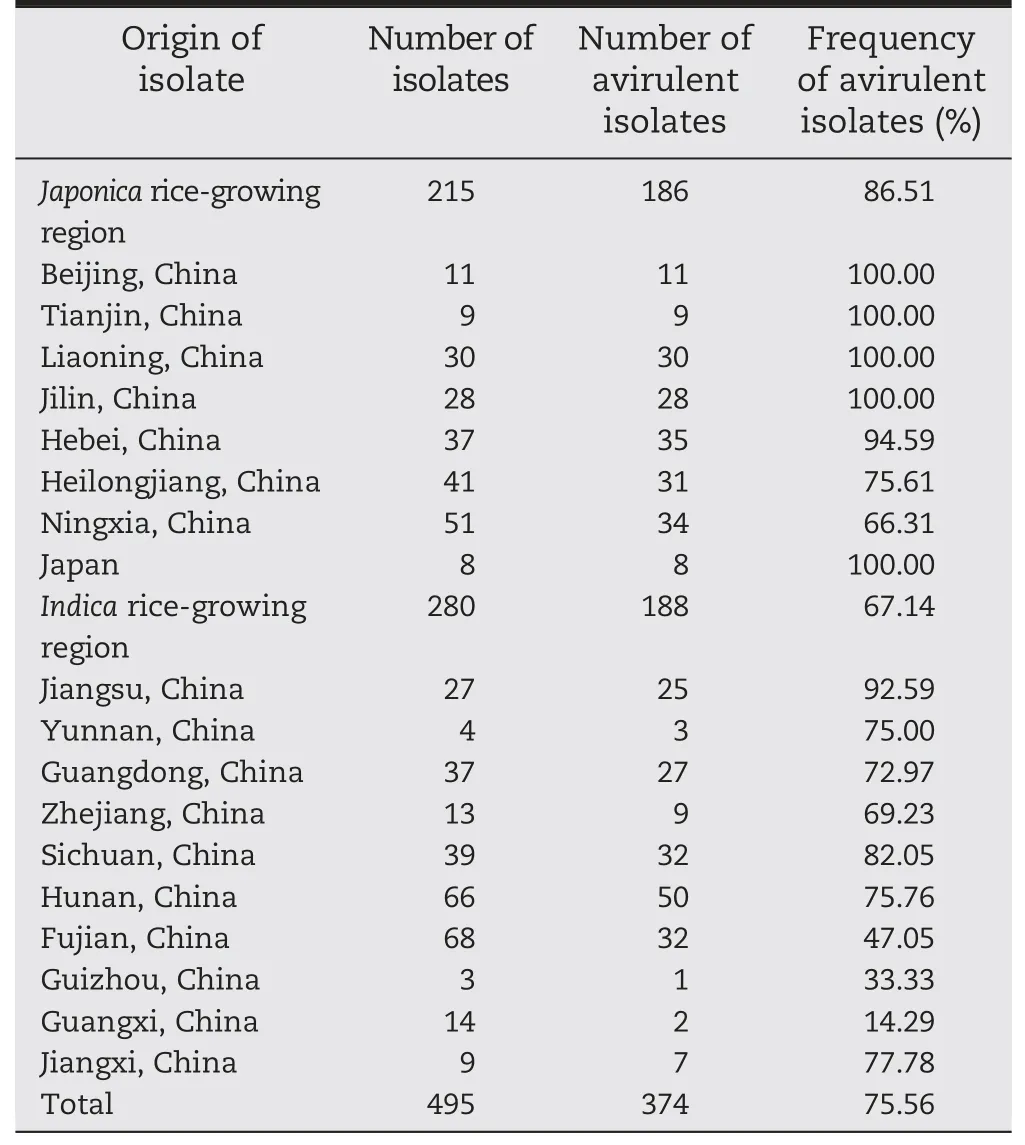
Table 5-Resistance spectra of cv.93-11 to 495 M.oryzae isolates from different rice-growing regions in China and from Japan.
3.3.Genetic mapping of Pi60(t) and Pi61(t)
Two hundred and twelve InDel and 290 SSR markers were screened for polymorphisms between parents 93-11 and LTH,and between the two sets of DNA bulks.Six InDel markers,viz.11-2,B3,C6,11-4,11-7 and S11-6-2 (Table 2),on chromosome 11 were polymorphic between both parents and DNA bulks for gene Pi60(t) (set 1); and InDel markers 12-1 and 12-6,and SSR markers RM101 and RM519,(Table 3) on chromosome 12,showed distinct polymorphisms between both parents and DNA bulks for gene Pi61(t)(set 2).These polymorphic markers were validated by genotyping individuals in the respective populations.For rough mapping of the Pi60(t) and Pi61(t)loci,160 001-99-1-susceptible F2individuals and 124 99-26-2-susceptible F2individuals were further subjected to linkage analysis with the above respective polymorphic markers for the two R genes.Pi60(t) was delimited to a 8.8 cM interval on the short arm of chromosome 11 by flanking markers B3(2.5 cM)and A4(5.3 cM)(Fig.1-a);and Pi61(t)was delimited to a 24.4 cM interval near the centromere of chromosome 12 by flanking markers G2(12.4 cM)and 12-6(12.0 cM)(Fig.2-a).
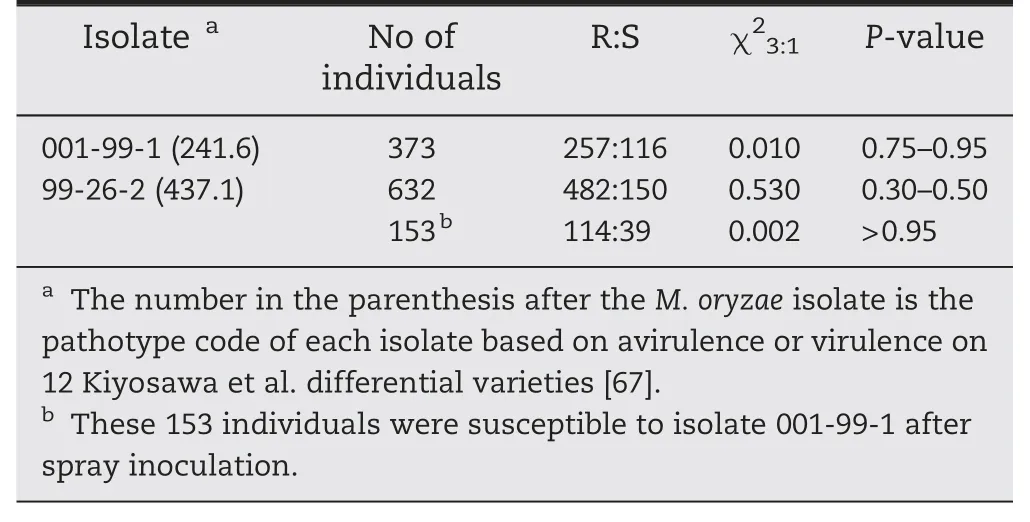
Table 6-Segregation of blast resistance to two isolates in F2 population of LTH × 93-11.

Fig.1-Genetic and physical map of the Pi60(t)region.a:Rough genetic map of Pi60(t)using 160 susceptible F2 plants.The number above the bold horizontal line:genetic distance(cM);CEN,centromere;b:Fine genetic map of map of Pi60(t)using 1629 susceptible F2 plants.In parentheses,numbers of recombinants; c:Marker genotypes and response phenotypes of five recombinants identified from 745 resistant F2 plants;d:Physical map of the Pi60(t)region based on the Nipponbare sequence.The colored horizontal block:BAC/PAC clone;vertical red dashed line: marker positions;red bold horizontal line:predicted gene;e:Physical map of the Pi60(t)region based on the 93-11 sequence.The colored horizontal block:scaffold;white block:gap in 93-11 sequence;vertical red dashed line: marker positions;red bold horizontal line:predicted gene.
For fine mapping of the Pi60(t) locus,1629 001-99-1-susceptible F2individuals were genotyped with the flanking markers B3 and C6,and 12 newly developed parental polymorphic InDel markers in the target interval of 8.8 cM,namely,K4-1,K2-1,K1-4,B1,Y10,E12,H6,H4,B14,C13,C7 and C6(Table 2).Pi60(t)was narrowed to a 0.58 cM interval(629 kb)on chromosome 11,flanked by InDel markers K1-4 (0.49 cM) and B14 (0.09 cM) and it co-segregated with five InDel markers (B1,Y10,E12,H6 and H4; Fig.1-b).To further reduce the Pi60(t)region,745 001-99-1-resistant F2individuals were also genotyped with the above 14 markers,and five individuals,P118,P6,P115,P11 and P2,were identified as distinct recombinants with at least one crossover between marker loci K1-4 and E12(Fig.1-c).The F3progenies derived from these five recombinants showed the expected segregating or homozygous resistant responses after challenge with isolate 001-99-1,completely corresponding to their genotypes at the two marker loci(Fig.1-c).Thus Pi60(t) was delimited to a 274 kb region flanked by InDel markers K1-4 and E12.
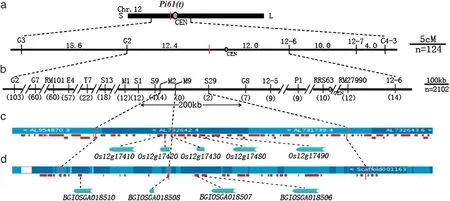
Fig.2-Genetic and physical map of the Pi61(t)region.a:Rough genetic map of Pi61(t)using 124 susceptible F2 plants.The numbers above the bold horizontal line,genetic distance(cM);CEN,centromere;b:Fine genetic map of map of Pi61(t)using 1911 susceptible F2 plants.The numbers in parentheses,number of recombinants;c: Physical map of the Pi61(t)region based on Nipponbare sequence.Colored horizontal block,BAC/PAC clone;vertical red dashed line,marker positions;red bold horizontal line,predicted gene;d:Physical map of the Pi61(t)region based on the 93-11 sequence.Colored horizontal block,scaffold;white block,gap in 93-11 sequence;vertical red dashed line,marker positions;red bold horizontal line,predicted gene.
3.4.Construction of physical maps of Pi60(t) and Pi61(t)
For Pi60(t),the target 274 kb region(6,374,147–6,648,601 bp)was covered by four PAC/BAC clones,including 48 putative genes annotated in the Gramene and TIGR databases(Fig.1-d);these included 8 intact NBS-LRR genes (Os11g11550,Os11g11580,Os11g11770,Os11g11790,Os11g11810,Os11g11940,Os11g11950 and Os11g11960),12 expressed proteins,16 hypothetical proteins and 12 retrotransposons.Sequence alignment of the NBS-LRR genes showed that 93-11 contained only six NBS-LRR genes,viz.BGIOSGA034264,BGIOSGA034263,BGIOSGA035032,BGIOSGA035036,BGIOSGA034259 and BGIOSGA034258,corresponding to Os11g11770,Os11g11790 (SasRGA4 allele of Pia),Os11g11810(SasRGA5 allele of Pia),Os11g11940,Os11g11950 and Os11g11960 at identity levels of 79.1%,89.5%,45.7%,96.4%,84.5%and 89.2%in protein sequence,respectively.
For Pi61(t),the target 200 kb region(9,924,675–10,124,186)in the Nipponbare sequence was covered by six PAC/BAC clones,including 44 putative genes annotated in the Gramene and TIGR database (Fig.2-c),viz.5 tandem NBSLRR type genes,Os12g17410,Os12g17420,Os12g17430,Os12g17480 and Os12g17490 in a 40 kb cluster,21 retrotransposons,1 transposon,11 hypothetical proteins and 6 expressed proteins.However,only four NBS-LRR genes can be amplified in cv.93-11 using the specific primers(Table 4),viz.BGIOSGA018510,BGIOSGA018508,BGIOSGA018507 and BGIOSGA018506,corresponding to Os12g17410,Os12g17430,Os12g17480 and Os12g17490 at identity levels of 68.7%,99.3%(2-amino acid differences),99.7% (3-amino acid differences)and 99.7% (3-amino acid differences) in protein sequences,respectively.
3.5.Allelism of Pi60(t) and Pia/PiCO39
Two other major blast R genes,Pi30(t) and cloned Pia/PiCO39,were previously mapped in the vicinity of Pi60(t) (6,374,147–6,648,601 bp) on chromosome 11 [11,37,38].Pi30(t) was roughly located within an interval of 6.1 Mb (441,392–6,578,785),and presumed to be Pia [59].Sequencing of the two Pia/PiCO39 alleles in 93-11 showed that the two alleles,viz.BGIOSGA034263 and BGIOSGA035032,corresponded with the dual gene functions required of Pia/PiCO39,SasRGA4 and SasRGA5,respectively,at identity levels of 100 and 99.19% in protein sequence[37,38].The variance of 0.81% (nine amino acids) between the protein sequences of BGIOSGA035032 and SasRGA5 was in the HMA domain (amino acids 1001–1070),C′-end (amino acids 1071–1116)and the NBS domain(amino acid 416:V →M)(Fig.3).The slight variance of the two Pia/PiCO39 alleles in cv.93-11 may not affect the function of Pia/PiCO39 because cultivar 104(Peh-kuh-tsao-tu) with the same two Pia alleles as 93-11 was earlier deduced to harbor just the Pia gene [37].In addition,the Pi60(t)-differential isolate 001-99-1 was avirulent to all four Pia/PiCO39-harboring lines,namely,IRBLa-A,IRBLa-C,Aichi Asahi and CO39 (Table 7).These results indicated that Pi60(t)could be Pia/PiCO39 or its allele.
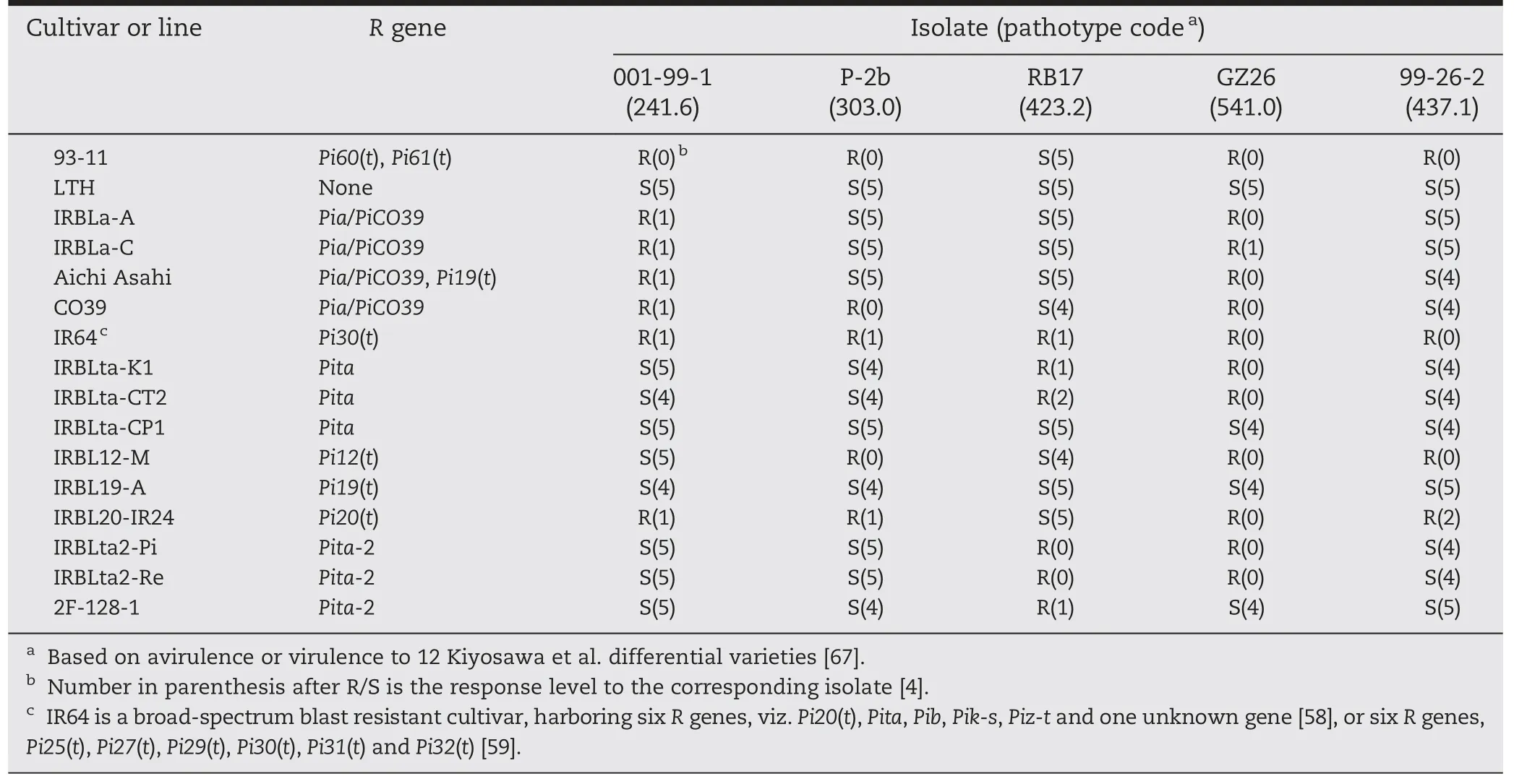
Table 7-Reactions of 16 rice genotypes to five M.oryzae differential isolates.
Differences in amino acids are marked in rectangular blocks.
3.6.Differentiation of Pi61(t) from other blast R genes
Eleven blast R genes,namely,Pita,Pita-2,Pi6(t),Pi12(t),Pi19(t),Pi20(t),Pi21(t),Pi39(t),Pi42(t),Pi58(t) and Pi157(t),are reported in the vicinity of Pi61(t) (9,924,675–10,124,186).Their target regions were roughly 5.6 kb (10,603,772–10,609,330),3.1 Mb(10,078,620–13,211,331),14.8 Mb (4,053,339–18,867,450),8.1 Mb(6,988,220–15,120,464),4.6 Mb (8,826,555–13,417,087),3.6 Mb(6,988,220–10,603,823),9.4 Mb (6,988,220–16,395,622),38 kb(10,614,346–10,652,094),4.2 Mb (8,073,819–12,248,913),3.4 Mb(7,461,555–10,900,056)and 9.2 Mb(8,826,555–18,050,447),respectively [11,57,68–71].To distinguish Pi61(t) from neighboring R genes,eight monogenic lines for Pita,Pita-2,Pi12(t),Pi19(t) and Pi20(t),i.e.,IRBLta-CT2,C104PKT,IRBLta2-Pi,IRBLta2-Re,F128-1,IRBL12-M,IRBL19-A and IRBL20-IR24,were tested with five differential isolates,001-99-1,P-2b,RB17,GZ26 and 99-26-2,and compared with the donor 93-11 (Table 7).Differential reactions were clearly observed among the eight lines,except for IRBL12-M and IRBL20-IR24 (Table 7),suggesting that Pi61(t)was different from Pita,Pita-2 and Pi19(t).
Pi39(t) was at least 490 kb (10,124,186–10,614,346) away from Pi61(t)according to the distances between markers most tightly linked to the two genes.In addition,Pi39(t)was mapped using the same differential isolate CHL724 as was Pi41,which also originated from cv.93-11 and delimited to 16,534,669–16,588,406 bp on chromosome 12 [47,69].This indicated that Pi39(t) could not be present in 93-11 together with Pi41.Therefore,we concluded that Pi61(t) was different from Pi39(t).As for Pi42(t),its co-segregating markers,including RRS63,were at least 0.19 cM from Pi61(t)(Fig.2-b).The target region of Pi42(t)contained six candidate NBS-LRR genes,and among them LOC_Os12g18374 was short-listed as a potential candidate of Pi42(t) based on restriction analysis of 11 candidate R genederived sequence tagged sequence (CRG-STS) markers [70].However,LOC_Os12g18374 (10,621,450–10,630,781) was at least 497 kb (10124186–10621450) from Pi61(t).This indicated that Pi61(t) was be different from Pi42(t).For the other six R genes,appropriate differential isolates (for Pi12(t) and Pi20(t))or mono-genic lines (for Pi6(t),Pi21(t),Pi58(t) and Pi157(t)) are lacking,and therefore we could not distinguish Pi61(t) from them.
4.Discussion
The availability of public sequence information for rice subspecies japonica cv.Nipponbare and the indica cv.93-11 has enabled the development of high density molecular markers,and accelerated fine mapping of blast R genes [21,40,69,70].In this study,we verified that the sequenced indica rice cv.93-11 conferred broad-spectrum resistance against multiple Chinese and Japanese M.grisea isolates,and identified two dominant blast R genes,Pi60(t) and Pi61(t),by using BSA-RCA linkage analysis combined with bioinformatics analyses.Pi60(t) was finely mapped to a 274 kb interval on chromosome 11,and Pi61(t) was finely mapped to a 200 kb interval on chromosome 12.Previously,Yang et al.[47]identified blast R gene Pi41 in cv.93-11,at least 6.3 Mb (10276467–16582733) away from Pi61(t).These results indicated that 93-11 possessed at least three blast R genes,viz.Pi60(t),Pi61(t)and Pi41.
Many relatively durable or broad-spectrum blast resistant rice cultivars possess more than two R genes.IR64 [58,59],Moroberekan [49,72–74],Suweon 365 [11,75],Teqing [76],Sanhuangzhan 2 [50],Digu[26,32,77]and Gumei 2 [51] possess at least 6,5,4,3,3,3 and 3 blast R genes,respectively.On the other hand,single genes,such as Pi9,Pi2 (Piz-5),Piz-t,Piz and Pigm,were reported to confer broad-spectrum resistance[21,24,25,78].In the case of 93-11,Pi41 was identified using isolates CHL724 and CHL743 from the cold japonica rice-growing region (Jilin of China)[47],whereas Pi60(t) was identified using isolate 001-99-1 from an indica cropping region (Jiangsu of China),and Pi61(t) was identified using isolate 99-26-2 from a temperate japonica region (Hebei of China).To test the resistance specificity of Pi60(t),Pi61(t) and Pi41(t),we inoculated F2populations of the cross LTH × 93-11 using 18 differential isolates(except 001-99-1 and 99-26-2)from different geographic origins,and genotyped 30–100 extremely susceptible F2individuals from 13 small population-isolate combinations segregating in 3R:1S ratios using tightly-linked markers for Pi60(t),Pi61(t)and Pi41.Pi60(t)conferred resistance to four isolates,including two indica-derived isolates (one from Jiangsu and the other from Hunan of China),and two Japanese japonica-derived isolates.Pi61(t) conferred resistance to six isolates,including two indica-derived isolates (one from Guangdong and the other from Fujian of China) and four japonica-derived isolates (one each from Liaoning,Heilongjiang,Hebei and Beijing of China).Pi41 conferred resistance to three isolates,one indica-derived isolate (from Guangdong of China) and two japonica-derived isolates (both from Beijing of China) (data not shown).Thus,the three R genes in 93-11 showed no obvious specificity to indica-or japonica-derived isolates,suggesting that their combined actions may constitute the broad-spectrum resistance in cv.93-11,particularly to Chinese japonica-derived isolates.
It is well established that two-thirds of over 70 major blast R genes mapped to date are clustered,especially on chromosomes 6,11 and 12[11–13].Considering the difficulties in testing for allelism between an unknown blast R gene and others mapping within a cluster [15,59,76],fine-scale mapping and differential pathotesting are regarded as alternatives for allelism tests[47,71].In this study Pi61(t)was mapped in the vicinity of 11 previously mapped R genes.Of these Pita,Pita-2 and Pi19(t)were considered to be different from Pi61(t)by comparing their reactions with that of 93-11 against differential isolates(Table 7).Pi39(t) was excluded due to its similarity to Pi41(t) in 93-11 and a physical distance of 490 kb from Pi61(t) [47,69].Pi42(t)could also be excluded according to the physical distance of at least 497 kb between its only short-listed potential candidate gene,LOC_Os12g18374,and Pi61(t) [70].We thus can conclude that Pi61(t) should be different from some nearby R genes,including Pita,Pita-2,Pi19(t),Pi39(t)and Pi42(t).However,we could not determine the differentiation of Pi61(t) from six other mapped genes (Pi6(t),Pi12(t),Pi20(t),Pi21(t),Pi58(t) and Pi157(t)) in this region due to their rough physical regions and unknown response spectra.In the Pi60(t)cluster,Pia and PiCO39 were both cloned and confirmed to be the same gene [37,38].Even though the SasRGA4 and SasRGA5 alleles in 93-11 are quite similar to the Pia/PiCO39 alleles in Sasanishiki and CO39,we could not completely exclude the possibility that Pi60(t)and Pia/PiCO39 had a more complex relationship due to the DNA sequence variations between the six NBS-LRR alleles in resistant cv.93-11 and those in susceptible cv.Nipponbare.So far,we have cloned the two Pia alleles (BGIOSGA034263 and BGIOSGA035032) in 93-11,and co-transformation of the two candidates is underway for clarification of the nature of Pi60(t).
It is notable that both the Pi60(t) and Pi61(t) clusters are embedded in recombination-suppressed regions [47,68,70].In the 0.58 cM interval(629 kb)of Pi60(t)delimited by markers K1-4 and B14,and the 0.15 cM interval(200 kb)of Pi61(t)delimited by markers M2 and S29,the physical/genetic distance ratios are 1084 kb cM-1and 1333 kb cM-1,respectively.The low recombination rates may be due to lack of sequence homology between the parental genotypes,abdundance of repetitive sequences near the centromeres and transposon/retrotransposon-rich regions [47,68,70].These situations further increase the difficulties of performing allelism tests between R genes within a cluster,even though large segregating populations were used.Therefore,fine mapping and cloning of each of the clustered R genes will be essential to elucidate their relationships and to dissect their detailed race-or isolate-specificities.
NBS-LRR proteins represent the largest class of R genes in plants,and nearly 500 NBS-LRR genes have been identified in both Nipponbare and 93-11 [31].Eighteen of 20 cloned blast R genes (except Pid2 and pi21) encode NBS-LRR proteins[21,26,35,40].Colocalizations of the NBS-LRR genes and blast resistance loci were identified through genetic analyses[14,59].Therefore,NBS-LRR genes are the most likely potential candidates for further blast R genes [70].In our study,eight intact NBS-LRR genes in the 274 kb region encompassing Pi60(t)were identified in the Nipponbare sequence,but only six intact NBS-LRR genes were identified in the 93-11 sequence in the Gramene database(Fig.1-d),including the two alleles of Pia/PiCO39 (SasRGA4 and SasRGA5).On the other hand,four NBS-LRR genes exist in the 200 kb target region of Pi61(t) in 93-11,and all of them showed differences in comparison with the corresponding NBS-LRR genes in Nipponbare.Therefore,it is difficult to shortlist candidate genes for Pi61(t).We need to further reduce the target interval of Pi61(t),or to transform all four NBS-LRR genes into susceptible cultivars for complementation tests.In addition,another blast R gene,Pi41(t),present in 93-11 was predicted as a NBS-LRR-type gene[47].Therefore,we postulate that the broad-spectrum blast resistance in 93-11 is mediated by multiple NBS-LRR genes,representing a molecular mechanism of broad-spectrum resistance different from Digu[77],and Pi2,Pi9 and Piz-t[79].
5.Conclusion
In summary,the broad-spectrum blast resistant cv.93-11 harbors at least three R genes,Pi60(t) on chromosome 11,and Pi61(t) and Pi41 on chromosome 12.Pi60(t) and Pi61(t) are both embedded in recombination-suppressed regions with several clustered NBS-LRR genes.We identified two tightly linked flanking markers,K1-4 and E12,and two co-segregating markers,Y10 and B1,for Pi60(t);and two tightly linked flanking markers G8 and M2,and one co-segregating marker M9 for Pi61(t).These markers should ensure rapid and accurate transfer of the two R genes from 93-11 into new breeding lines through MAS.The delineation of physical positions and the short-listed candidate genes of the two blast R loci have set solid foundations for positional cloning of Pi60(t)and Pi61(t).
We thank Dr.Yulin Jia,USDA-ARS Dale Bumpers National Rice Research Center,Stuttgart,Arkansas,USA for helpful discussion.This work was supported by grants from the National Natural Science Foundation of China(Grant No.30871606),the Special Fund for Agro-scientific Research in the Public Interest Program of China (Grant No.20120314),and the Major Science and Technology Project to Create New Crop Cultivars using Gene Transfer Technology(Grant No.2011ZX08001-002).
[1] D.Siluê,J.L.Notteghem,D.Tharreau,Evidence of a gene-for-gene relationship in the Oryza sativa-Magnaporthe grisea pathosystem,Phytopathology 82(1992) 577–580.
[2] R.S.Zeigler,J.Tohme,R.Nelson,M.Levy,F.J.Correa Victoria,Lineage exclusion: a proposal for linking blast population analysis to resistance breeding,in: R.S.Zeigler,S.A.Leong,P.S.Teng (Eds.),Rice Blast Disease,CAB international,Wallingford,UK,1994,pp.267–292.
[3] Y.Jia,G.T.Bryan,L.Farrall,B.Valent,Natural variation at the Pi-ta rice blast resistance locus,Phytopathology 93(2003)1452–1459.
[4] D.J.Mackill,J.M.Bonman,Inheritance of blast resistance in near-isogenic lines of rice,Phytopathology 82(1992) 746–749.
[5] E.Zhou,Y.Jia,J.Correll,F.N.Lee,Instability of the Magnaporthe oryzae avirulence gene AVR-Pita alters virulence,Fung Genet.Biol.44(2007) 1024–1034.
[6] Y.Dai,Y.Jia,J.Correll,X.Wang,Y.Wang,Diversification and evolution of the avirulence gene AVR-Pita1 in field isolates of Magnaporthe oryzae,Fung Genet.Biol.47(2010) 973–980.
[7] S.Hittalmani,A.Parco,T.V.Mew,R.S.Zeigler,N.Huang,Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice,Theor.Appl.Genet.100 (2000) 1121–1128.
[8] S.A.Goff,D.Ricke,T.H.Lan,G.Presting,R.Wang,M.Dunn,J.Glazebrook,A.Sessions,P.Oeller,H.Varma,D.Hadley,D.Hutchison,C.Martin,F.Katagiri,B.M.Lange,T.Moughamer,Y.Xia,P.Budworth,J.Zhong,T.Miguel,U.Paszkowski,S.Zhang,M.Colbert,W.L.Sun,L.Chen,B.Cooper,S.Park,T.C.Wood,L.Mao,P.Quail,R.Wing,R.Dean,Y.Yu,A.Zharkikh,R.Shen,S.Sahasrabudhe,A.Thomas,R.Cannings,A.Gutin,D.Pruss,J.Reid,S.Tavtigian,J.Mitchell,G.Eldredge,T.Scholl,R.M.Miller,S.Bhatnagar,N.Adey,T.Rubano,N.Tusneem,R.Robinson,J.Feldhaus,T.Macalma,A.Oliphant,S.Briggs,A draft sequence of the rice genome (Oryza sativa L.ssp.japonica),Science 296 (2002) 92–100.
[9] J.Yu,S.Hu,J.Wang,G.K.Wong,S.Li,B.Liu,Y.Deng,L.Dai,Y.Zhou,X.Zhang,M.Cao,J.Liu,J.Sun,J.Tang,Y.Chen,X.Huang,W.Lin,C.Ye,W.Tong,L.Cong,J.Geng,Y.Han,L.Li,W.Li,G.Hu,X.Huang,W.Li,J.Li,Z.Liu,L.Li,J.Liu,Q.Qi,J.Liu,L.Li,T.Li,X.Wang,H.Lu,T.Wu,M.Zhu,P.Ni,H.Han,W.Dong,X.Ren,X.Feng,P.Cui,X.Li,H.Wang,X.Xu,W.Zhai,Z.Xu,J.Zhang,S.He,J.Zhang,J.Xu,K.Zhang,X.Zheng,J.Dong,W.Zeng,L.Tao,J.Ye,J.Tan,X.Ren,X.Chen,J.He,D.Liu,W.Tian,C.Tian,H.Xia,Q.Bao,G.Li,H.Gao,T.Cao,J.Wang,W.Zhao,P.Li,W.Chen,X.Wang,Y.Zhang,J.Hu,J.Wang,S.Liu,J.Yang,G.Zhang,Y.Xiong,Z.Li,L.Mao,C.Zhou,Z.Zhu,R.Chen,B.Hao,W.Zheng,S.Chen,W.Guo,G.Li,S.Liu,M.Tao,J.Wang,L.Zhu,L.Yuan,H.Yang,A draft sequence of the rice genome (Oryza sativa L.ssp.indica),Science 296 (2002) 79–92.
[10] IRGSP,The map-based sequence of the rice genome,Nature 436 (2005) 793–800.
[11] E.Ballini,J.B.Morel,G.Droc,A.Price,B.Courtois,J.L.Notteghem,D.Tharreau,A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance,Mol.Plant Microbe Interact.21(2008) 859–868.
[12] Y.Koide,N.Kobayashi,D.Xu,Y.Fukuta,Blast resistance genes and their selection markers in rice (Oryza sativa L.),JIRCA Work Rep.63(2009) 95–122.
[13] J.Liu,X.Wang,T.Mitchell,Y.Hu,X.Liu,L.Dai,G.Wang,Recent progress and understanding of the molecular mechanisms of the rice-Magnaporthe oryzae interaction,Mol.Plant Pathol.11(2010) 419–427.
[14] R.J.Wisser,Q.Sun,S.H.Hulbert,S.Kresovich,R.J.Nelson,Identification and characterization of regions of the rice genome associated with broad-spectrum,quantitative disease resistance,Genetics 169 (2005) 2277–2293.
[15] Y.Deng,X.Zhu,Y.Shen,Z.He,Genetic characterization and fine mapping of the blast resistance locus Pigm(t) tightly linked to Pi2 and Pi9 in a broad-spectrum resistant Chinese variety,Theor.Appl.Genet.113 (2006) 705–713.
[16] S.Chen,L.Wang,Z.Que,R.Pan,Pan QGenetic and physical mapping of Pi37(t),a new gene conferring resistance to rice blast in the famous cultivar St.No.1,Theor.Appl.Genet.111(2005) 1563–1570.
[17] X.Liu,L.Wang,S.Chen,F.Lin,Q.Pan,Genetic and physical mapping of Pi36(t),a novel rice blast resistance gene located on rice chromosome 8,Mol.Genet.Genomics 274(2005)394–401.
[18] R.Fjellstrom,C.A.Conaway-Bormans,A.M.McClung,M.A.Marchetti,A.R.Shank,W.D.Park,Development of DNA markers suitable for marker assisted selection of three Pi Genes conferring resistance to multiple Pyricularia grisea pathotypes,Crop.Sci.44 (2004) 1790–1798.
[19] R.Fjellstrom,A.M.McClung,A.R.Shank,SSR markers closely linked to the Pi-z locus are useful for selection of blast resistance in a broad array of rice germplasm,Mol.Breed.17(2006) 149–157.
[20] K.Hayashi,H.Yoshida,I.Ashikawa,Development of PCR-based allele-specific and InDel marker sets for nine rice blast resistance genes,Theor.Appl.Genet.113(2006)251–260.
[21] L.Hua,J.Wu,C.Chen,W.Wu,X.He,F.Lin,L.Wang,I.Ashikawa,T.Matsumoto,L.Wang,Q.Pan,The isolation of Pi1,an allele at the Pik locus which confers broad spectrum resistance to rice blast,Theor.Appl.Genet.125 (2012)1047–1055.
[22] Z.Wang,M.Yano,U.Yamanouchi,M.Iwamoto,L.Monna,H.Hayasaka,Y.Katayose,T.Sasaki,The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes,Plant J.19(1999) 55–64.
[23] G.T.Bryan,K.Wu,L.Farrall,Y.Jia,H.P.Hershey,S.McAdams,R.Tarchini,G.Donaldson,K.Faulk,B.Valent,A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta,Plant Cell 12(2000)2033–2045.
[24] S.Qu,G.Liu,B.Zhou,M.Bellizzi,L.Zeng,L.Dai,B.Han,G.L.Wang,The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice,Genetics 172 (2006)1901–1914.
[25] B.Zhou,S.Qu,G.Liu,M.Dolan,H.Sakai,G.Lu,M.Bellizzi,G.L.Wang,The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea,Mol.Plant Microbe Interact.19 (2006) 1216–1228.
[26] X.Chen,J.Shang,D.Chen,C.Lei,Y.Zou,W.Zhai,G.Liu,J.Xu,Z.Ling,G.Cao,B.Ma,Y.Wang,X.Zhao,S.Li,L.Zhu,A B-lectin receptor kinase gene conferring rice blast resistance,Plant J.46(2006) 794–804.
[27] X.Liu,F.Lin,L.Wang,Q.Pan,The in silico map-based cloning of Pi36,a rice coiled-coil nucleotide-binding site leucine-rich repeat gene that confers race-specific resistance to the blast fungus,Genetics 176 (2007) 2541–2549.
[28] F.Lin,S.Chen,Z.Que,L.Wang,X.Liu,Q.Pan,The blast resistance gene Pi37 encodes a nucleotide binding site-leucine-rich repeat protein and is a member of a resistance gene cluster on rice chromosome 1,Genetics 177(2007) 1871–1880.
[29] I.Ashikawa,N.Hayashi,H.Yamane,H.Kanamori,J.Wu,T.Matsumoto,K.Ono,M.Yano,Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance,Genetics 180 (2008) 2267–2276.
[30] S.K.Lee,M.Song,Y.S.Seo,H.K.Kim,S.Ko,P.Cao,J.P.Suh,G.Yi,J.H.Roh,S.Lee,Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil–nucleotide-binding–leucine-rich repeat genes,Genetics 181 (2009) 1627–1638.
[31] J.Shang,Y.Tao,X.Chen,Y.Zou,C.Lei,J.Wang,X.Li,X.Zhao,M.Zhang,Z.Lu,J.Xu,Z.Cheng,J.Wan,L.Zhu,Identification of a new rice blast resistance gene,Pid3,by genome wide comparison of paired nucleotide-binding site-leucine-rich repeat genes and their pseudogene alleles between the two sequenced rice genomes,Genetics 182 (2009) 1303–1311.
[32] J.Chen,Y.Shi,W.Liu,R.Chai,Y.Fu,J.Zhuang,J.Wu,A Pid3 allele from rice cultivar Gumei2 confers resistance to Magnaporthe oryzae,J.Genet.Genomics 38(2011) 209–216.
[33] K.Hayashi,H.Yoshida,Refunctionalization of the ancient rice blast disease resistance gene Pit by the recruitment of a retrotransposon as a promoter,Plant J.57(2009) 413–425.
[34] A.Takahashi,N.Hayashi,A.Miyao,H.Hirochika,Unique features of the rice blast resistance Pish locus revealed by large scale retrotransposon-tagging,BMC,Plant Pathol.10(2010) 175,(http://www.biomedcentral.com/1471-2229/10/175).
[35] S.Fukuoka,N.Saka,H.Koga,K.Ono,T.Shimizu,K.Ebana,N.Hayashi,A.Takahashi,H.Hirochika,K.Okuno,M.Yano,Loss of Function of a proline-containing protein confers durable disease resistance in rice,Science 325 (2009) 998–1001.
[36] N.Hayashi,H.Inoue,T.Kato,T.Funao,M.Shirota,T.Shimizu,H.Kanamori,H.Yamane,Y.Hayano-Saito,T.Matsumoto,M.Yano,H.Takatsuji,Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication,Plant J.64 (2010) 498–510.
[37] Y.Okuyama,H.Kanzaki,A.Abe,K.Yoshida,M.Tamiru,H.Saitoh,T.Fujibe,H.Matsumura,M.Shenton,D.C.Galam,J.Undan,A.Ito,T.Sone,R.Terauchi,A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes,Plant J.66(2011) 467–479.
[38] S.Cesari,G.Thilliez,C.Ribot,V.Chalvon,C.Michel,S.Rivas,L.Alaux,H.Kanzaki,Y.Okuyama,J.B.Morel,E.Fournier,D.Tharreau,T.Kroja,The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding,Plant Cell 25(2013)1463–1481.
[39] A.K.Rai,S.P.Kumar,S.K.Gupta,N.Gautam,N.K.Singh,T.R.Sharma,Functional complementation of rice blast resistance gene Pi-kh(Pi54) conferring resistance to diverse strains of Magnaporthe oryzae,J.Plant Biochem.Biot.20(2011) 55–65.
[40] C.Zhai,F.Lin,Z.Dong,X.He,B.Yuan,X.Zeng,L.Wang,Q.Pan,The isolation and characterization of Pik,a rice blast resistance gene which emerged after rice domestication,New Phytol.189 (2011) 321–334.
[41] B.Yuan,C.Zhai,W.Wang,X.Zeng,X.Xu,H.Hu,F.Lin,L.Wang,Q.Pan,The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes,Theor.Appl.Genet.122 (2011) 1017–1028.
[42] Y.Jia,Z.Wang,P.Singh,Development of dominant rice blast Pi-ta resistance gene markers,Crop.Sci.42(2002)2145–2149.
[43] Y.Jia,Z.Wang,R.G.Fjellstrom,K.A.K.Moldenhauer,M.A.Azam,J.Correll,F.N.Lee,Y.Xia,J.N.Rutger,Rice Pi-ta gene confers resistance to the major pathotypes of the rice blast fungus in the United States,Phytopathology 94(2004)296–301.
[44] Z.Dai,B.Zhao,X.Liu,G.Xia,C.Tan,B.Zhang,H.Zhang,A new middle-mature indica rice variety Yangdao 6 with high yielding,high quality and multiple disease resistance,Jiangsu Agric.Sci.4(1997) 13–14,(in Chinese with English abstract).
[45] Z.Dai,A.Li,L.Huang,G.Liu,C.Zhou,H.Zhang,Utilization of an elite rice germplasm –Yangdao 6 with high combining ability created by60Co-γirridation,Acta Agric.Nucl.Sin.20(2)(2006) 83–86,(in Chinese with English abstract).
[46] H.Zhang,Z.Dai,B.Zhao,X.Kong,G.Xia,A.Li,C.Pan,C.Zhou,B.Wang,N.Huang,C.Tan,Z.Zhu,Construction of ideal plant type of an elite indica rice cultivar Yangdao 6 with high yield and good grain quality,J.Yangzhou Univ.(Agric.Life Sci.Ed.)29 (3)(2008) 1–6,(in Chinese with English abstract).
[47] Q.Yang,F.Lin,L.Wang,Q.Pan,Identification and mapping of Pi41,a major gene conferring resistance to rice blast in the Oryza sativa subsp.indica reference cultivar 93-11,Theor.Appl.Genet.118 (2009) 1027–1034.
[48] D.Huang,Characterization and SSR mapping of rice blast resistance genes,Postdoctoral Research Working Report of the Graduate School of Chinese Academy of Agricultural Sciences,Beijing,2005.78 ,(in Chinese with English abstract).
[49] G.L.Wang,D.J.Mackill,J.M.Bonman,S.R.McCouch,M.C.Champoux,R.J.Nelson,RFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivar,Genetics 136 (1994) 1421–1434.
[50] B.Liu,S.Zhang,X.Zhu,Q.Yang,S.Wu,M.Mei,R.Mouleon,J.Leach,T.New,H.Leung,Candidate defense genes as predictors of quantitative blast resistance in rice,Mol.Plant Microbe.Interact.17(2004) 1146–1152.
[51] J.Wu,Y.Fan,D.Li,K.Zheng,J.Zhuang,H.Leung,Genetic control of rice blast resistance in the durably resistant cultivar Gumei 2 against multiple isolates,Theor.Appl.Genet.111 (2005) 50–56.
[52] J.Wu,X.Liu,L.Dai,G.Wang,Advances on the identification and characterization of broad spectrum blast resistance genes in rice,Chin.Bull.Life Sci.19(2007) 233–238.
[53] X.Zhu,S.Chen,J.Yang,S.Zhou,L.Zeng,J.Han,J.Su,L.Wang,Q.Pan,The identification of Pi50(t),a new member of the rice blast resistance Pi2/Pi9 multigene family,Theor.Appl.Genet.124 (2012) 1295–1304.
[54] Z.Ling,T.New,J.Wang,C.Lei,N.Huang,Development of near-isogenic lines of rice and their differential ability to the blast pathogen,Int.Rice Res.Notes 20(1995) 13–14.
[55] Z.Ling,T.New,J.Wang,C.Lei,N.Huang,Development of Chinese near isogenic lines of rice and their differential ability of pathogenic races of blast fungus,Sci.Agric.Sin.33(4) (2001) 1–8,(in Chinese with English abstract).
[56] N.Kobayashi,M.J.Telebanco-Yanoria,H.Tsunematsu,H.Kato,T.Imbe,Y.Fukuta,Development of new sets of international standard differential varieties for blast resistance in rice (Oryza sativa L.),Jpn.Agric.Res.Q 41 (2007)31–37.
[57] N.Hayashi,I.Ando,T.Imbe,Identification of a new resistance gene to a Chinese blast fungus isolate in the Japanese rice cultivar Aichi Asahi,Phytopathology 88(1998)822–827.
[58] N.Kobayashi,L.A.Ebron,D.Fujita,Y.Fukuta,Identification of blast resistance genes in IRRI-bred rice varieties by segregation analysis based on a differential system,JIRCAS Work Rep.63(2009) 69–86.
[59] C.Sallaud,M.Lorieux,E.Roumen,D.Tharreau,R.Berruyer,P.Svestasrani,O.Garsmeur,A.Ghesquiere,J.L.Notteghem,Identification of five new blast resistance genes in the highly blast-resistant rice variety IR64 using a QTL mapping strategy,Theor.Appl.Genet.106 (2003) 794–803.
[60] D.Chen,R.S.Zeigler,S.W.Ahn,R.J.Nelson,Phenotypic characterization of the rice blast resistance gene Pi2(t),Plant Dis.80(1996) 52–56.
[61] C.Lei,J.Wang,S.Mao,L.Zhu,Z.Ling,Gene analysis of blast resistance in indica variety Zhaiyeqing 8,Chin.J.Genet.23(1996) 273–277.
[62] M.G.Murray,W.F.Thompson,Rapid isolation of high molecular weight plant DNA,Nucl.Acids Res.8(1980)4321–4325.
[63] R.W.Michelmore,I.Paran,R.V.Kesseli,Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations,Proc.Natl.Acad.Sci.U.S.A.88 (1991) 9828–9832.
[64] Q.Zhang,B.Z.Shen,X.K.Dai,M.H.Mei,Saghai Maroof M A,Li Z B.Using bulked extremes and recessive class to map genes for photoperiod-sensitive genic male sterility in rice,Proc.Natl.Acad.Sci.U.S.A.91(1994) 8675–8679.
[65] C.J.Sanguinetti,N.E.Dias,A.J.Simpson,Rapid silver staining and recovery of PCR products separated on polyacrylamide gels,Biotechniques 17(1994) 914–921.
[66] K.Gu,D.Tian,F.Yang,L.Wu,C.Sreekala,D.Wang,G.L.Wang,Z.Yin,High-resolution genetic mapping of Xa27(t),a new bacterial blight resistance gene in rice,Oryza sativa L,Theor.Appl.Genet.108 (2004) 800–807.
[67] S.Kiyosawa,H.Ikehashi,H.Kato,Z.Ling,Pathogenicity test of Philippine isolates of blast fungus using two sets of rice varieties,Jpn.J.Breed 31(1981) 367–376.
[68] W.Li,C.Lei,Z.Cheng,Y.Jia,D.Huang,J.Wang,X.Zhang,N.Su,X.Guo,H.Zhai,J.Wan,Identification of SSR markers for a broad-spectrum blast resistance gene Pi20(t) for marker-assisted breeding,Mol.Breed.22(2008) 141–149.
[69] X.Liu,Q.Yang,F.Lin,L.Hua,C.Wang,L.Wang,Q.Pan,Identification and fine mapping of Pi39(t),a major gene conferring the broad-spectrum resistance to Magnaporthe oryzae,Mol.Genet.Genomics 278 (2007) 403–410.
[70] P.K.Kumar,S.Pathania,P.Katoch,T.R.Sharma,P.Plaha,R.Rathour,Genetic and physical mapping of blast resistance gene Pi-42(t) on the short arm of rice chromosome 12,Mol.Breed.25(2010) 217–228.
[71] Y.Koide,M.J.Telebanco-Yanoria,Y.Fukuta,N.Kobayashi,Detection of novel blast resistance genes,Pi58(t)and Pi59(t),in a Myanmar rice landrace based on a standard differential system,Mol.Breed.(2013)(online),http://dx.doi.org/10.1007/s11032-013-9865-5.
[72] T.Inukai,R.S.Zeigler,S.Sarkarung,M.Bronson,L.V.Dung,T.Kinoshita,R.J.Nelson,Development of pre-isogenic lines for rice blast-resistance by marker-aided selection from a recombinant inbred population,Theor.Appl.Genet.93(1996)560–567.
[73] N.I.Naqvi,B.B.Chattoo,Molecular genetic analysis and sequence characterized amplified region-assisted selection of blast resistance in rice,Rice Genet.3(1996)570–576.
[74] D.Chen,dela Viña M,Inukai T,Mackill D J,Ronald P C,Nelson R J.Molecular mapping of the blast resistance gene,Pi44(t),in a line derived from a durably resistant rice cultivar,Theor.Appl.Genet.98(1999) 1046–1053.
[75] S.N.Ahn,Y.K.Kim,H.C.Hong,S.S.Han,S.J.Kwon,H.C.Choi,H.P.Moon,S.R.McCouch,Molecular mapping of a new gene for resistance to rice blast (Pyricularia grisea Sacc),Euphytica 116 (2000) 17–22.
[76] R.E.Tabien,Z.Li,A.H.Paterson,M.A.Marchetti,J.W.Stansel,S.R.M.Pinson,Mapping of four major rice blast resistance genes from ‘Lemont' and‘Teqing'and evaluation of their combinatorial effect for field resistance,Theor.Appl.Genet.101 (2000) 1215–1225.
[77] X.Chen,J.Shang,C.Lei,J.Xu,S.Li,L.Zhu,Genetic and molecular analysis of blast resistance in a universal blast resistant variety,Digu,in: G.L.Wang,B.Valent (Eds.),Advances in Genetics,Genomics and Control of Rice Blast,Springer of Congress + Business Media B.V.,USA,2009,pp.149–159.
[78] Y.Deng,X.Zhu,J.Xu,H.Chen,Z.He,Map-based cloning and breeding application of a broad-spectrum resistance gene Pigm to rice blast,in: G.L.Wang,B.Valent (Eds.),Advances in Genetics,Genomics and Control of Rice Blast,Springer of Congress + Business Media B.V.,USA,2009,pp.161–171.
[79] J.Wu,H.Mizuno,M.Hayashi-Tsugane,Y.Ito,Y.Chiden,M.Fujisawa,S.Katagiri,S.Saji,S.Yoshiki,W.Karasawa,Physical maps and recombination frequency of six rice chromosomes,Plant J.36(2003) 720–730.
- The Crop Journal的其它文章
- Difference between resistant and susceptible maize to systematic colonization as revealed by DsRed-labeled Fusarium verticillioides
- Expression of an(E)-β-farnesene synthase gene from Asian peppermint in tobacco affected aphid infestation
- Anatomical and chemical characteristics associated with lodging resistance in wheat
- Zea mays(L.)P1 locus for cob glume color identified as a post-domestication selection target with an effect on temperate maize genomes
- Genome-wide association of 10 horticultural traits with expressed sequence tag-derived SNP markers in a collection of lettuce lines
- Dissection of two quantitative trait loci for grain weight linked in repulsion on the long arm of chromosome 1 of rice(Oryza sativa L.)

