Molecular dynamics simulations of A-DNA in bivalent metal ions salt solution
Jingjing Xue(薛晶晶), Xinpeng Li(李新朋), Rongri Tan(谈荣日), and Wenjun Zong(宗文军)
Department of Physics,Jiangxi Science and Technology Normal University,Nanchang 330013,China
Keywords: transition of DNA structure, bivalent metal ions, molecular dynamics simulations, effect of concentration
1. Introduction
Deoxyribonucleic acid (DNA) is an important nucleic acid in biological cell.[1]DNA,which stores genetic information,is an essential and necessary biological molecule for daily activities of living organisms.[2,3]Human biological characteristics are determined by DNA in the cell nucleus. By transcription and translation,the genetic information in DNA can guide the process of protein synthesis,to ensure normal physiological activities.DNA is double helix structure consisting of two antiparallel polydeoxynucleotide chains.[4]Deoxyribonucleotide is the basic unit of DNA,including a nitrogenous base,a phosphate group and a deoxyribose,as shown in Fig.1(a).[5]Four kinds of nitrogen-containing nucleobases are cytosine,guanine,adenine and thymine.The folded form of sugar group is called sugar pucker.[6]In a real environment, sugar moiety is not a standard plane, C atom will always offset from the plane of sugar moiety to form the sugar pucker.[7]When the C3 atom and C5 atom offset from the plane of sugar moiety in the same direction, the sugar pucker is C3’-endo, which is the representative sugar pucker of A-form DNA. The sugar pucker type will be defined as C3’-endo, when the phase angle of pseudorotation range is 0°-36°. As shown in Fig.1(b),it means that the C2 atom and C5 atom offset from the plane of sugar moiety in the same direction and its phase angle of pseudorotation range is 144°-180°, when the sugar pucker is C2’-endo.[8]It can be seen from Fig. 1(c), sugar-phosphate backbone and base stacking will cause the formation of minor groove and major groove. Three typical DNAs are A-DNA,B-DNA, Z-DNA.[9]B-DNA, which is the commonest DNA,has C2’-endo sugar pucker. For B-DNA,the major groove is wider than minor groove.[10]The structure of A-DNA,similar to the B-DNA, has a right-handed double helix. A-DNA, in which the width of major groove is close to the minor groove,has more significant C3’-endo sugar pucker than C2’-endo. ZDNA is a left-handed double helix,its whole helical structure is a zigzag pattern.[11]Based on the difference of DNA structure characteristics,the DNA type can be defined.
Understanding the dynamics of DNA structure and its interactions with other molecules becomes crucial for the studies of gene regulation,protein expression.The studies of DNA structure have significant meaning for the fields of DNA drugs and gene therapy which are developing rapidly now.[12-14]Except for the biological fields, DNA structure researches also are of importance to the DNA nanotechnology fields. For example,functionalized gold nanoparticles can be used for DNA sensing by using the characteristics of A/B form DNA conformational changes.[15]In pure water,B-form DNA is the stable configuration of DNA.[16]While A-DNA is only seen under low-humidity environments,sometimes a phenomenon can be found such that A-DNA is bound to certain proteins. Most importantly,the structure of DNA is sensitive to the presence of salt. Experimental studies on DNA structure have shown that A/B form transitions can be induced by changing DNA environments.[17]DNA structure always adopts the B-form in high relatively humidity conditions,but DNA structure can be reversibly driven to the A-form in certain low humidity environments. When some certain organic solvents are introduced as a cosolvent, DNA structure will exhibit reversible A/B transitions.[18]
In the previous years,Cheatham and Kollma investigated the transition from A-DNA to B-form DNA in aqueous solution by MD simulations.[19]Many researchers made series related discussion about the transform of A-DNA structure.From both experimental and theoretical aspects,these studies show that A-form structure could be stabilized in salt solution such as KCl, NaCl solution or under dehydrating conditions such as with the addition of ethanol.[20,21]A majority of studies of DNA structure are carried out on the interplay between B-DNA and monovalent cations, while relatively little research is implemented on the A-DNA structure and divalent ions like Mg2+ions.[22,23]As a matter of fact, divalent ions have stronger interactions with DNA chains compared with univalent ions.
In this work, a molecular dynamics (MD) simulation method is employed to analyze the transition of A-form DNA in different solvents.[24]The conformation of A-DNA structure will be investigated in pure water and different salt solutions, such as MgCl2, ZnCl2, CaCl2. The detailed structural properties of A-DNA will be provided and the rule of the interaction of divalent metal ions with A-DNA will also be disclosed.

Fig. 1. (a) The plane structure of deoxyribonucleotide. (b) The C2’-endo sugar pucker and the C3’-endo sugar pucker.(c)The major groove and the minor groove of DNA structure. (d)The double helix structures of A-DNA(left)and B-DNA(right).
2. Computational procedures
The initial state of A-DNA duplex with a nucleotide sequence of (CCCGGCCGGG) comes from the entry 1ZEX in the Protein Data Bank (PDB).[25]The reason why chose this DNA structure is high CG content. As known to all,high CG will contribute to stabilize A-DNA conformations.[26,27]
All MD simulations were carried out by GROMACS 5.0.7 program with interaction potential taken from AMBER bsc0 force field.[28,29]During the simulations, the standard molecular dynamics techniques were used,including periodic boundary conditions and the velocity Verlet integrator. Longrang electrostatic interactions were calculated by the particle mesh Ewald summation method to avoid cutoff effects.[30]The size of the simulation cubic box(57 °A×57 °A×57 °A)is large enough to ensure that the DNA does not interact with its periodic images, containing one A-DNA, metal ions and water molecules.[31]All of simulations are implemented at constant temperature 300 K and constant pressure 1 bar. The time step is 2 fs and each of the simulations process runs 50 ns. First,DNA structure is solvated in the simulation cubic box, and equilibrated by energy minimization of the solvent to ensure that the system has no steric clashes or inappropriate geometry.Experiencing NVT equilibration, the temperature can be stabilized at the desired value. Then the equilibration of pressure is conducted under an NPT ensemble. After the two equilibration phases, MD simulation is run at the desired temperature and pressure.
After simulations, DNA structural parameters were analyzed by the program 3DNA, which can generate the typical A-DNA and the typical B-DNA structure with the (CCCGGCCGGG)nucleotide sequence. Among the structure parameters,groove width and sugar pucker type are crucial factors and essential standards to determine DNA type. Minor and major groove widths use directly the P-P distance,which takes into account the direction of the sugar-phosphate backbones.[32]The sugar pucker type is determined by the phase angle of pseudorotation.
3. Results and discussion
3.1. The transition of A-DNA in aqueous solution
The MD simulations of A-DNA in aqueous solution have been performed as a reference. There are 5506 water molecules and 9 Mg2+counter ions in this system.
As is known to all, the root-mean-square deviation(RMSD) can represent the changes of structure and a little change of DNA structure can cause large fluctuating RMSD values.[33]From Fig. 2(a), it can be seen that RMSD values present large fluctuation relative to the initial 1ZEX structure and gradually decrease in comparison to the typical B-form DNA. This means that the structure of DNA gives rise to a great change during the process of evolution. Compared with the previous work by Cheatham and Kollma,[19]the RMSD values of A-DNA structure is 3.1 °A to 3.6 °A away from the B-DNA after 1 ns simulations in their experimental data. In Fig.2(a),the RMSD values relative to the typical B-DNA are around 0.3 nm in the initial phase, which are in agreement well with the previous data,and gradually stabilize at 0.2 nm.After experiencing 49.5 ns MD simulations, we collected the last 500 ps data to analyze the change of DNA sequence in comparison with the initial structure,as shown in Fig.2(b).

Fig. 2. (a) The RMSDs of the DNA trajectories with respect to the 1ZEX structures(black)and to the typical B-form DNA(red)during 50 ns in aqueous solution. (b) The 1ZEX structure and the overall facade structure of DNA in aqueous solution.
It can be seen obviously that the last structure of DNA sequence is rather extended than compact compared to the initial state. Phosphate groups are hydrophilic, so the binding of water molecules to the phosphate group is more favored than to base. In pure water, the water activity is high,the phosphate group can be fully and separately hydrated. In consequence, the phosphate groups of DNA are far apart and are independently hydrated, tending to the form of compact DNA structure.[34]The structure parameters of DNA include the types of sugar pucker,the width of minor groove and major groove,X-displacement(X-disp),helical rise(H-rise)and slide,as shown in Table 1 and Table 2.[35,36]The sugar puckers of DNA starting structure are all C3’-endo,which is the typical characteristic of A-DNA.The width of minor groove is wider and close to that of major groove for the starting DNA.After evolution of time,most of the sugar pucker types have become C1’-exo and C2’-endo,which are the typical characteristics of B-DNA.[37]Its major groove width is much wider than minor groove. In addition,the other structure data are rather close to typical B-form DNA than A-form DNA.[38]The structure parameters are close to those in the experiment by Cheatham and Kollma,where the minor groove width was reduced to around 13 °A,and the X-disp rised was up to around 2.5 °A.
Therefore,it can be identified basically that the last structure of DNA appears B-like features. It can be easily drawn a conclusion that the structure of A-DNA will transfer into B-like form in aqueous solution, which is consistent with the previous work.[39]
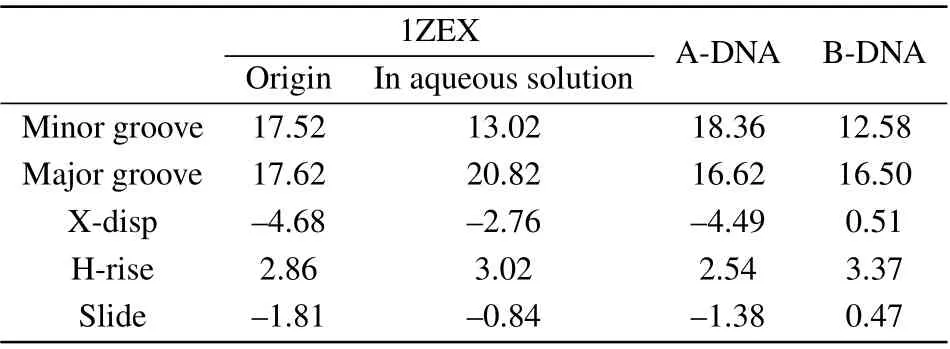
Table 1.The DNA structural parameters,including the DNA in aqueous solution,1ZEX,A-form DNA and B-form DNA.
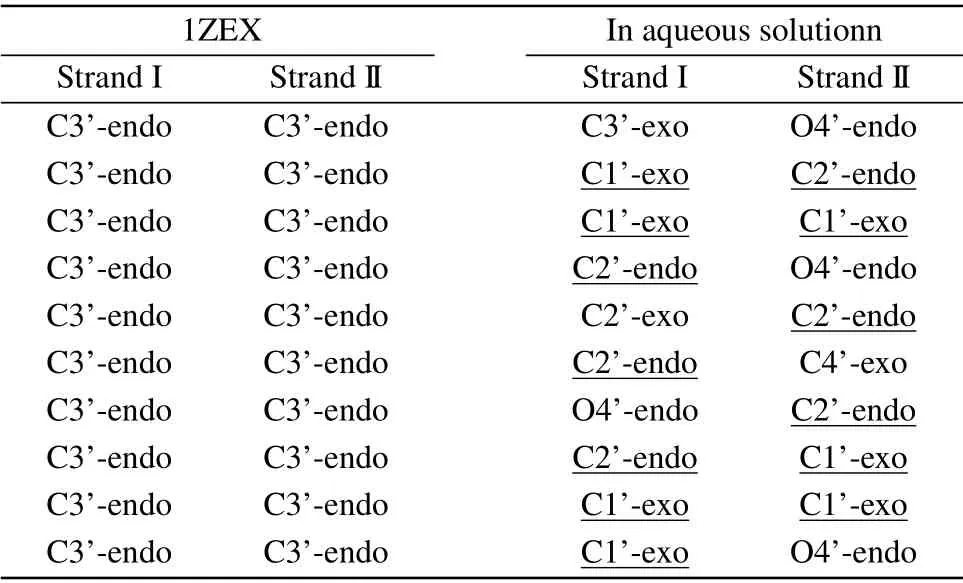
Table 2. The base pair sugar puckers of DNA structure,including DNA structure in aqueous solution and the starting DNA structure 1ZEX.
3.2. Metal ions concentration and A-DNA conformation
In pure water, the structure of A-DNA is unstable and experiences a transition from A-form to B-form. However,the previous experimental studies supported that the structure of A-DNA structure can be held in Na+ions solution with high concentration.[40]A deep investigation was implemented to examine the influence of different concentrations of MgCl2salt solution on the structural stability of A-DNA.Figures 3(a)and 3(b) present respectively the RMSDs of the DNA trajectories with respect to the starting 1ZEX structure and to the typical B-form DNA in the different concentrations of MgCl2solution, such as 1 M, 3 M and 5 M. It can be seen clearly from Figs.3(a)and 3(b)that the RMSD values exhibit a great considerably fluctuation and the largest value with respect to the 1ZEX structure when the concentration is 1 M, while the corresponding RMSD values is the smallest compare to BDNA among three different concentrations of 1 M, 3 M and 5 M.Furthermore, all of the RMSD curves are smoother and smoother with the increase of Mg2+ions concentration. The RMSD values hardly change when the concentration is 5 M.It indicates that the structure of A-form DNA is more stable in the solution with higher concentration. We can see that the RMSD values with the respect to the 1ZEX are around 0.1 nm when the concentration of MgCl2solution is 5 M during the process of evolution. It demonstrates that the structural changes of DNA are comparatively small and the DNA exhibits the state of A-form characters in the MgCl2solution with 5 M.
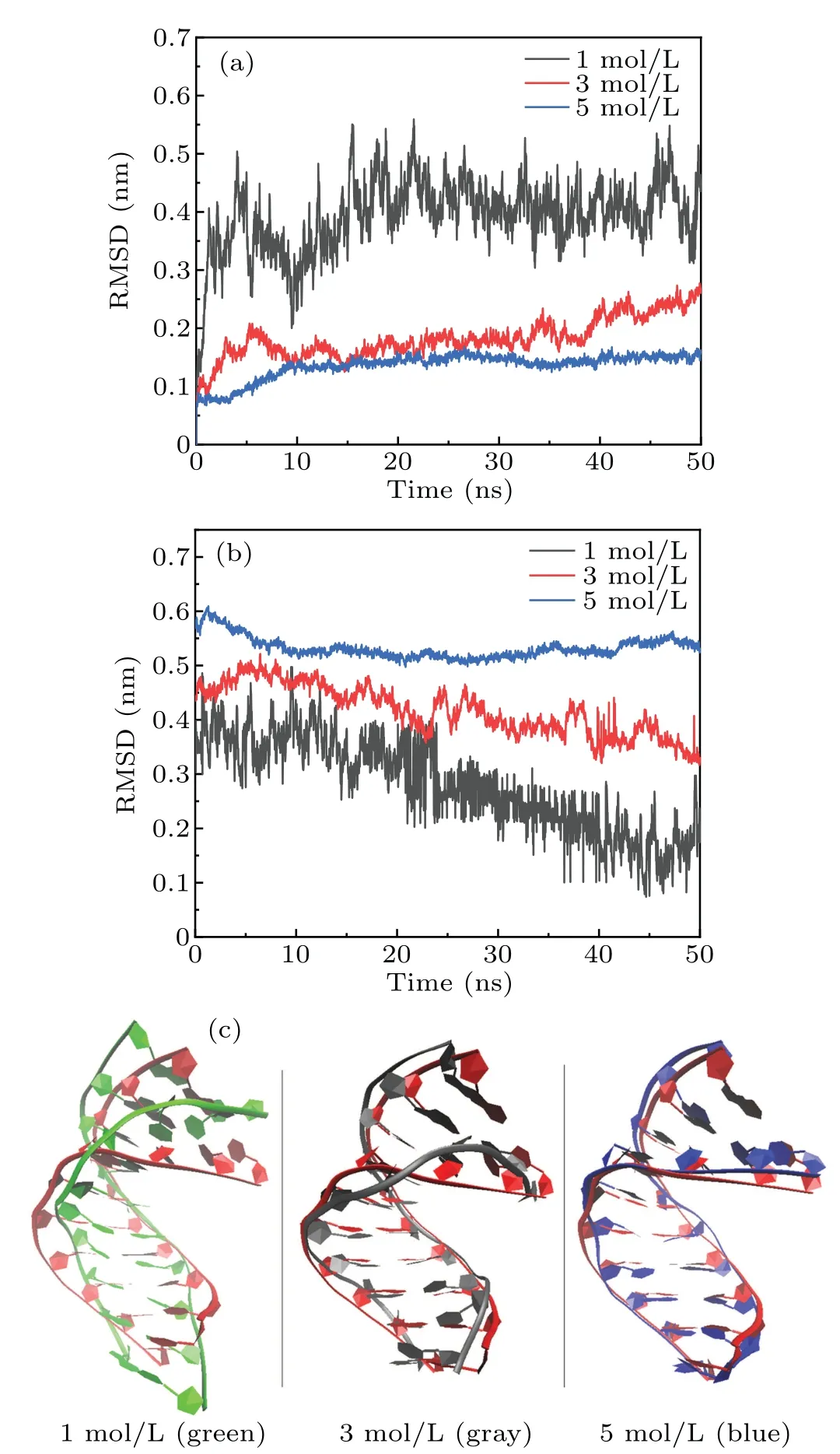
Fig. 3. (a) The RMSDs of the DNA trajectories with respect to the 1ZEX and (b) to the typical B-form DNA in three concentrations (including 1 M, 3 M, 5 M) of MgCl2 solution. (c) The overall facade structures of 1ZEX (red) and the DNA in MgCl2 salt three concentration(including 1 M(green),3 M(gray),5 M(blue))MgCl2 solution.
Figure 3(c) shows the overall facade structures of DNA in MgCl2salt solution with different concentrations. It can be easily observed that the averaged DNA structures becomes closer and closer to the starting 1ZEX structure with the increasing concentration.Combining with the RMSF(root mean square fluctuation) of DNA chains shown in Fig. 4(a), the changes of the DNA structure tend to happen in both ends of DNA chains instead of the center part of DNA chains. As the quantity of Mg2+increases, this kind of change becomes much weaker.
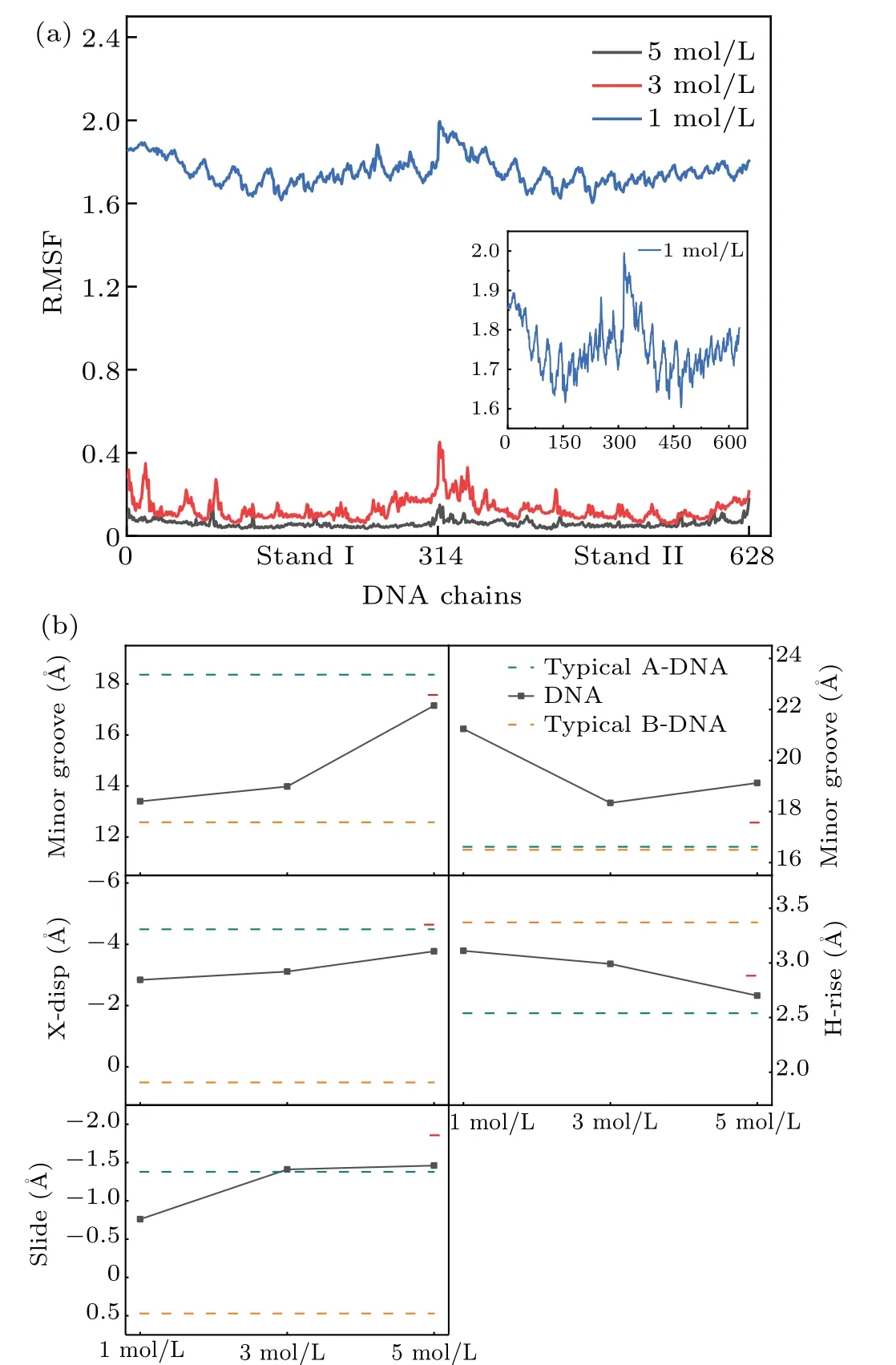
Fig.4. (a)The RMSF of DNA chains in three concentration(including 1 M,3 M,5 M)MgCl2 solution. The inset shows the enlarged view of the 1 M MgCl2 system. (b) The structural parameters of DNA structures in three concentration(including 1 M,3 M,5 M)MgCl2 solution and the horizontal dashed lines indicate the reference values of typical A(cyan)and B(orange)forms and the short red line shows the parameters of the 1ZEX.
The DNA structure parameters are provided in details for different concentrations of MgCl2, as shown in Table 3 and Table 4. From Table 3, it can be seen clearly that the major groove width becomes narrower from 21.24 °A to 19.12 °A and the minor groove is wider and wider from 13.40 °A to 17.18 °A with the increasing concentration from 1 M to 5 M.In the environment of higher concentration,the difference of width between major groove and minor groove is only 1.94 °A, which is similar to 1.74 °A in the typical A-DNA.On the other hand,we can describe the type of A/B-form DNA from the three characteristic quantity such as X-disp, H-rise and slide. The value of X-disp decreases from-2.84 °A to-3.77 °A and the change trend of the H-rise and slide value is similar to the X-disp when the concentration increases. It is worth mentioning that the H-rise value comes to 2.70 °A at 5 M, which is fairly close to 2.86 °A of 1ZEX and 2.54 °A of typical A-form.The change trend of different parameters are also depicted in Fig. 4(b). From these data, it is reasonable to believe that the structure of DNA tends to become the starting structure of 1ZEX or the typical A-DNA with the increasing concentration of metal ions. From Table 4,DNA structures appear more and more C3’-endo sugar pucker with the increasing concentration.In the environment of low concentration,the C3’-endo sugar puckers did not appear. Here,we calculated additionally some data in the concentration with 0.9 M to explain clearly the change of the structure. When the concentration of MgCl2up to 1 M, the C3’-endo sugar puckers begin to appear and Mg2+ions begin to have a direct effect on the stability of Aform DNA.The number of C3’-endo reaches 8 in 5 M MgCl2solution. In comparison with the previous work by Panet al.,[41]there were less C3’-endo sugar pucker in their simulations,most phase angles were larger in higher concentration of salt solution. In 4M NaCl solution, most phase angles of DNA structure are around 100°-160°,i.e.,the phase angles of C2’-endo and C1’-exo.
From the comparison of the above results, it is not difficult to find that the bivalent Mg2+ions work more effectively on the stability of A-DNA structure than the monovalent Na+ions. Even in the same concentration, this conclusion can be drawn. We can draw the conclusion that the presence of bivalent metal ions contributes to the stability of A-DNA in solution. The reason why the bivalent metal ions have great influences on the structure of A-DNA can be explained in the following.
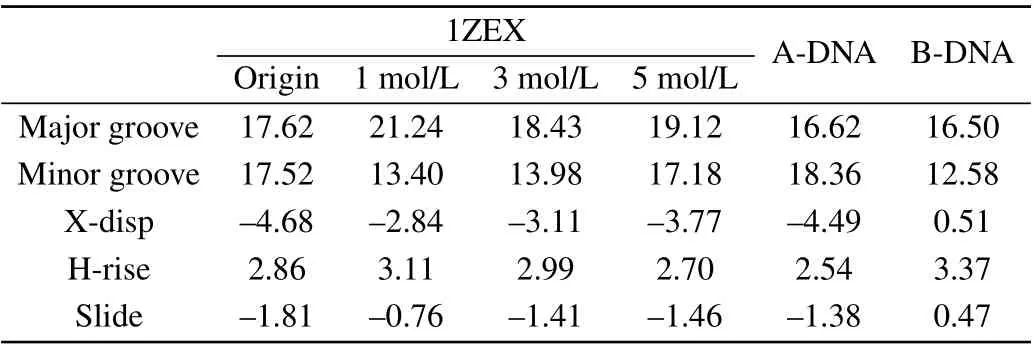
Table 3. The structural parameters of DNA structures in three concentrations(including 1 M,3 M,5 M)of MgCl2 solution.
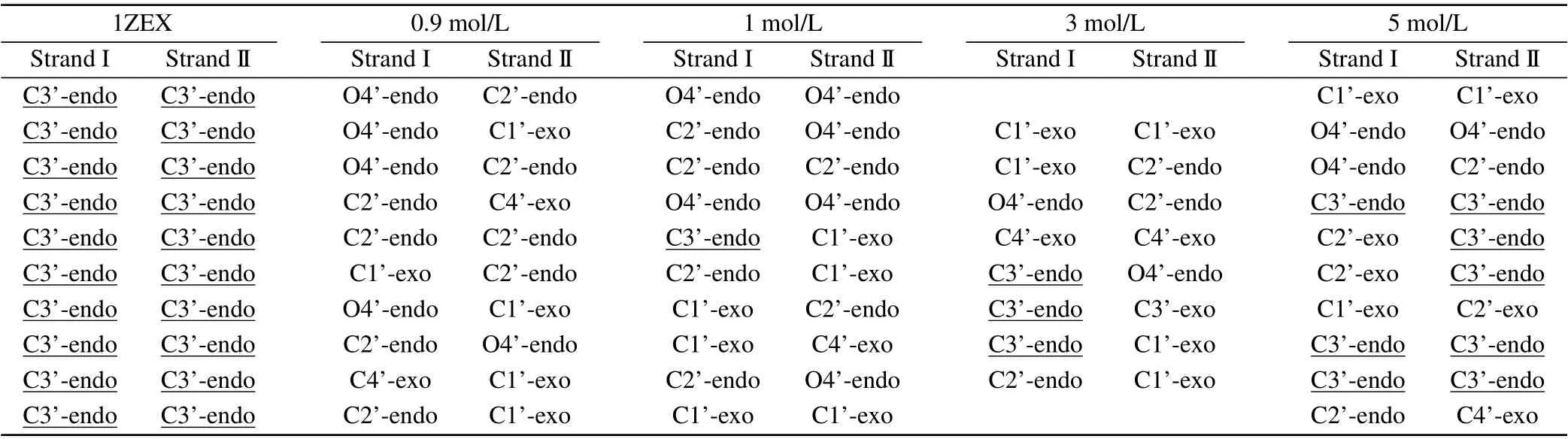
Table 4. The base pair sugar pucker of DNA structures in three concentrations of MgCl2 solution(including 1 M,3 M and 5 M).
Generally speaking,DNA molecule can hydrate with water molecules in both pure water and solution, so the hydration shells will be formed around phosphate groups of DNA,the space distribution and the volume slice of water molecules around phosphate groups of DNA are given in Figs. 5(a) and 5(b). Then, the water ridge will also be formed in both DNA major grooves and minor grooves. As the presence of metal ions in solution, in fact, the ion atmosphere will be formed around DNA strands.Metal ions can enter the deep and narrow major groove of A-form helix to clamp the two adjacent phosphate backbones, and consequently can stabilize more compact A-form structure.[40,42-44]For electronegative phosphate groups, electropositive metal ions can enter into solvation shells of phosphate groups, and interact with electronegative phosphate groups. It contributes to decline the electrostatic repulsion between DNA chains,which is beneficial to the formation of shorter and more compact DNA structure.[45,46]As we know,the radial distribution function(RDF)describes how the density of matter varies as a function of distance from a reference point, which is a power tool to describe the change of molecular micro structure. Figures 6(a)and 6(b)show the RDFs of Mg2+ions and oxygen atoms of water around electronegative oxygen atoms of phosphate groups. It can be seen obviously from Fig.6(a)that the first peak of the oxygen atoms of water is tall and narrow,which means that the first solvation shell is formed around the phosphate groups. While the Mg2+ions tend to be distributed at 0.18-0.19 nm from phosphate groups and have higher intensity,as shown in Fig.6(b). From the comparison of the RDFs of Mg2+with water, the radial distances of first peak are very close, almost 0.17-0.19 nm.It indicates that the distribution of Mg2+almost overlaps the first solvation shell around DNA phosphate groups. With the concentration increasing, the first peak of RDFs of Mg2+becomes higher and higher,while the density of Mg2+is stronger around phosphate groups. The number of Mg2+ions and water molecules in the first solvation shell are given in Fig.6(c),which indicates that the number of Mg2+ions around phosphate groups increases while that of water decreases with the increasing concentration of MgCl2solution. Certainly, the electrostatic interaction between Mg2+and phosphate groups are also gradually strengthen as the concentration increasing.Therefore, it is not difficult to understand that the metal ions can easily enter into the solvation shells around phosphate groups of DNA and interact with DNA chains.
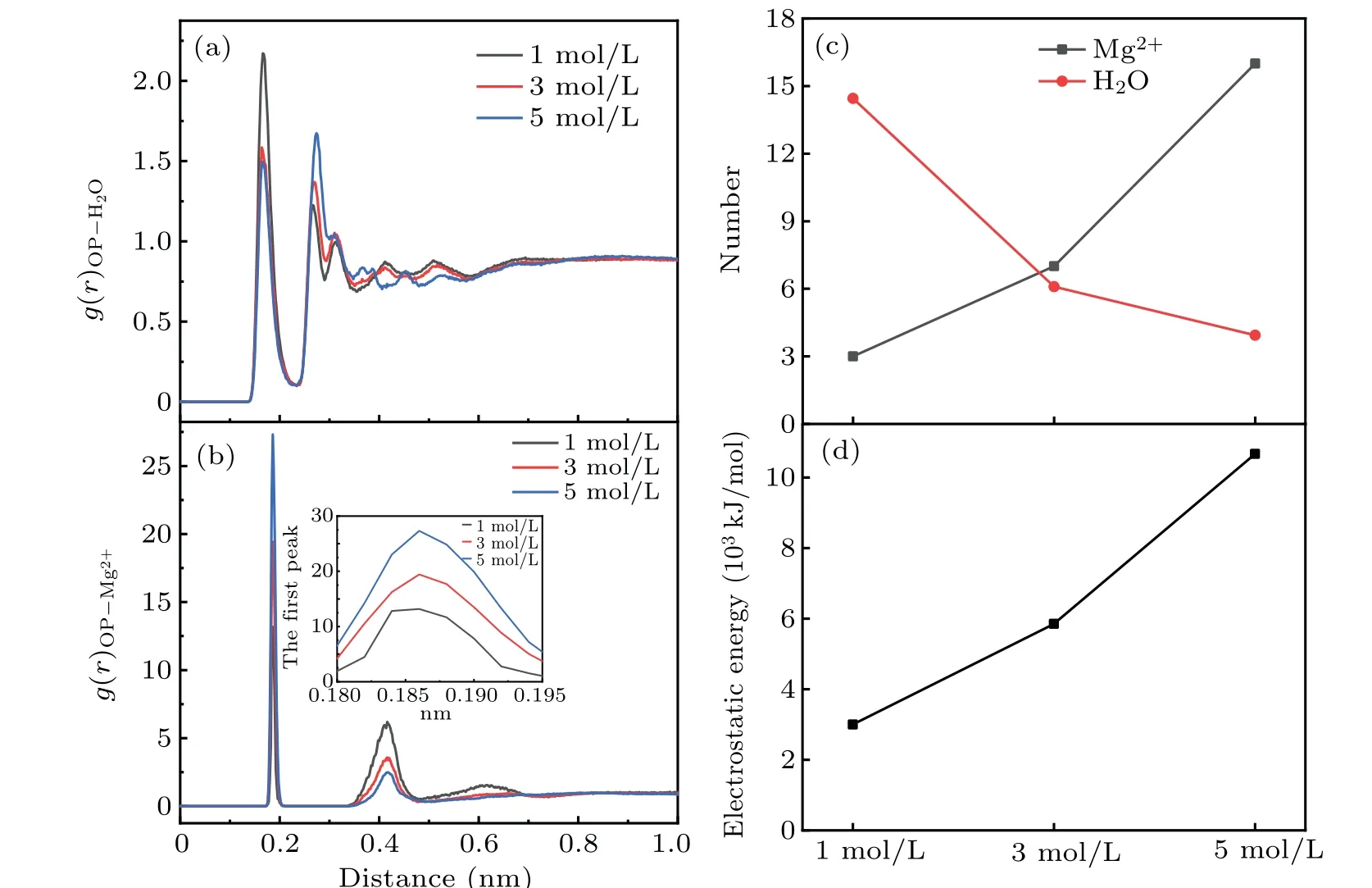
Fig.6. (a)The RDFs of Mg2+ ions and(b)water oxygen atoms around electronegative oxygen atoms of phosphate groups. (c)The number of Mg2+ ions and water molecules around 0.2 nm of phosphate groups. (d)The electrostatic interaction between Mg2+ and phosphate groups of DNA structure.
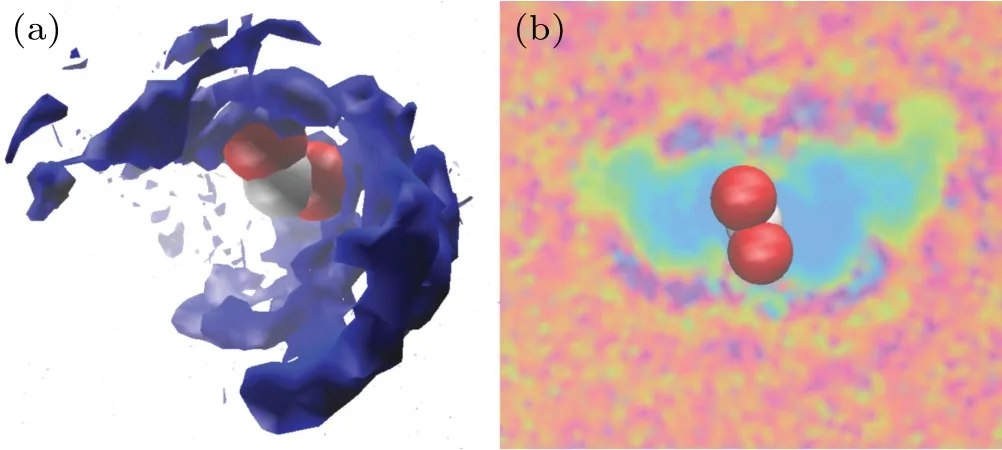
Fig.5. (a)The space distribution of water molecules around phosphate group.(b)The volumeslice of 3D view of water molecules around phosphate group.
3.3. Different bivalent metal cation and A-DNA
To understand deeply and comprehensively the influence of metal ions on the stability of A-DNA structure, we made some investigations in the presence of the various bivalent metal cation such as Mg2+, Zn2+and Ca2+ions. Here, we made a choice of 3 M strength according to the calculations above. Because it is easy to form cluster phenomenon if the concentration of metal ions is too high. Sometimes, too high concentration will cause the break of the hydrogen bond,in turn, producing a great influence on the stability of DNA structure.[47]It will be impossible for us to analyze correctly the physical process. The method and process in Subsections 3.1 and 3.2 are the same.The RMSDs of the DNA trajectories with respect to the 1ZEX structure and typical B-DNA in the three various bivalent metal cation salt solutions at 3 M strength are given in Figs.7(a)and 7(b),they are the solutions of Mg2+,Zn2+and Ca2+ions. Compared to Zn2+and Mg2+ions, the RMSD values of the Ca2+ions solution have larger ranges of fluctuation. This suggests that the 1ZEX structure is unstable in Ca2+ions solution. Figure 7(c)shows the final overall averaged facade structures of DNA in the salt solution of MgCl2, ZnCl2and CaCl2. Although there are slight differences among three averaged DNA structures, the ends of DNA chains in MgCl2and ZnCl2solutions are much more in accord with the initial structure. The similar parameters of DNA structure are given in Tables 5 and 6 detailedly in the three kinds of bivalent ions salt solution. From Table 5,it can be observed clearly that the sugar puckers of base pairs in the solutions of MgCl2and ZnCl2are almost same and hardly appear in the solution of Ca2+. As a reference,the widths of the major and minor grooves, X-disp, H-rise and slide are more close to A-form DNA in the solution of MgCl2and ZnCl2,but it seems to be exception in the solution of CaCl2, where the width of minor groove is 12.56 °A as same as 12.58 °A of Bform DNA. From this perspective, it seems that the structure of DNA is more unstable in the CaCl2solution than the other two kinds of solution. This phenomenon may be caused by the increase in the radius of calcium ions. Comprehensively,it indicates that Zn2+and Ca2+ions have the same effect on the stability of DNA structure as Mg2+ions from the changed characteristics of the RMSD values and DNA structure parameters.
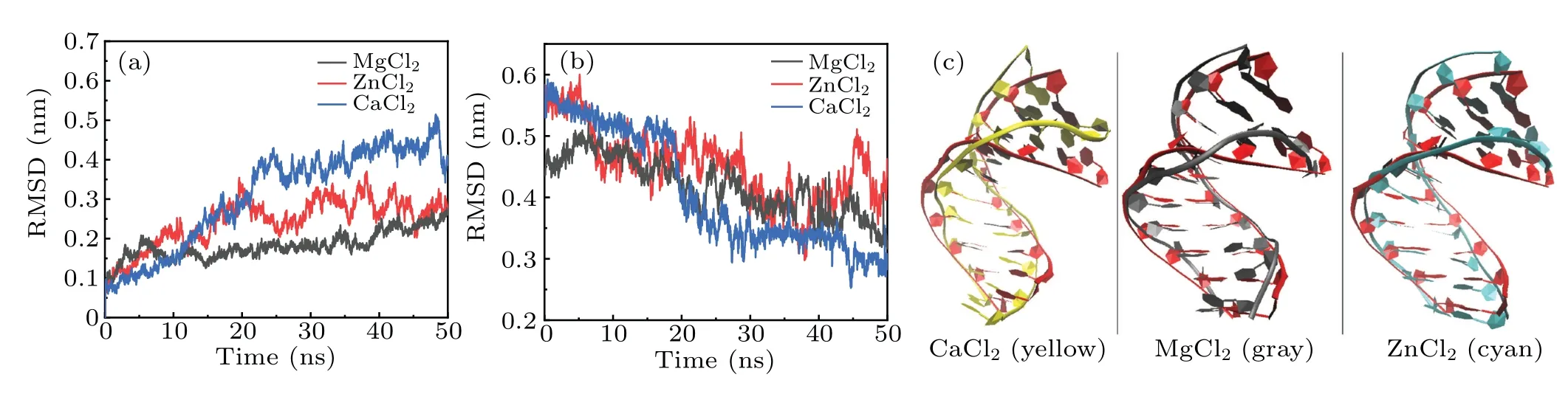
Fig.7. (a)The RMSDs of the DNA trajectories with respect to 1ZEX and(b)to the typical B-DNA in three 3 M salt solutions(including MgCl2,ZnCl2 and CaCl2). (c)The overall facade structures of 1ZEX(red)and the DNA in 3 M salt solutions of MgCl2,ZnCl2 and CaCl2.
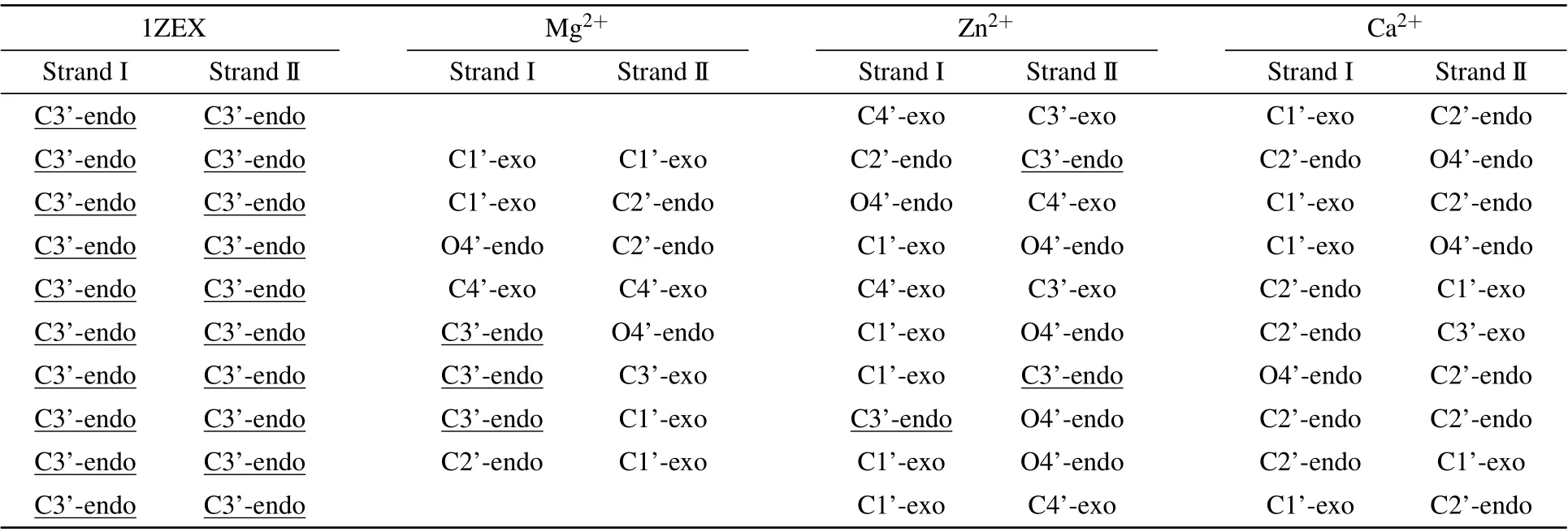
Table 5. The base pair sugar pucker of DNA structure in three 3 M salt solutions(MgCl2,ZnCl2 and CaCl2).
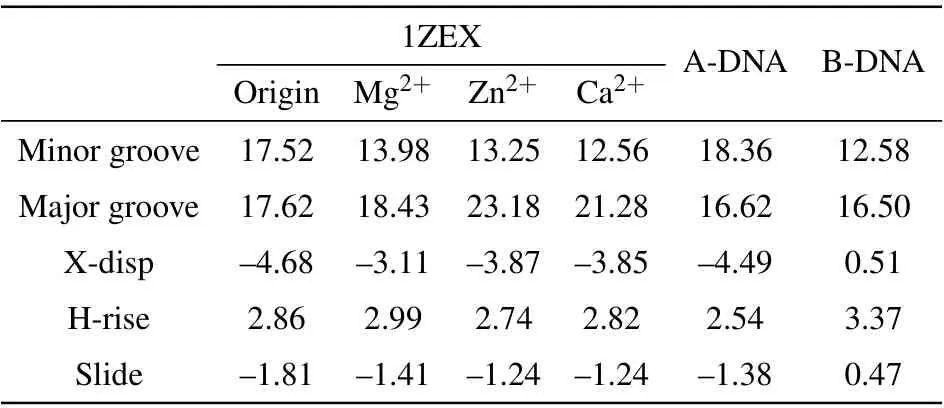
Table 6. The structural parameters of DNA structures in 3 M salt solutions of MgCl2,ZnCl2 and CaCl2.
It is obvious that different metal ions have different influences on DNA structure even in metal ions solution with same concentration and valency, which is related to metal ions inherent nature and characteristics. Because of the electrostatic interaction between metal ions and DNA chains, the metal ions are not distributed homogeneously in solution, most of the metal ions distribute around the DNA chains. In salt solutions,ions will form ionic atomsphere around DNA.The size of metal ions in salt solutions is an important factor to affect their distribution around DNA chains,which means that these ions with larger radii are difficult to enter the major and minor grooves of DNA.[48]We can see clearly from Fig.8(a)that the peaks of RDF appear at the position of 0.18 nm for the solution of Mg2+and Zn2+ions while the corresponding position at 0.26 nm for the solution of Ca2+ions.It indicates that Mg2+and Zn2+ions bind with phosphate groups more compact and their distribution is closer to phosphate groups. As a fact,the radius of Zn2+ions is very similar to Mg2+ions,while the radius of Ca2+ions is greater than Mg2+and Zn2+ions. Thus,Ca2+ions are harder to enter into the grooves of DNA double helix. The interaction force is also conducted to analyze the influence on the configuration of DNA due to the effect of metal ions. As shown in Fig. 8(b), the electrostatic force plays a major role in these systems,where their proportion is almost as high as 95%. The energy of interaction does not present a rule of increasing or decreasing when the ions radius increases.Here,the energy of interaction displays a maximum in the solution of Zn2+ions with the radius of 74 pm,the energy of interaction presents a minimum in the solution of Ca2+ions with radius of 100 pm. In fact,the radius of Mg2+is the smallest one, 72 pm, but the energy of interaction does not appear the maximum.
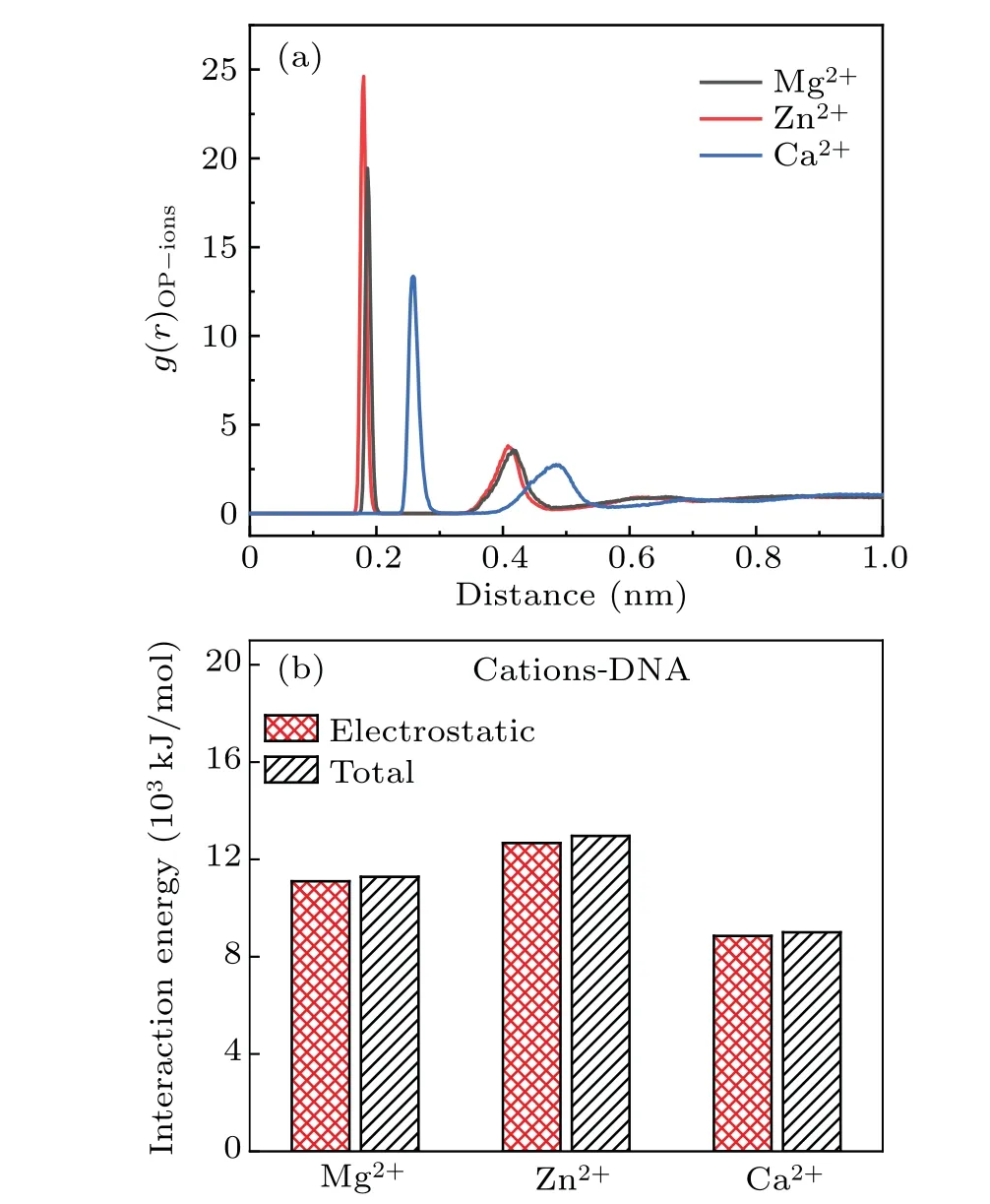
Fig.8. (a)The RDF of metal ions around electronegative oxygen atoms of phosphate groups in 3 M salt solution of MgCl2, ZnCl2 and CaCl2.(b) The electrostatic energies and total interaction energies between metal ions and DNA chains in 3 M salt solutions of MgCl2,ZnCl2 and CaCl2.
4. Conclusion
In summary,we have studied the structural transitions and the relevant theory of A-DNA in different metal ionic solutions using the molecular dynamics simulations. Based on the data from simulations,some conclusions can be drawn as follows.In aqueous solutions, the structure of A-DNA will eventually stabilize at the B-like DNA structure over time.However,with the addition of bivalent ions (included Mg2+, Zn2+, Ca2+)in solutions, A-form DNA can have relatively stable structure and is not transformed to B-form. The important reason should be placed on the interaction force between electropositive metal ions and electronegative phosphate groups. Most metal ions distribute around the DNA chains in high concentration salt solutions. Metal cations can enter into the solvation shells around the phosphate groups of DNA and generate a great electrostatic interaction with the electropositive DNA molecule. The metal ions in groove can connect neighboring phosphate groups along the DNA chains, which is conducive to the stability of more compact DNA structure. In other words, these metal cations can weaken electrostatic repulsion force of DNA chains and finally lead to the formation of shorter and wider A-DNA structure. Furthermore,the higher the ion concentration,the stronger the interaction force,and the more stable the structure of A-DNA.
Furhter,an important conclusion can be drawn that different cations have different influences on DNA though under the condition of same concentrations. The calculations indicate that the structure of A-DNA is less stable in CaCl2solution than in ZnCl2and MgCl2solutions. The radius of Ca2+ions is larger than Zn2+and Mg2+ions, therefore, it is more difficult for Ca2+ions to enter into the grooves of DNA chains.From the RDFs of metal ions, it is very clear that Zn2+and Mg2+ions would like to combine with phosphate groups and their distributions is more close to phosphate groups.
In fact, the transition of DNA structure is a complex process.[49-53]In the future, we will continue to investigate deeply the influence of metal ions on A-DNA structure under different conditions of temperature and concentration.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 11564015), the Foundation of Educational Committee of Jiangxi Province, China(Grant No.GJJ211112),and the Fund for Distinguished Young Scholars of Jiangxi Science&Technology Normal University(Grant No.2015QN-BJRC002).
- Chinese Physics B的其它文章
- Helium bubble formation and evolution in NiMo-Y2O3 alloy under He ion irradiation
- Dynamics and intermittent stochastic stabilization of a rumor spreading model with guidance mechanism in heterogeneous network
- Spectroscopy and scattering matrices with nitrogen atom:Rydberg states and optical oscillator strengths
- Low-overhead fault-tolerant error correction scheme based on quantum stabilizer codes
- Transmembrane transport of multicomponent liposome-nanoparticles into giant vesicles
- Evaluation on performance of MM/PBSA in nucleic acid-protein systems

