南海链霉菌SCSIO 1635中抗霉素类化合物的分离鉴定
李苏梅,田新朋,牛四文,张文军,张 偲,鞠建华,张长生
中国科学院南海海洋研究所海洋生物资源可持续利用重点实验室海洋微生物研究中心广东省海洋药物重点实验室,广州510301
南海链霉菌SCSIO 1635中抗霉素类化合物的分离鉴定
李苏梅,田新朋,牛四文,张文军,张 偲,鞠建华,张长生*
中国科学院南海海洋研究所海洋生物资源可持续利用重点实验室海洋微生物研究中心广东省海洋药物重点实验室,广州510301
我们从南海海底沉积环境分离了一株放线菌SCSIO 1635,经16S r DNA的序列分析将该株菌鉴定为链霉菌属。我们从该菌的发酵液中分离得到了4个化合物,经质谱和核磁共振波谱解析,确定为抗霉素类化合物:异构体antimycin A1a和A1b(1)、deisovalerylblastmycin(2)、kitamycin A(3)和antimycin A9(4)。
南海沉积环境;海洋放线菌;海洋微生物天然产物;链霉菌;抗霉素
Abstract:The strain SCSIO 1635 was isolated from marine sediment of the South China Sea,and identified as a member of Streptomyces according to the 16S rDNA sequence.Four compounds were isolated from Streptomyces sp.SCSIO 1635. The compounds were found to be structurally-related to antimycins by detailed analyses ofMS and NMR spectroscopic data,and were identified as antimycin A1aand A1b(1),deisovalerylblastmycin(2),kitamycin A(3),and antimycin A9(4),respectively.
Key words:marine sediments of South China Sea;marine actinomycetes;marine microbial natural products;Streptomyces;antimycins
Introduction
Marine microorganisms are rich in species diversity,genetic diversity and functionality diversity due to their unique living environment.Especially,marine actinomycetes are emerging as the most important new sources for novel drug discovery and have produced lots of secondary metabolites possessing excellent bioactivities, such as the anticancer compound salinosporamide A (NPI-0052)and the antibacterial compound abyssomicin C[1,2].The unusual productivity of novel compounds from marine actinomycetes shifts great interests of microbiologists and chemists from the land to the ocean[3].In recent years,more and more new genera and species of marine actinobacteria were describe from the South China Sea[4-6],suggesting South China Sea to be a rich source for exploring new species and new compounds.During our search of bioactive secondary metabolites from marine actinomycetes isolated from oceanic bottom sediments of South China Sea,the strain SCSIO 1635 was found to exhibit potent inhibitory activities to several tumor cells(data not shown).Herein we describe the identification of the strain as the genus of Streptomyces,and the isolation and structural elucidation of 4 antimycin-related compounds from this strain.Thus,we provided a new marine Streptomyces species capable of producing antimycins.
Experimental
General
1H,13CNMR and 2D NMR spectra were recorded on a Bruker AV-500 MHz NMR spectrometer with tetramethylsilane(TMS)as internal standard.Mass spectral data were obtained on an LCQDECA XP HPLC/MS spectrometer for ESIMS.Materials for column chromatography(CC)were silica gel(100-200 mesh;300-400 mesh;Jiangyou Silica gel development,Inc.,Yantai,P.R.China),Sephadex LH-20(40-70μm;Amersham Pharmacia Biotech AB,Uppsala,Sweden),Preparative Thin-Layer-Chromatography(TLC,0.3-0.4 mm)was conducted with precoated glass plates(silica gel GF254,Yantai).
Stra in isolation
The strain SCSIO 1635 was isolated from a sed iment sample collected fromSouth China Sea(E 113° 441320′,N 21°14.773′)at the depth of 65 m in September 2006,and deposited in the type culture collection of RNAM Center for Marine Microbiology,CAS, South China Sea Institute of Oceanology,Chinese A-cademy of Sciences,Guangzhou,China.
Identification of the stra in SCSI O 1635
Genomic DNA isolation,PCR amplification of 16S r DNA,sequence comparison,and phylogenetic tree construction were performed as described previously[6].
Fermentation and isolation
The strain SCSIO 1635 was cultured at 28℃for two days in 30 mL AM4 medium(soybean 20 g/L,peptone 2 g/L,glucose 20 g/L,starch 5 g/L,yeast extract 2 g/ L,NaCl 4 g/L,K2HPO40.5 g/L,MgSO4·7H2O 0.5 g/L,CaCO32 g/L,sea salt 30 g/L,adjusted to pH 718)in a 250 mL triangular flask(with a total of 30 such flasks),then the 30 mL precultures were transferred to 250 mL AM4 medium in a 1-L triangular flask (with a total of 30 such flasks),and were cultured at 28℃for 7 days.The fermentation broths(7.5 L) were filtrated by gauze.The mycelia were first extracted 3 times by acetone(3 L in total)and then by butanone(2 L in total)for 4 times.The filtrates were exhaustively extracted 5 times by butanone(25 L in total).The butanone phases were combined and dried by vacuum to obtain the crude extracts(12.7 g).The crude extracts were subjected to silica gel column chromatography(100-200 mesh,200 g)and were eluted with chlorofor m/methanol(100∶0 to 0∶100)to produce 6 fractions(Fr.1 to Fr.6).Fr.3(121.6 mg) was subjected to Sephadex LH-20 column chromatography(30 g)and was eluted with chloroform/methanol (1∶1)to afford 2 fractions(Fr.3-1 and Fr.3-2).Fr. 3-2(44.8 mg)was subjected to preparative TLC on a silica gel plate(20×20 cm)and was developed with chloroform/methanol(10∶1)to obtain 1(4.3 mg). Fr.4(247.8 mg)was chromatographed on a silica gel column(300-400 mesh,100 g)and was eluted with petroleum ether/ethyl acetate(8∶1 to 1∶1)to get 3 fractions of Fr.4-1,Fr.4-2 and Fr.4-3.Fr.4-2(75.6 mg)was separated by Sephadex LH-20(30 g)and was eluted with chloroform/methanol(1∶1)to afford Fr.4-2-1(10.1 mg).Fr.4-2-1 was loaded on silica gel plate(20×20 cm)and was developed with chlorofor m/methanol(10∶1)to afford 2(3.6 mg).Fr.4-1 (29.3 mg)was subjected to Sephadex LH-20(20 g) and eluted with chloroform/methanol(1∶1)to afford Fr.4-1-1(15.1 mg).Fr.4-1-1 was further separated on a silica gel plate(20×20 cm),developing with chloroform/methanol(10∶1),to afford compounds 3 (2.6 mg)and 4(6.6 mg).
Results and Discussion
The partial 16S rDNA sequence of the strain SCSIO 1635 was PCR amplified,sequenced and submitted to GenBank.The accession number is HM116843.NCB I Blast searching results showed that the new isolate had highest similarities to Streptomyces violascensISP 5183 (9917%),Streptomyces hydrogenans NBRC 13475T (9914%),Streptomyces odorifer DS M40347T (9912%),Streptomyces albidoflavusDS M40455T (99.2%).The phylogenetic tree generated by a neighbor-joining method clearly revealed the evolutionary relationship of the strain SCSIO 1635 to a group of Streptomyces species(Fig.1),indicating this strain to be a member of theStreptomyces genus.
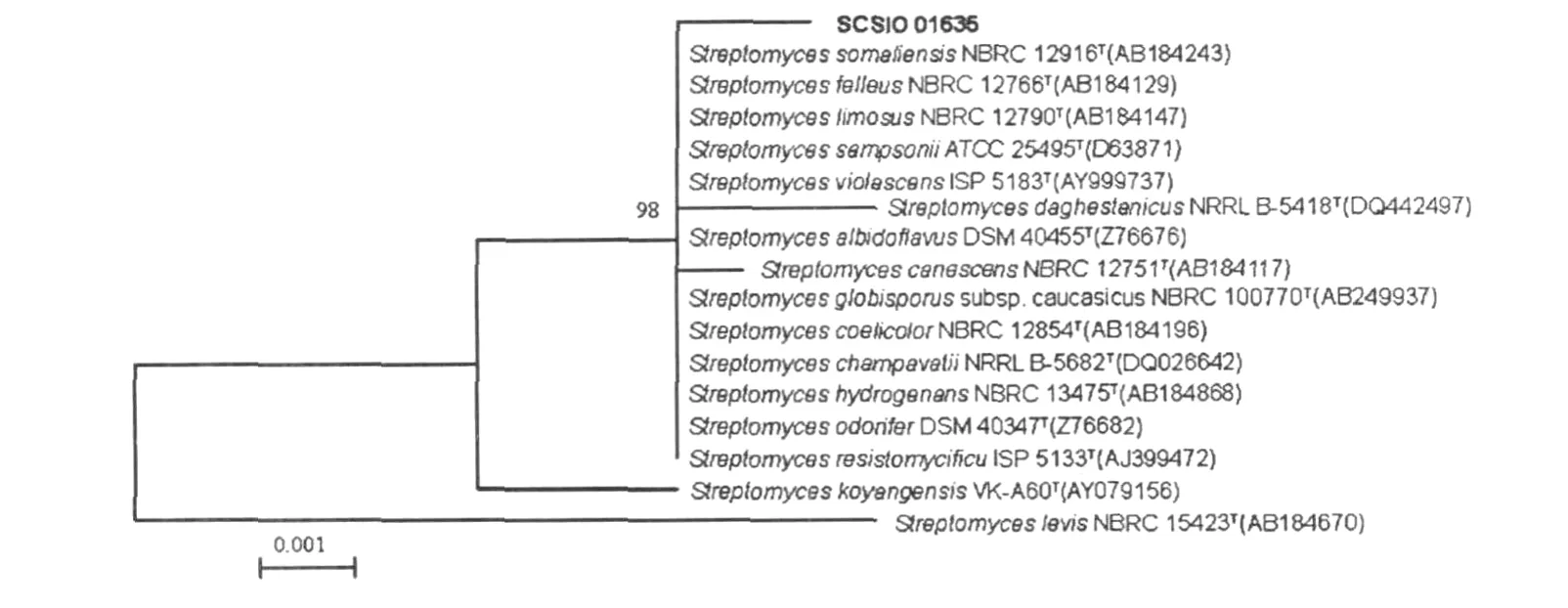
Fig11 Phylogenetic dendrogram of the stra in SCSI O 1635Tand its closest relatives reconstructed by the neighbor-join ing method based on 16S rDNA sequences
ture of 1a and 1b,Fig.2)was established by spectroscopic data of MS and 2D NMR and comparisons with reference[7].And compounds 2-4(Fig.2)were identified by directly comparing NMR data with known compounds in literatures[8-12].
Compound 1Colorless oil.The molecular for mula was deduced asC28H40N2O9on the basis of ESI-MS data(m/z571[M+Na]+and m/z547[M-H]-).The 1D and 2D NMR data of 1 were shown in Table 1. Comparing our data with the reference[7],we found that 1 was almost identical to antimycin A1,with a slight difference in the proportion of A1aand A1b.The percentage of A1bwas 20%in the reference,however, the value was 42%in compound 1.
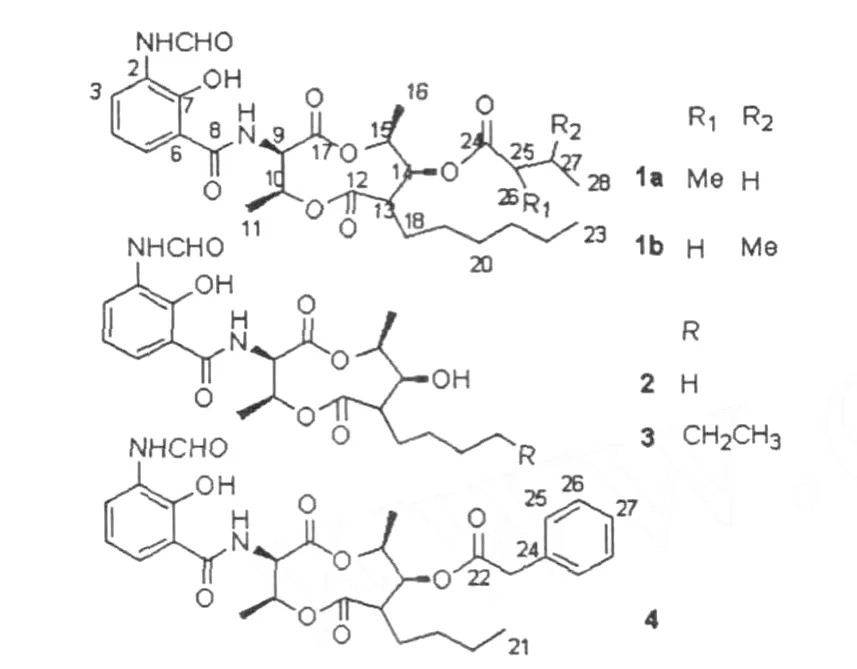
Fig12 Structures of compounds 1-4
Structure of the complex compound 1(an is omermix

Table 1 1D and 2D NM R(500 M Hz)spectral data for ant imycin A1aand A1bin CDCl3(δppm,JHz)
Compound 2Colorless oil.Its molecular for mular was deduced asC21H28N2O8on the basis of ESI-MS data(m/z459[M+Na]+and m/z435[M-H]-)11HNMR(CD3OD,500MHz)δ:8137(1H,s,H-1),8128 (1H,d,J=810 Hz,H-3),7172(1H,d,J=810 Hz, H-5),6195(1H,t,J=810 Hz,H-4),5165(1H,t,J =710 Hz,H-9),5134(1H,d,J=710 Hz,H-10), 4179(1H,dt,J=610,915 Hz,H-15),3143(1H,t, J=1010 Hz,H-14),2133(1H,dt,J=310,1010 Hz, H-13),1165(2H,m,H-18),1143(3H,d,J=610 Hz,H-16),1133(2H,m,H-20),1125(2H,m,H-19),0194(3H,t,J=715 Hz,H-21)113C NMR (CD3OD,125 MHz)δ:16212(d,C-1),12811(s,C-2),12617(d,C-3),11917(d,C-4),12413(d,C-5),11519(s,C-6),15214(s,C-7),17013(s,C-8),5513 (d,C-9),7214(d,C-10),1818(q,C-11),17617(s,C-12),5317(d,C-13),7811(d,C-14),7719(d,C-15), 1514(q,C-16),17112(s,C-17),2918(t,C-18),2317 (t,C-19),3017(t,C-20),1412(q,C-21)1The1H and13CNMR data of compound 2 were identical to those of deisovalerylblastmycin in literatures[8,9],thus 2was identified as deisovalerylblastmycin 1

?
Compound 3Colorless oil1Its molecular for mular was deduced as C23H32N2O8on the basis of ESI-MS data(m/z487[M+Na]+and m/z463[M-H]-)11HNMR(CDCl3,500 MHz)δ:12185(1H,s,OH-6), 8155(1H,d,J=810 Hz,H-3),8150(1H,s,H-1), 7194(1H,s,NH-1),7125(1H,d,J=810 Hz,H-5), 7109(1H,d,J=715 Hz,OH-8),6192(1H,t,J=810 Hz,H-4),5171(1H,penta,J=710 Hz,H-10),5125 (1H,t,J=718 Hz,H-9),4187(1H,dd,J=615,915 Hz,H-15),3160(1H,m,H-14),2135(1H,dt,J= 218,1013 Hz,H-13),1159(2H,brs,H-18),1146 (3H,d,J=615 Hz,H-16),1131(3H,d,J=618 Hz, H-11),1127-1130(8H,m,H-19-22),0187(3H,t,J =710 Hz,H-23)113C NMR(CDCl3,125 MHz)δ: 15910(d,C-1),12714(s,C-2),12418(d,C-3), 11910(d,C-4),12012(d,C-5),11217(s,C-6),15017(s,C-7),16915(s,C-8),5318(d,C-9),7017 (d,C-10),1510(q,C-11),17410(s,C-12),5212 (d,C-13),7714(d,C-14),7617(d,C-15),1814 (q,C-16),17011(s,C-17),2819(t,C-18),2215 (t,C-19),2712(t,C-20),3116(t,C-21),2911(t, C-22),1410(q,C-23)1Comparing the1H and13C NMR data with references[10],3 was determined to be kitamycin A1
Compound 4Colorless oil1Its molecular for mular was deduced as C29H34N2O9on the basis of ESI-MS data(m/z577[M+Na]+and m/z553[M-H]-)11HNMR(CDCl3,500 MHz)δ:8155(1H,d,J=810 Hz,H-3),8150(1H,s,H-1),7134(1H,d,J=618 Hz,H-26,28),7129(1H,d,J=618 Hz,H-25,27, 29),7124(1H,d,J=810 Hz,H-5),6192(1H,t,J= 810 Hz,H-4),5173(1H,dd,J=618,1410 Hz,H-10),5129(1H,t,J=715 Hz,H-9),5108(1H,t,J= 918 Hz,H-14),4198(1H,dd,J=615,918 Hz,H-15),3166(2H,s,H-23),2149(1H,dd,J=218, 1110 Hz,H-13),1159(1H,m,H-18,19),1130(1H, d,J=618 Hz,H-11),1116(1H,m,H-20),1115 (3H,d,J=615 Hz,H-16),1115(1H,m,H-19), 1106(1H,m,H-20),0181(3H,t,J=710 Hz,H-21).13CNMR(CDCl3,125MHz)δ:15910(d,C-1), 12715(s,C-2),12419(d,C-3),11910(d,C-4), 12011(d,C-5),11216(s,C-6),15017(s,C-7), 16914(s,C-8),5317(d,C-9),7110(d,C-10), 1510(q,C-11),17219(s,C-12),5011(d,C-13), 7518(d,C-14),7418(d,C-15),1718(q,C-16), 17011(s,C-17),2812(t,C-18),2913(t,C-19), 2214(t,C-20),1318(q,C-21),16916(s,C-22), 4116(t,C-23),13311(s,C-24),12912(d,C-25, 29),12818(d,C-26,28),12716(d,C-27)1Compound 4 was identified as antimycin A9by comparing the1H and13C NMR data with references[11,12]1
Antimycins are a series of antibiotics possessing antibacterial,anti-fungal,anthelmintic,anti-tumor and mitochondrial respiration inhibiting bioactivities[13]1 Antimycin A1(1)exhibits angiogenesis inhibiting activities and has been used as a tool in studying cell apoptosis[14].Antimycin A1(1)was also commercially available as a fish eradicant[15,16].Deisovalerylblastmycin(2)was weakly active against some bacteria and fungi[8].Kitamycin A(3)was found to inhibit plant growth[10].Antimycin A9(4)showed potent nematocidal and insecticidal activities[11].Thus,antimycinlike compounds have very wide applications.In the past 60 years,over 30 antimycin-like compounds have been described from microorganis ms,however,most producerswere isolated from soil samples[8,10,11,17-27]. Until very recently,3 marine-derived or mangrove endophytic actinomycetes were also found to produce antimycins[25,27].Our findings in this paper presented another new marinemicrobial source capable of producing this important class of bioactive compounds.
AcknowledgementsWe are grateful to Ms.Xiao,Ms. Sun,and Mr.Li of South China Sea Institute ofOceanology,CAS for recording NMR data,and Mr.Liu of South China Botanical Garden,CAS forMS data.
1 Feling RH,Buchanan GO,Mincer TJ,et al.Salinosporamide A:a highly cytotoxic proteasome inhibitor from a novelmicrobial source,a marine bacterium of the new genus Salinospora.Angew Chem Int Ed Engl,2003,42:355-357.
2 BisterB,Bisch off D,StrobeleM,et al.Abyssomicin C-A polycyclic antibiotic from a marine Verrucosispora strain as an inhibitor of the p-aminobenzoic acid/tetrahydrofolate biosynthesis pathway.Angew Chem Int Ed Engl,2004,43:2574-2576.
3 Williams PG.Panning for chemical gold:marine bacteria as a source of new therapeutics.Trends B iotechnol,2009,27(1): 45-52.
4 Tian XP,Tang SK,Dong JD,et al.Marinactinospora ther motoleransgen.nov,sp nov,a marine actinomycete isolated from a sediment in the northern South China Sea.Int J Syst EvolM icrobiol,2009,59:948-952.
5 Tian XP,Zhang YQ,Li QX,et al.Streptomyces nanshensis sp.nov.,isolated from the Nansha Islands in the South China Sea.Int J Syst EvolM icrobiol,2009,59:745-749.
6 Tian XP,Zhi XY,Qiu YQ,et al.Sciscionella m arinagen. nov.,sp.nov.,a marine actinomycete isolated from a sediment in the northern South China Sea.Int J Syst EvolM icrobiol,2009,59(Pt2):222-228.
7 Kim H,Esser L,Hossain MB,et al.Structure of antimycin A1,a specific electron transfer inhibitor of ubiquinol-cytochrome coxidoreductase.J Am Chem Soc,1999,121:4902-4903.
8 Ishiyama T,Endo T,Otake N,et al.Deisovalerylblastmycin produced by Streptomyces sp.J Antibiot,1976,29:804-808.
9 Aburaki S,KinoshitaM.Synthesis of deisovalerylblastmycin. Chem Lett,1976,7:701-704.
10 Hayashi K,Nozaki H.Kitamycins,new antimycin antibiotics produced byStreptomyces sp.J Antibiot,1999,52:325-328.
11 Shiomi K,Hatae K,Hatano H,et al.A new antibiotic,ant imycin A9,produced by Streptomyces sp.K01-0031.J Antibiot, 2005,58(1):74-78.
12 Nishii T,InaiM,Kaku H,et al.A practical total synthesis of (+)-ant imycin A9.J Antibiot,2007,60(1):65-72.
13 Wu Y,Yang YQ.An expeditious enantioselective synthesis of ant imycin A3b.J O rg Chem,2006,71:4296-4301.
14 MaedaM,Hasebe Y,Egawa K,et al.Inhibition of angiogenesis and H IF-1alpha activity by ant imycin A1.B iol Pharm Bull,2006,29:1344-1348.
15 Derse PH,Strong FM.Toxicity of Antimycin to Fish.Nature, 1963,200:600-601.
16 Greselin E,Herr F.Further toxicity studies with ant imycin,a fish eradicant.J Agric Food Chem,1974,22:996-998.
17 Dunshee BR,Leben C,Keitt G W,et al.The isolation and properties of antimycin A.J Am Chem Soc,1949,71:2436-2437.
18 Watanabe K,Tanaka T,Fukuhara K,et al.Blas tmycin,a new antibiotic from Streptomyces sp.J Antibiot,1957,10(2):39-45.
19 Liu WC,Strong FM.The chemistry of antimycin A.V I.separation and properties of antimycin A subcomponents1,2.J Am Chem Soc,1959,81:4387-4390.
20 KluepfelD,Sehgal SN,Vezina C.Antimycin A components. I.Isolation and biological activity.J Antibiot,1970,23(2): 75-80.
21 Barrow CJ,Oleynek JJ,MarinelliV,et al.Ant imycins,inhibitors ofATP-citratelyase,from a Streptomyces sp.J Antibiot, 1997,50:729-733.
22 Chen G,Lin B,Lin Y,et al.A new fungicide produced by a Streptomyces sp.GAAS7310.J Antibiot,2005,58:519-522.
23 HosotaniN,Kumagai K,Nakagawa H,et al.AntimycinsA10-A16,seven new antimycin antibioticsproduced by Streptomyces spp.SPA-10191 and SPA-8893.J Antibiot,2005,58: 460-467.
24 Liu N(刘宁),Zhang H(张辉),Zheng W(郑文),et al.Bioactivity of endophytic actinomycetes from medicinal plants and secondary metabolites from strain D62.Acta Microbiologica Sinica(微生物学报),2007,47:823-827.
25 Strangman WK,Kwon HC,Broide D,et al.Potent inhibitors of pro-inflammatory cytokine production produced by a marine-derived bacterium.J M ed Chem,2009,52:2317-2327.
26 Yan LL,Han NN,Zhang YQ,et al.Antimycin A(18)produced by an endophytic Streptomyces albidoflavus isolated from a mangrove plant.J Antibiot,2010,doi:10.1038/ja. 2010.1021.
27 Xu LY(许琳雅),Wang C(王成),Xu W(徐文),et al. Chemical constituents of antifungal actinomycete strain(No. H74-18)from the mangrove zone of South China Sea.Nat Prod Res Dev(天然产物研究与开发),2010,22:189-192.
Antimycins from Marine Strep tomycessp.SCSIO 1635 from the South China Sea
LI Su-mei,TIAN Xin-peng,NIU Si-wen,ZHANGWen-jun,ZHANG Si,JU Jian-hua,ZHANG Chang-sheng*
CAS Key Laboratory of Marine B io-resources Sustainable U tilization,RNAM Center forM arineM icrobiology,CAS,Guangdong Key Laboratory of Marine Materia Medica,South China Sea Institute of Oceanology,Chinese Academy of Sciences,Guangzhou 510301,China
Q936;R284.1
A
1001-6880(2011)01-0010-06
Received May 10,2010;Accepted July 30,2010
Foundation item:This research was supported by the Funds of the Chinese Academy of Sciences for Key Topics in Innovation Engineering (KZCX2-Y W-216,KSCX2-Y W-G-065,KSCX2-Y W-G-073),Natural Science Funds of South China Sea Institute of Oceanology for Young Scholar(SQ200903),Funds of Frontier Areas of SCSIO (LYQY200805),Open Project Program of Key Laboratory of Marine Bio-resources Sustainable Utilization(LMB091013).
*Corresponding authorTel:86-20-89023038;E-mail:czhang2006@ gmail.com

