DNA Repair Gene Polymorphisms in the Nucleotide Excision Repair Pathway and Lung Cancer Risk: A Meta-analysis
Chao-rong Mei, Meng Luo, Hong-mei Li, Wen-jun Deng, Qing-hua Zhou,*
1Tianjin Key Laboratory of Lung Cancer Metastasis and Tumor Microenvironment, Tianjin Lung Cancer Institute, Tianjin Medical University General Hospital, Tianjin 300052, China;2West China Hospital, Sichuan University, Chengdu 610041, China;
3Cancer Center, the Affiliated Hospital, Qingdao University Medical College, Qingdao 266003, China
DNA Repair Gene Polymorphisms in the Nucleotide Excision Repair Pathway and Lung Cancer Risk: A Meta-analysis
Chao-rong Mei1, Meng Luo2*, Hong-mei Li3, Wen-jun Deng2, Qing-hua Zhou1,2**
1Tianjin Key Laboratory of Lung Cancer Metastasis and Tumor Microenvironment, Tianjin Lung Cancer Institute, Tianjin Medical University General Hospital, Tianjin 300052, China;2West China Hospital, Sichuan University, Chengdu 610041, China;
3Cancer Center, the Affiliated Hospital, Qingdao University Medical College, Qingdao 266003, China
Objective: A number of studies have reported the association of “XPA”, “XPC”, “XPD/ERCC2” gene polymorphisms with lung cancer risk. However, the results were conflict. To clarify the impact of polymorphisms of“XPA”, “XPC”, “XPD/ERCC2”, on lung cancer risk, a meta-analysis was performed in this study.
Methods: The electronic databases PubMed and Embase were retrieved for studies included in this meta-analysis by “XPA”, “XPC”, “XPD/ERCC2”, “lung”, “cancer/neoplasm/tumor/carcinoma”, “polymorphism” (An upper date limit of October, 31, 2009). A meta-analysis was performed to evaluate the relationship among XPA, XPC and XPD polymorphism and lung cancer risks.
Results: A total of 31 publications retrieved from Pubmed and Embase included in this study. XPC A939C CC genotype increased lung cancer risk in total population (recessive genetic model: OR=1.23, 95% CI:1.05-1.44; homozygote comparison: OR=1.21,95%CI:1.02-1.43and CC vs. CA contrast: OR=1.25,95%CI:1.06-1.48), except in Asians. XPD A751C, 751C allele and CC genotype also increased lung cancer risk in total population and in Caucasians (recessive genetic model: Total population: OR=1.20, 95%CI:1.07-1.35). No significant correlation was found between XPD A751C and lung cancer risk in Asians and African Americans. XPD G312A AA genotype increased lung cancer risk in total population, in Asians and Caucasians(recessive genetic model: Total population: OR=1.20, 95%CI: 1.06-1.36). No significant association was found between XPA G23A, XPC C499T, XPD C156A and lung cancer risk.
Conclusion: Our results suggest that the polymorphisms in XPC and XPD involve in lung cancer risks. XPA polymorphisms is less related to lung cancer risk.
Nucleotide excision repair; Polymorphism; Lung cancer; Meta-analysis
INTRODUCTION
Mutations are early events in carcinogenesis and defective DNA repair is a risk factor for many cancer[1]. The maintenance of genome integrity is very important for the survival of all organisms and DNA repair systems play a crucial role. The reduction in DNA repair capacity likely leads to birth defects, cancer and reduced lifespan[2]. At least three syndromes are associated with inborn defects in NER: Xeroderma pigmentosum(XP), Cockayne syndrome(CS) and trichothiodystrophy (TTD), all characterized by exquisite sun sensitivity. The prototype repair disorder, xeroderma 1111111111111111pigmentosum exhibits a dramatic >1000-fold incidence of sun-induced skin cancer[3]. DNA repair consists at least, four types of damaged DNA-nucleotide excision repair (NER), base excision repair (BER), double strand break repair (DSBR) and mismatchrepair (MMR)[4]. Among these, NER is a highly versatile and sophisticated DNA damage removal pathway that counteracts with the deleterious effects of a multitude of DNA lesions, including major types of damage induced by environmental sources. In eukaryotic cells, the process requires more than 30 proteins to perform at different steps[5]. Production by the shortwave UV component of sunlight and numerous bulky chemical adducts are eliminated by this process[6]. There are two subpathways of NER: global genome NER (GG-NER) and transcription- coupled NER (TC-NER). Damage to DNA that occurs anywhere in the genome is recognized by the XPC and XPE (or UV-DDB) protein complexes, which are specific components of the GG-NER system. Damage that actually blocks transcription [e.g., cyclobutanepyrimidine dimers (CPDs) resulting from exposure to UV radiation]is detected by the TC-NER system, which involves the CSB and CSA proteins. The DNA helix is opened by the XPB and XPD helicases of the repair and transcription factor IIH (TFIIH), allowing damage verification by the XPA protein. Single-strand binding protein RPA prevents reannealing and dual incisions in the damaged strand are caused by the ERCC1-XPF and XPG endonucleases, excising the damage as part of a piece of 25 to 30 bases. The single-strand gap is filled by the replication machinery, and the final nick sealed by DNA ligase[3,7].
XPA (xeroderma pigmentosum, complementation group A) locates at 9q22.3 and encodes a zinc finger protein with affinity for various DNA damages involved in DNA excision repair[8]. XPA has many interactions with other NER components, such as the single-strand binding complex replication protein A (RPA), the TFIIH complex and the ERCC1/XPF endonucleases. XPA may orchestrate the repair machinery around the DNA lesion when XPC/HHR23B with or without the help of TFIIH has locally opened the helix6. XPA G23A (reference SNP ID: rs1800975) is an important polymorphism in XPA that is a A to G (or G to A) transition at fourth nucleotide upstream from the ATG start codon in 5’-UTR.
XPC (xeroderma pigmentosum, complementation group C) is located at 19q13.3. It encodes a component of the NER pathway and plays an important role in the early steps of GG-NER. In GG-NER, XPC and HR23B form XPC-HR23B complex to recognize DNA damage and initiate to combine with other NER factors to accomplish the NER[5,9]. The important polymorphisms of XPC are XPC C499T, XPC A939C and XPC PAT. XPC C499T (XPC Ala499Val; reference SNP ID: rs2228000) is a C-to-T transition in exon 8, resulting in the change Ala499—> Val. XPCA939C(XPC Lys939Gln: reference SNP ID: rs2228001) is a A-to-C transition in exon 15, leading to the change Lys939—>Gln. Poly-AT(XPC PAT) consisting of an insertion of 83 bases of A and T[poly(AT)] and a 5 base deletion within intron910. As XPC A939C was found to be in strong linkage disequilibrium with the XPC PAT[10,11]. We just recruited XPC A939C in our meta-analysis in the two polymorphisms. XPD (ERCC2, excision repair cross-complementing rodent repair deficiency, complementation group 2) is located at 19q13.3. XPD protein involves in transcription-coupled nucleotide excision repair and is an integral member of the basal transcription factor BTF2/TFIIH complex. The gene product has ATP-dependent DNA helicase activity and belongs to the RAD3/XPD subfamily of helicases. Thepolymorphisms of XPD are XPD A751C, XPD G312A and XPD C156A.XPD A751C (XPD Lys751Gln; reference SNP ID: rs13181) is an A-to-C transition in exon23, resulting in the change Lys751—> Gln.
XPD G312A (XPD Asp312Asn; reference SNP ID: rs1799793) is a G-to-A transition in exon10, resulting in the change Asp312—> Asn. XPD C156A (XPD Arg156Arg; reference SNP ID: 238406) is a C-to-A transition in exon6 and is a silent polymorphism.
In recent years, an increasing body of epidemiologic studies on lung cancers demonstrated that the potential role of polymorphisms of genes in NER pathway. However, the results showed controversial. Herein, we perform a meta- analysis to evaluate the associations between the polymorphisms of XPA, XPC and XPD genes in NER pathway and lung cancer risk.
MATERIALS AND METHODS
Search Strategy and Identification of Eligibility of Studies
To examine the association of XPA, XPC, XPD polymorphisms with lung cancer, we conducted a computerized literature search of Pubmed database and EMBASE( last search updated in October 2009) using the keywords and subject terms “XPA”, “XPC”, “XPD/ ERCC2”, “lung” “cancer/neoplasm/tumor/carcinoma”,“polymorphism” in published references. The studies included in our meta-analysis all met the following criteria: The study use an unrelated case-control design and had genotype frequency available.
Data Extraction
Data were collected on the genotype of XPA G23A(rs1800975), XPC C499T (rs2228000), XPC A939C (rs2228001), XPD A751C(rs13181), XPD G312A(rs1799793) and XPD C156A(rs238406), We collected following data from each studies: first author's surname, year of publication, ethnicity, country of origin, source of controls, genotyping method, number of case and control groups, minor allele frequency of controls. The following categories for ethnicity in our meta-analysis were listed: Caucasian; Asian; African Americans; Mexican American; Latinos; Mixed. The details of these studies in the meta-analysis were in Table 1.
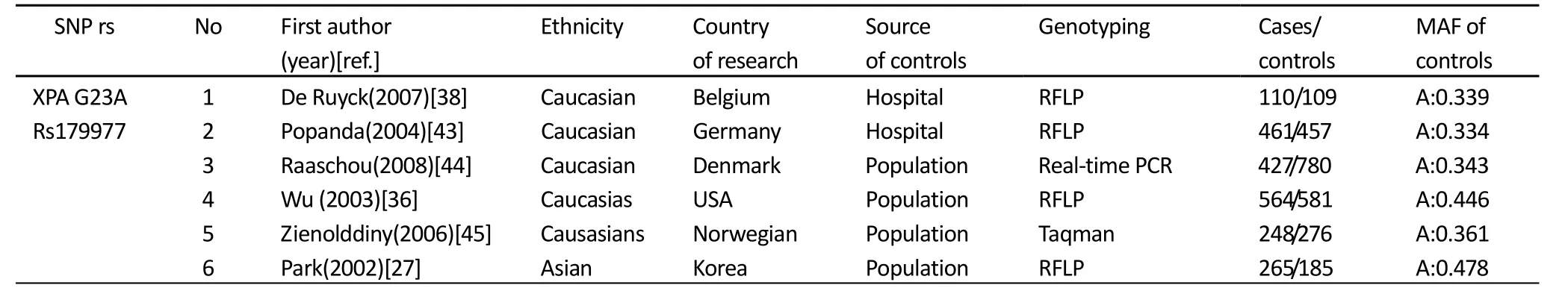
Table 1.Charaacteristics of studies included in the meta-analysis

(Continued)
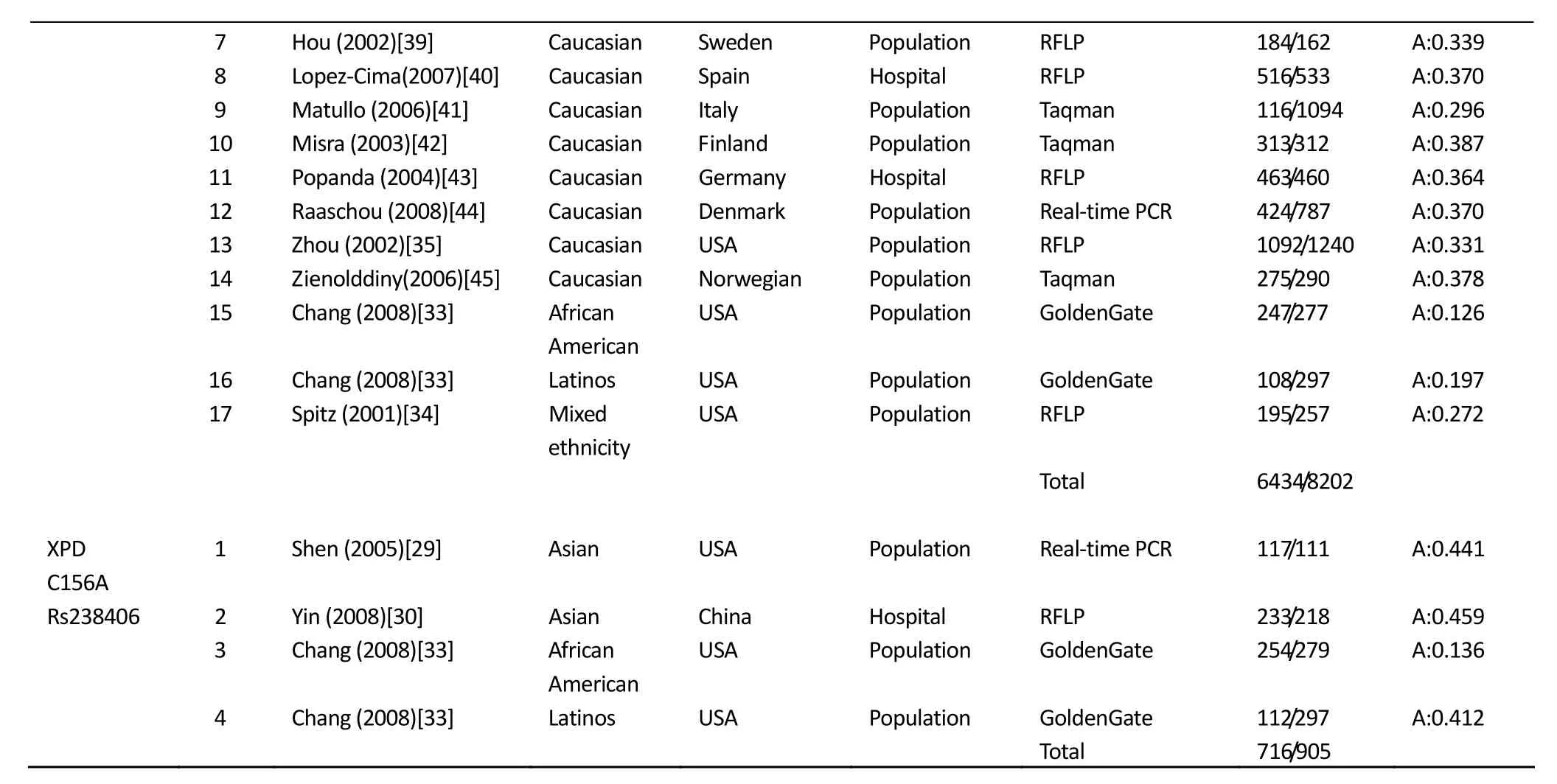
7 Hou (2002)[39] Caucasian Sweden Population RFLP 184/162 A:0.339 8 Lopez-Cima(2007)[40] Caucasian Spain Hospital RFLP 516/533 A:0.370 9 Matullo (2006)[41] Caucasian Italy Population Taqman 116/1094 A:0.296 10 Misra (2003)[42] Caucasian Finland Population Taqman 313/312 A:0.387 11 Popanda (2004)[43] Caucasian Germany Hospital RFLP 463/460 A:0.364 12 Raaschou (2008)[44] Caucasian Denmark Population Real-time PCR 424/787 A:0.370 13 Zhou (2002)[35] Caucasian USA Population RFLP 1092/1240 A:0.331 14 Zienolddiny(2006)[45] Caucasian Norwegian Population Taqman 275/290 A:0.378 15 Chang (2008)[33] African American USA Population GoldenGate 247/277 A:0.126 16 Chang (2008)[33] Latinos USA Population GoldenGate 108/297 A:0.197 17 Spitz (2001)[34] Mixed ethnicity USA Population RFLP 195/257 A:0.272 Total 6434/8202 XPD C156A 1 Shen (2005)[29] Asian USA Population Real-time PCR 117/111 A:0.441 Rs238406 2 Yin (2008)[30] Asian China Hospital RFLP 233/218 A:0.459 3 Chang (2008)[33] African American USA Population GoldenGate 254/279 A:0.136 4 Chang (2008)[33] Latinos USA Population GoldenGate 112/297 A:0.412 Total 716/905
Meta Analysis
The summary odds ratio (OR) and 95% confidence internal (95%CI) were used to estimate the association between the six polymorphisms of NER genes and lung cancer risk. For each polymorphism, we used individuals with the homozygous common genotype as the reference group and calculated OR for those with the heterozygous genotype and homozygous rare genotype. We also used individuals with the heterozygous genotype as the reference group and calculated OR for individuals with the homozygous rare genotype. The association between rare allele of gene polymorphisms and lung cancer risks were evaluated using dominant and recessive genetic models. Moreover, studies were divided into subgroups according to the ethnicity of samples. Heterogeneity among combined studies was assessed with the chi-square based Q-test[12].P<0.01 was considered significant difference. I2was also used to evaluate for heterogeneity. The closer to zero I2is more likely no difference. Less than 0.25 is considered mild, between 0.25 and 0.5 was considered moderate and greater than 0.75 was considered high degree of heterogeneity[13]. When no heterogeneity existed, fixed effect model with Mantel-Haenszel method was used to combine data. When heterogeneity existed, random effect model with Inverse Variance method was used. The pool OR was analyzed for statistical significance by the Z test. The same analysis method was applied to the subgroups divided according to ethnicity. Potential publication bias was evaluated by using funnel plots and Egger’s test[14]. Hardy-Weinberg equilibrium (HWE) for genotype frequencies was tested by Chi-Square test. Data mentioned above were performed using STATA version 11.0 (STATA Corporation) and Review Manager 5.0(Oxford, England) software. All P values were two-sided.
RESULTS
The Studies and Meta-analysis Databases
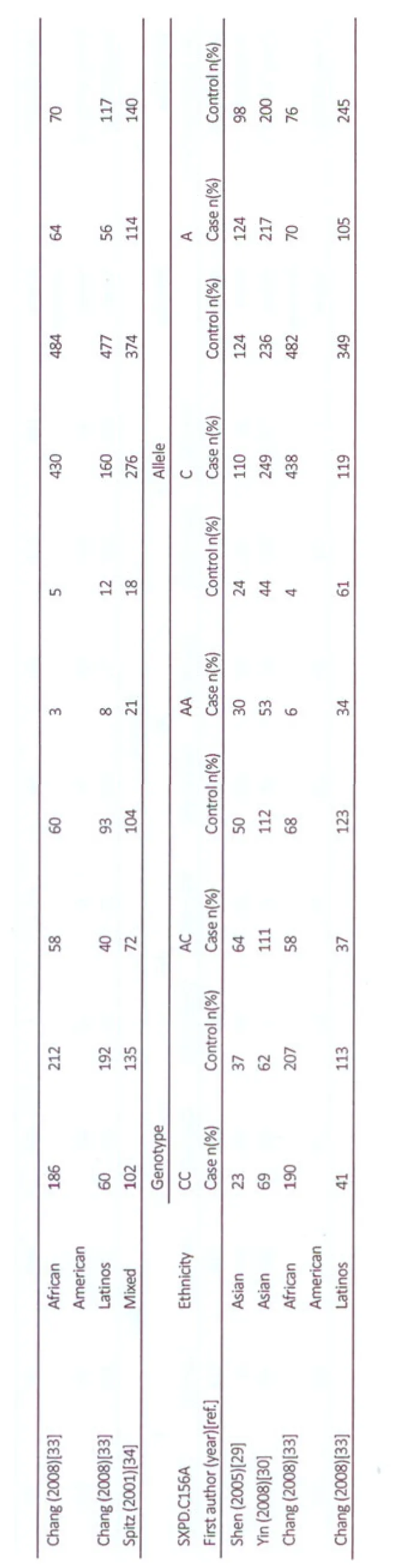
A total of 31 publications retrieved from Pubmed and EMBASE met our inclusion criteria. Among it, six publications were excluded because of reported publication[15], and same population that had been reported[16-20]. Thus, 25 publications were included in our meta-analysis, including 10 publications in Asian[21-30], 6 publications in the United States[31-36]and 9 publications in European countries[37-45]. Three publications recruited subjects of different racial descents. David et al[31]focused their recruitment on African Americans and Caucasian subjects. Chang et al[33]provided data on subjects of Latinos and African Americans. Wu[36]provided data on subjects of Caucasians, African Americans and Mexican Americans. The different races in the publications were considered as separate studies. Spitz et al[34]provided genotyping data of American population and which was defined as “mixed”ethnic. One study[33]used Illumina’s GoldenGate genotyping platform and the others used PCR-based methods to genotype. In five studies, the distribution of genotypes frequencies in control group was not in Hardy-Weinberg equilibrium[29,30,33,36,45]. All of them were published in English. Among the 25 publications, 6 publications (8 studies) investigated XPA G23A[27,36,38,43-45]. 4 studies studied XPC C499T[21,23,25,29]. 6 publications (7 studies) focused on XPC A939C[21,23,25,29,33,44]. 19 publications(21 studies) tested XPD A751C[22,24,26,28-35,38-45]. 16 publications (17 studies) examined XPD G312A[24,26,29,30,33-35,37-45], and 3 publications (4 studies) analyzed XPD C156A[29,30,33]. The distribution of the alleles and genotypes among cases and controls are shown in Table 2.


XPA and Lung Cancer Risk
XPA, coding for zinc-finger DNA binding protein and plays a central role at an early stage in the NER pathway for it plays in DNA lesion recognition and its interaction with other NER repair proteins. Variations of XPA gene probably affect protein function and NER pathway[46]. XPA G23A was found in the 5’ non-coding region of the XPA gene, a A-to-G substitution in the nucleotide –4 from ATG start codon[47]. 5' UTR might be essential for gene expression through transcriptional and post-transcriptional control mechanisms[48]. Polymorphisms in this area proximal to the start codon could have implications for the binding of the 40S ribosomal subunit and as a result influence protein levels in the cell[49].
The pooled analysis for XPA G23A included 2206 cases and 2502 controls. In the meta-analysis, the 23A allele frequency was varied from 0.334 to 0.361 in Caucasians[38,43-45]except for a outlier 0.446[36], which was reported in a study that showed a evident HWE deviation. And it was 0.478 in Asians[27], 0.299 in African Americans and 0.394 in Mexican-American[36]. In all controls of the recruited studies, XPA 23G allele was more common than 23A allele. We assumed G allele was wild allele and a allele as variant allele. Overall, significant between-study heterogeneity in 8 studies was found when we analysis the association between A allele and lung cancer risk, compared with G allele. Random-effect model was used to combine data. No significant association was found between the 23A allele and lung cancer risk in all subjects (P=0.29, OR=1.07, 95%CI: 0.94-1.22, Pheterogeneity=0.06). No heterogeneity existed in the 8 studies when investing AG vs GG, AA vs GG, AA vs AG, AA vs AG+GG and AA+AG vsGG. Using the fixed-effects model to combine data, carriers with AA genotype significantly elevated lung cancer risk under the recessive genetic model(P=0.003, OR=1.27, 95%CI:1.08-1.48, Pheterogeneity=0.36), homozygote comparison (P=0.02, OR=1.22, 95%CI: 1.03-1.46, Pheterogeneity=0.29) and in AA vsAG contrast (P=0.002, OR=1.31, 95%CI:1.10-1.55, Pheterogeneity=0.43). In stratified analysis by ethnicity, we performed meta-analysis on Caucasians and found carriers with AA genotype significantly elevated lung cancer risk under the recessive genetic model (P=0.02, OR=1.23, 95%CI:1.03-1.47, Pheterogeneity=0.45) and in AA vsAG contrast (P=0.005, OR=1.31,95% CI:1.08-1.58, Pheterogeneity=0.65). In the 8 studies, a study of Wu et al[36]in Caucasians, the distribution of genotypes frequencies in control group was not in Hardy-Weinberg equilibrium. We excluded this study and performed meta-analysis once again. However, no significant association was found between XPA G23A and lung cancer risk in total population and in Caucasians.
Pool analysis was done in the 8 studies using individuals with the AA genotype as the reference group and calculated summary OR for those with the AG or GG genotype. Moreover, we evaluated the relationship between GG genotype and lung cancer risk in comparing with the combination of AA and AG genotype. In total population, no heterogeneity existed in the 8 studies, under fix-effect model, comparing with AA, carriers with AG or GG both reduced the risk of lung cancer (P=0.002, OR=0.76, 95%CI:0.65-0.91, Pheterogeneity=0.43;P=0.02, OR=0.82, 95%CI: 0.69-0.97, Pheterogeneity=0.29). In Caucasian, the protective effect wasobserved in AG vs AA (P=0.005, OR=0.76, 95%CI:0.63-0.92, Pheterogeneity=0.65). However, excluding the study of[36], which showed a evident HWE deviation, no association was found between XPA G23A and lung cancer risk in total population and in Caucasians.
XPC Polymorphisms and Lung Cancer Risk
XPC binds to HR23B forming XPC-HR23B complex and play a crucial role in the recruitment of TFIIH to damaged DNA and initiate and combine with other NER factors in global genome repair[5,9,50]. XPC also plays an unexpected and multifaceted role in cell protection from oxidative DNA damage[51]. 100% of mice with deleted XPC develop spontaneous lung tumors. XPC allelic loss may be an etiological factor in lung tumorigenesis in humans[52]. XPC A939C and XPC C499T were two key polymorphisms in XPC. XPC A939C is a A-to-C transition in exon 15, resulting in the change Lys939—>Gln. XPC C499T is a C-to-T transition in exon8, resulting in the change Ala499—> Val. XPC C499T were found in LD with XPC A939C and XPC PAT[25,29,53].
The association studies between XPC A939C, C499T and lung cancer risk were mostly from Asians. Most of studies[21,29,33,44]reported there have no association between the two polymorphisms with the lung cancer risk except for two studies in Asians.Hu et al[23]reported 939C and 499T variant genotype respectively and combined to elevate lung cancer risk and more pronounced among smokers. Lee et al[25]revealed 939C were associated with a decreased risk of small cell carcinoma (adOR=0.58, 95%CI:0.35-0.97). No significant interaction between the XPC Lys939Gln polymorphism and smoking and intake of fruit/vegetables was found[25,44].
XPC C499T and Lung Cancer Risk
The pooled analysis for XPC C499T included 1862 cases and 1854 controls. The 499T allele frequency was varied from 0.284 to 0.332 in Asian[21,23,25,29]. All of the 4 studies of XPC C499T recruited in the meta-analysis were from Asian population. Except for the CTvsCC contrast and the CT+TTvsCC contrast (dominate genetic model), no significant heterogeneity was observed between the 4 studies in any genetic model. Using random-effect model on the CTvsCC contrast and CT+TTvsCC contrast and using fix-effect model on the rest genetic model, no evidence of an association between XPC C499T and lung cancer risk was found. To identify sources of heterogeneity further, we exclude each study in turn and found after excluding the study of Hu[23], the heterogeneity between studies was not significant any more. Meanwhile, there were also no significant association between XPC C499T and lung cancer risk.
XPC A939C and Lung Cancer Risk
There are 2650 cases and 3211 controls included in the pooled analysis for XPCA939C. In the meta-analysis, the 939C allele frequency was varied from 0.326 to 0.394 in Asians[21,23,25,29]. And it was 0.368 in Caucasians[44], 0.307 in African Americans and 0.29 in Latinos[33].
In total population, no significant heterogeneity was observed between the 7 studies and the fix-effect model Carriers with CC genotype significantly increased lung cancer risk under the recessive genetic model(P=0.003, OR=1.27, 95%CI:1.09-1.48, Pheterogeneity=0.39), homozygote comparison (P=0.01, OR=1.24, 95% CI:1.05-1.46, Pheterogeneity= 0.33) and CCvsCA contrast (P=0.002, OR=1.29, 95%CI: 1.10-1.52, Pheterogeneity=0.44). In stratified analysis by ethnicity in Asians. A close association between CC genotype and lung cancer was also found in the three genetic model(recessive genetic model:P=0.02, OR=1.26, 95% CI:1.04-1.52, Pheterogeneity=0.26, homozygotecomparison:P=0.05, OR=1.23, 95%CI:1.00-1.51, Pheterogeneity=0.28, CCvs. CA contrast (P=0.02, OR=1.28, 95%CI:1.05-1.56, Pheterogeneity= 0.25).
In total population, except for the CA+CCvsAA, no heterogeneity was detected between the 6 studies in any genetic models. Using random-effect model on the CA+CCvsAA and using fix- effect model on the rest genetic models, a close association of CC genotype and increased lung cancer risk was verified in the recessive genetic model (P=0.01, OR=1.23, 95%CI:1.05-1.44, Pheterogeneity=0.69), homozygote comparison (P=0.03, OR=1.21, 95%CI: 1.02-1.43, Pheterogeneity=0.43) and CC vs. CA contrast (P=0.009, OR=1.25, 95%CI:1.06-1.48, Pheterogeneity=0.82). In Asians, no heterogeneity was observed among the 6 studies. However, after excluded the study of Shen[29], no significant association was found between XPC A939C and lung cancer risk.
In Caucasians[44], the XPC A939C CC genotype was associated with a non-significant 41% higher IRR for lung cancer than the XPC A939C AA wild type. In African Americans and Latinos, no remarkable association was found between XPC A939C and lung cancer risk[33].
XPD A751C and Lung Cancer Risk
There are 7385 cases and 9453 controls included in the pooled analysis for XPD A751C. In the meta-analysis, the751C allele frequency was varied from 0.04 to 0.111 in Asians[24,26,27,29,30]except for a outlier 0.403[22]. The 751C allele frequency was varied from 0.273 to 0.407 in Caucasians[32,31,35,38-45]. It was varied from 0.225 to 0.25 in African Americans[31,33], 0.219 in Latinos[33]and 0.333 in the mixed ethnicity in the United States[34].
Overall, no heterogeneity was observed between the 21 studies and the fix-effect model was used to combine data. Carriers with CC genotype significantly elevated lung cancer risk under the recessive genetic model (P=0.0003, OR=1.23, 95%CI:1.10-1.37, Pheterogeneity=0.59), dominate genetic model (P=0.003, OR=1.11, 95%CI:1.04-1.19, Pheterogeneity=0.64), homozygote comparison (P<0.0001, OR=1.28, 95%CI:1.14-1.44, Pheterogeneity=0.56) and CC vs. CA contrast (P=0.006, OR=1.18, 95%CI:1.05-1.32, Pheterogeneity= 0.8). Carriers with C allele significantly increased risk of lung cancer compared with carriers with A allele. (P<0.0001, OR=1.11, 95% CI:1.06-1.17, Pheterogeneity=0.29). In Caucasians, under fix-effect model, significant increase of lung cancer risk was also found in C vs. A (P=0.0002, OR=1.13, 95%CI:1.06-1.20, Pheterogeneity=0.81), homozygote comparison (P=0.0002, OR=1.28, 95%CI:1.12-1.45, Pheterogeneity=0.94), CC vs. CA contrast (P=0.02, OR=1.16, 95% CI:1.02-1.32, Pheterogeneity=0.97), recessive genetic model (P=0.001, OR=1.22, 95%CI:1.08-1.37, Pheterogeneity=0.95) and dominate genetic model (P=0.004, OR=1.14, 95%CI: 1.04-1.24, Pheterogeneity=0.89). In Asians,except for the dominate genetic model and C vs. A, no heterogeneity was found in any genetic model. Using the random-effect model on the dominate genetic model and C vs. A and using fix-effect model on the rest genetic model, no significant association between XPD A751C and lung cancer risk was found in Asians. Further identifying sources of heterogeneity in Asians, we found the heterogeneity decreased after excluding the study of Chen et al[22]and even disappeared after excluding the study of Shen et al[29]. In African Americans, no heterogeneity was found between the studies. Under fix-effect model, no association between XPD A751C and lung cancer risk was found.
In the 21 recruited studies, in a study of Zienolddiny et al[45]in Caucasians, the distribution of genotypes frequencies in control group was not in HWE. We excluded the study and performed meta-analysis once again. No heterogeneity was found between 20 studies in total population and in Caucasians. Under fix-effect model, the association of CC genotype and C allele with elevation of lung cancer risk was also observed in the recessive genetic model (Total population:P=0.002, OR=1.20, 95%CI:1.07-1.35, Pheterogeneity=0.59; Caucasian:P=0.008, OR=1.19, 95%CI: 1.05-1.35, Pheterogeneity=0.97), dominate genetic model (Total population:P=0.009, OR=1.10, 95%CI:1.02-1.18, Pheterogeneity=0.69; Caucasian:P=0.02, OR=1.12, 95%CI: 1.02-1.23, Pheterogeneity=0.93), homozygote comparison (Total population:P=0.0004, OR=1.25, 95%CI:1.11-1.42, Pheterogeneity=0.57; Caucasian:P=0.002, OR=1.24, 95%CI: 1.08-1.43, Pheterogeneity=0.97) and CCvsCA contrast (Total population:P=0.01, OR=1.17, 95%CI:1.03-1.32, Pheterogeneity= 0.76; Caucasian:P=0.05, OR=1.15, 95%CI:1.00-1.31, Pheterogeneity=0.96) and CvsA (Total population:P=0.0005, OR=1.10, 95%CI:1.04-1.16, Pheterogeneity=0.4; Caucasian:P=0.002, OR=1.11, 95%CI:1.04-1.18, Pheterogeneity=0.95).
XPD G312A and Lung Cancer Risk
A total of 6434 cases and 8202 controls were included in the pooled analysis of XPD G312A. In the meta-analysis, the 312A allele frequency was varied from 0.057 to 0.065 in 3 studies of Asians[24,26,29]. Except for a very low figure 0.006, which from a study of Yin et al[30]. The distribution of genotypes in the study was not in HWE. The 312A allele frequency was varied from 0.296 to 0.436 in Caucasians[35,37-45]. It was 0.126 in African Americans[33], 0.197 in Latinos[33].
In total population, no heterogeneity was observed between the 17 studies and the fix-effect model was used to combine data, carriers with AA genotype increase risk of lung cancer under recessive genetic model (P=0.005, OR=1.20, 95%CI:1.05-1.36, Pheterogeneity=0.38), homozygote comparison (P=0.01, OR=1.18, 95%CI: 1.03-1.35, Pheterogeneity= 0.36) and AAvsAG contrast (P=0.004, OR=1.22, 95%CI: 1.07-1.39, Pheterogeneity=0.31).
In the subgroup analysis by ethnicity, Carriers in Asian with AA genotype also significantly increased lung cancer risk in above genetic model (recessive genetic mode: P=0.01, OR=4.34, 95%CI:1.37-13.71, Pheterogeneity=0.16. AAvs.GG:P=0.01, OR=4.34, 95%CI:1.37-13.75, Pheterogeneity=0.16. AA vs. AG:P=0.01, OR=4.79, 95%CI: 1.36-16.86, Pheterogeneity= 0.18). In Caucasian, Carriers with AA genotype significant increased lung cancer risk in two genetic model (recessive genetic mode:P=0.04, OR=1.15, 95%CI:1.01-1.31, Pheterogeneity=0.66. AA vs. AG:P=0.02, OR=1.17, 95%CI: 1.02-1.35, Pheterogeneity=0.49).
In the 16 recruited studies, the distribution of genotypes frequencies in control group was not in HWE. We performed meta-analysis once again. No significant heterogeneity was observed between the 16 studies in total population and in Asians. Under fix-effect model, the association of AA genotype with lung cancer elevation was also observed in the recessive genetic model (Total population:P=0.005, OR=1.20, 95%CI:1.06-1.36, Pheterogeneity= 0.36; Asians:P=0.007, OR=7.66, 95%CI:1.75-33.54, Pheterogeneity=0.50), homozygote comparison (Total population:P=0.01, OR=1.19, 95%CI: 1.04-1.36, Pheterogeneity= 0.34; Asians:P=0.007, OR=7.68, 95%CI:1.75-33.62, Pheterogeneity=0.51), AAvsAG contrast (Total population:P=0.003, OR=1.22, 95%CI:1.07-1.40, Pheterogeneity=0.31; Asians:P=0.008, OR=7.51, 95%CI:1.71-33.04, Pheterogeneity=0.47).
XPD C156A and Lung Cancer Risk
A total of 716 cases and 905 controls were included in the pooled analysis of XPD C156A. In the meta-analysis, the 156A allele frequency was varied from 0.441 to 0.459, 0.136 and 0.412 in Asian[29,30], in African Americans[33]and in Latinos[33]respectively..
Overall, no heterogeneity was observed between the 4 studies when analysis all the genetic models. Using fix-effect model to combine data. In total population, carriers with the AA genotype significantly increased the risk of lung cancer in the genetic recessive model (P=0.04, OR=1.36, 95%CI: 1.02-1.81, Pheterogeneity=0.71) and homozygote comparison (P=0.04, OR=1.42, 95%CI: 1.02-1.97, Pheterogeneity=0.58). In the subgroup analysis by ethnicity, in Asian, except for the analysis of dominate genetic model and AC vs. CC contrast, no heterogeneity was observed. The random-effect model was used to combine data in the two genetic models and the fix-effect model to the rest genetic model. No association was found between XPD C156A and lung cancer risk.
Publication Bias
We performed the funnel plot and Egger’s test to evaluate publication bias of XPA G23A, XPC A939C, XPD A751C, XPD G312A. As limited data in XPC C499T and XPD C156A, we didn’t evaluate their publication bias with the same method. No significant publication bias was found in these polymorphisms (Comparing AvsG for XPAG23A: t=0.66,P=0.532; Comparing CvsA for XPCA939C: t=0.22,P=0.835; Comparing C vs A for XPD A751C: t=-0.59,P=0.561; Comparing AvsG for XPD G312A: t=-1.76,P=0.099) r C vs. A allele comparison in XPD A751C; Funnel plot for A vs. G allele comparison in XPD G312A.
DISCUSSION
Nucleotide excision repair is a highly versatile DNA damage repair pathway by which the vast majority of DNA damage can be removed through incisions on both sides of the lesion. In humans, the NER system is an importantdefense mechanism against two major carcinogens, sunlight and cigarette smoke[60]. It has two branches: global genome repair, which probes the genome for strand distortions and transcription-coupled repair, which removes distorting lesions that block elongating RNA polymerases[3]. NER involves more than 30 proteins[61]. We analysis the association between 6 common polymorphisms of 3 NER genes and lung cancer risk in the meta-analysis. A total of 25 publications (61 studies) were included. Following polymorphisms were examined: XPA G23A, XPC C499T, XPC A939C, XPD A751C, XPD G312A and XPD C156A .We found XPA G23AAA genotype elevated lung cancer risk in total population and in Caucasians. AG or GG genotype decreased lung cancer risk in total population, AG genotype decreased lung cancer risk in Caucasians. However, after excluding a study in Caucasians, which was not in HWE[36]. The result showed no association between XPA G23A and lung cancer risk in total population and in Caucasians. XPC A939C CC genotype elevated lung cancer risk in total population and in Asians. However, after excluding a study in Asian[29], which was not in HWE. CC genotype elevated lung cancer risk was verified in total population but not in Asians. All studies recruited in the meta-analysis are about Asians and no association was found between XPC C499T and lung cancer risk in Asians. In the case of XPD A751 C, 751C allele and CC genotype both elevated lung cancer risk in total population and in Caucasians. The result was verified even after excluding a study in Caucasians[45], which was not in HWE. No association was found between XPD A751C and lung cancer risk in Asians and African Americans. XPD G312A AA genotype elevated risk of lung cancer in total population, in Asians and Caucasians. The result was verified even after excluding a study in Asians[30], which was not in HWE. XPD C156A AA genotype increased the risk of lung cancer in total population but had no association with lung cancer risk in Asians. However, after excluding a study in Latinos[33], which showed a evident HWE deviation, the C156A was found have no association with lung cancer risk in total population. XPC A939C was in linkage disequilibrium with XPC PAT and IVS11-5C->A(IVS11-5C->A was a SNP discovered in the XPC intron11 splice acceptor)[25,62].Khan et al found the three polymorphisms are consistent with a haplotype of PAT-/intron 11C/exon15 A in -60% of the donors and PAT+/intron11A/exon15C in-40%of the donors. The A allele of intron11 splice acceptor, which has a lower information content than the C allele and is associated with a higher frequency of deletion of exon12. This exon 12 deleted XPC mRNA isoform has reduced DNA repair activity. Thus this might increase cancer susceptibility[62].Utilizing post-UV host cell reactivation to assess DNA repair capacity of polymorphic alleles and found similar DNA repair with XPC939A and XPC939C[10]. XPC939C variant could affect the irradiation-specific DNA repair rates[63]. A recent study[53]revealed based on the mutagen-challenged comet assay, compared with their wild allele, the variant allele C of A939C decreased DRC and the variant allele T of C499T elevated DRC. Most of the functional studies revealed the polymorphisms of XPD modified the DNA repair capacity. Duell et al[64]reported XPD Lys751Gln was unrelated to sister chromatid exchange frequencies or DNA adduct level by a less than 80 subjects study. Some studies revealed the variant 751Gln and 312Asn genotypes were associated with less optimal DRC to repair BPDE or UV-induced DNA damage compared with wild-type alleles in the host cell reactivation assay[34,65,66]and were associated with higher DNA adducts[39,67,68,58]. They were also reported to have significantly association with increased chromosome aberrations (CAs) following exposure to UV light, but not to X-rays, which suggested these variant genotypes appear to be defective in nucleotide excision repair[32,69]. On the other hand, Lunn et al[70]. Compared XPD genotype at codon312 and 751 with DNA repair proficiency in 31 women and found the Lys/Lys751 genotype was associated with sub-optimal repair of DNA damage induced by X-irradiation. The Asp312Asn polymorphism did not appear to affect DNA repair proficiency. Vodicka et al[63]revealed that 751A allele(AA and AC) genotypes elevated the levels of chromosomal aberrations (CAs) and may increase the risk of SSB formation in peripheral lymphocytes.
The combination or interaction analysis between gene and gene in NER pathway showed The polymorphisms in XPC and XPD increased the risk of developing lung cancer and the association was particularly important for ever smokers and patients with adenocarcinomas[40]. Popanda[43]found a >5-fold increased risk for SCC (squamous cell carcinoma) in individuals with three variant NER alleles[XPA(-4A/A) and the XPD 312Asn/Asn and 751Gln/Gln] indicating that combinations of variant alleles within a specific pathway can affect the risk for SCC much stronger than the single variants.
Populations in different genetic background might account for the different association with lung cancer risk. In our meta-analysis, the prevalence of the variant allele in XPD A751C and G312A varies markedly among Caucasians and Asians. The 751C allele frequency was varied from 0.273 to 0.407 in Caucasians.The751C allele frequency was varied from 0.04 to 0.111 in Asians except for a evident outlier 0.403, which was reported by Chen et al[22]. The distribution of genotype frequency in the study varies markedly from the other Asian studies and it is probably some genotyping error in it. After excluding this study, however, the result of meta-analysis were not affected significantly (data not shown). The 312A allele frequency was varied from0.296 to 0.436 in Caucasians and varied from 0.057 to 0.065 in Asians except for a very low figure of 0.006, which from a study of Yin et al[30]. The 312A allele frequency in the study showed an evident HWE deviation. XPD G312A was observed to be in linkage disequilibrium with A751C[26,34,39,40,17,35,37]. Moreover, studies in Caucasians found G312A is in linkage disequilibria with both C156A and A751C71, C156A and A751C are tightly association[72]. However, the haplotype structure and frequency between populations were significant different in European, African and Asian individuals. In both the African and Asian populations, C156A was not significantly linked to A751C[73]. In our meta-analysis, the 156A allele frequency was varied from0.441 to 0.459 in Asian and very different from African Americans (0.136). No relative reports in Caucasians were recruited.
A number of association studies aimed to identify genetic variant that influence susceptibility to lung cancer. But the reports were not always consistently reproducible. The inconsistency may be due to too small samples, not population-based and among different ethnicity background, and so on. Meta-analysis, a statistical tool for combining results across studies, is becoming popular as a method for resolving discrepancies in genetic association studies[74]. It not only increases the power to detect an association but also helps make sense of conflicting results. It plays an important role in assessing that replication and in providing an estimate of the size of the genetic effect[75]. In our meta-analysis, total 25 publications were recruited and extracted data to pool analysis, which greatly increase the statistical power. We evaluated the role of NER genetic polymorphisms just in lung cancer rather than in all site cancer. It is because of that when examining the role of polymorphisms in DNA repair genes on cancer susceptibility, it is important to consider the importance of‘gene-environment’ interaction, which are crucial to characterize low-penetrance genes[76]. Carcinogenic mechanism may differ by different tumor sites. A genotype have elevation association with cancer in this site might have reduction association with cancer in that site[76]. Recently, there were 3 meta-analysis of the association between XPC polymorphisms(Lys939Gln; Ala499Val) and cancer risk[76-78].They all observed 939Gln homozygote significantly elevate the risk of lung cancer in all subjects. Two meta-analysis[79,80]of the association between XPD polymorphisms (Lys751Gln, Asp312Asn) and cancer or lung cancer risk were published in past two years. Kiyohara[80]found 751Gln/Gln genotype was associated with increasing risk of lung cancer and did not found 312Asn/ Asn genotype was significant associated with lung cancer risk in all subject. But another meta-analysis[79]reported 751Gln/ Gln and 312Asn/Asn were both associated with increasing risk of lung cancer in all subjects. Kiyohara et al[80]found a protective effect of the XPA 23G/G genotype in all subjects. For meta-analysis of genetic association studies, there was no consensus on whether to recruit the study that was not in Hardy-Weinberg equilibrium. Zhang et al[77]and Wang et al[79]excluded studies that was not in Hardy-Weinberg equilibrium. Qiu et al[78]and Kiyohara et al[80]recruited all relative studies regardless of study that was in Hardy-Weinberg equilibrium or not. Attia et al[81]suggested that sensitivity analysis should be performed, pooling with and without studies not in HWE, to test the robustness of the results.
In our meta-analysis, we focused on the association between polymorphisms of NER genes and lung cancer risk. We recruited more updated publications and polymorphisms of NER, as well as gave more detailed results on the base of ethnicity. Moreover, we estimated the deviations from Hardy-Weinberg equilibrium in the controls for each studies and make meta-analysis before and after excluding the study that was not in HWE. Thus, we assessed the sensitivity of the results by meta-analysis. We found that excluding the studies were not in HWE, the result of meta-analysis in XPD A751C and XPD G312A were not change. XPD A751C. C allele and CC genotype elevated lung cancer risk was also verified in total population and in Caucasians. No association was found between XPD A751C and lung cancer risk in Asians and African Americans. XPD G312A, AA genotype elevated risk of lung cancer in total population in Asians and Caucasians. However, the results of following polymorphisms of NER showed some difference after excluding the studies were not in HWE. For XPCA939C, CC genotype elevated lung cancer risk in total population but not in Asian population. C156A was no association with lung cancer risk in total population and in Asian population. It is noteworthy that after excluding the study was not in HWE, the result showed no association between XPAG23A and lung cancer risk in total population and in Caucasians, which is very different with the result when we recruited all studies. There is some limitation in our meta-analysis. First, some studies have suffered from a relatively small sample size or in some subgroup of ethnicity, the number of studies available were limited, which result in lacking enough statistical power to detect the true association between the polymorphism and lung cancer. Second, the available data from the individual studies are not detailed enough and we can not perform the pooled analysis to examine the combined effect of multiply polymorphisms or environmental factors. So did the linkage disequilibrium and haplotype analysis. Third, subjects were briefly divided in smoker and non-smoker may not reflect the true association between the polymorphism and lung cancer risk. In the meanwhile, Smoking statue defined different in different studies. We can not perform the pooled analysis to evaluate the interaction between polymorphisms and smoke statue. The last, the number of studies in XPA G23A and XPD C156A are limited and we did not perform the funnel plot and Egger’s test to evaluate their publication bias.
In conclusion, the meta-analysis indicates that the polymorphisms of NER genes XPC and XPD may play a role in the evaluating the risk of lung cancer. XPD A751C C allele and CC genotype elevate lung cancer risk in total and Caucasians population. No significant association was found between XPD A751C and lung cancer risk in Asians and African Americans. XPD G312A AA genotype increases risk of lung cancer in the total, Asians and Caucasians populations. XPC A939C CC genotype increased lung cancer risk in total population and but not in Asians. No significant association between XPA G23A, XPC C499T, XPD C156A and lung cancer risk was found. Lung cancer is a complex disease and influenced by gene-gene, gene-environment interaction. Multiply genes and environment factors might interact and contribute to the occurrence of lung cancer. Future study needs to be done to confirm the relationship among multiply polymorphisms of candidate gene, environment factors and lung cancer risk. sd.
Acknowledgements
We are grateful to Dr. Shou-wei Han from Emory University (USA) for comments on the manuscript.
REFERENCES
1. Ronen A, Glickman B. Human DNA repair genes. Environ Mol Mutagen2001; 37: 241-83.
2. Wei Q, Spitz MR. The role of DNA repair capacity in susceptibility to lung cancer: A review. Cancer Metastasis Rev 1997; 16: 295-307.
3. Hoeijmakers J. DNA Damage, Aging, and Cancer. N Engl J Med 2009; 361: 1475-86.
4. Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev 2002; 11: 1513-30.
5. van Hoffen A, Balajee AS, van Zeeland AA, et al. Nucleotide excision repair and its interplay with transcription. Toxicology 2003; 193: 79-90.
6. de Boer J, Hoeijmakers J. Nucleotide excision repair and human syndromes. Carcinogenesis 2000; 21: 453-60.
7. de Laat WL Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes dev 1999; 13: 768-85.
8. Asahina H, Kuraoka I, Shirakawa M, et al. The XPA protein is a zinc metalloprotein with an ability to recognize various kinds of DNA damage. Mutat Res 1994; 315: 229.
9. Sugasawa K, Ng JM, Masutani C, et al. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol cell 1998; 2: 223-32.
10. Khan SG, Metter EJ, Tarone RE, et al. A new xeroderma pigmentosum group C poly(AT) insertion/deletion polymorphism. Carcinogenesis 2000; 21: 1821-5.
11. Sak SC, Barrett JH, Paul AB, et al. The polyAT, intronic IVS11-6 and Lys939Gln XPC polymorphisms are not associated with transitional cell carcinoma of the bladder. Br J Cancer 2005; 92: 2262-5.
12. Lau J, Ioannidis JP, Schmid C. Quantitative synthesis in systematic reviews. Ann Intern Med 1997; 127: 820-6.
13. Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. Br Med J 2003; 327: 557-60.
14. Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. Br Med J 1997; 315: 629-34.
15. Butkiewicz D, Popanda O, Risch A, et al. Association between the risk for lung adenocarcinoma and a (-4) G-to-A polymorphism in the XPA gene. Cancer Epidemiol Biomarkers Prev 2004; 13: 2242-6.
16. Vogel U, Overvad K, Wallin H, et al. Combinations of polymorphisms in XPD, XPC and XPA in relation to risk of lung cancer. Cancer Lett 2005; 222: 67-74.
17. Vogel U, Laros I, Jacobsen NR, et al. Two regions in chromosome 19q13.2-3 are associated with risk of lung cancer. Mutat Res 2004; 546: 65-74.
18. Xing DY, Qi J, Tan W, et al. Association of genetic polymorphisms in the DNA repair gene XPD with risk of lung and esophageal cancer in a Chinese population in Beijing. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2003; 20: 35-8.
19. Xing D, Tan W, Wei Q, et al. Polymorphisms of the DNA repair gene XPD and risk of lung cancer in a Chinese population. Lung Cancer 2002; 38: 123-9.
20. Yin J, Li JC, Ma Y, et al. The DNA repair gene ERCC2/XPD polyrnorphism Arg 156Arg (A22541C) and risk of lung cancer in a Chinese population. Cancer Letters 2005; 223: 219-26.
21. Bai Y, Xu L, Yang XB, et al. Sequence variations in DNA repair gene XPC is associated with lung cancer risk in a Chinese population: a case-control study. BMC Cancer 2007; 7: 81.
22. Chen S, Tang D, Xue K, et al. DNA repair gene XRCC1 and XPD polymorphisms and risk of lung cancer in a Chinese population. Carcinogenesis 2002; 23: 1321-5.
23. Hu Z, Wang Y, Wang X, et al. DNA repair gene XPC genotypes/haplotypes and risk of lung cancer in a Chinese population. Int J Cancer 2005; 115: 478-83.
24. Hu Z, Xu L, Shao M, et al. Polymorphisms in the two helicases ERCC2/XPD and ERCC3/XPB of the transcription factor IIH complex and risk of lung cancer: a case-control analysis in a Chinese population. Cancer Epidemiol Biomarkers Prev 2006; 15: 1336-40.
25. Lee GY, Jang JS, Lee SY, et al. XPC polymorphisms and lung cancer risk. Int J Cancer 2005; 115: 807-13.
26. Liang G, Xing D, Miao X, et al. Sequence variations in the DNA repair gene XPD and risk of lung cancer in a Chinese population. Int J Cancer 2003; 105: 669-73.
27. Park JY, Park SH, Choi JE, et al. Polymorphisms of the DNA repair gene xeroderma pigmentosum group A and risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev 2002; 11: 993-7.
28. Park JY, Lee LY, Jeon HS, et al. Lys751Gln polymorphism in the DNA repair gene XPD and risk of primary lung cancer. Lung Cancer 2002; 36: 15-6.
29. Shen M, Berndt SI, Rothman N, et al. Polymorphisms in the DNA nucleotide excision repair genes and lung cancer risk in Xuan Wei, China. Int J Cancer 2005; 116: 768-73.
30. Yin J, Vogel U, Ma Y, et al. Haplotypes of nine single nucleotide polymorphisms on chromosome 19q13.2-3 associated with susceptibility of lung cancer in a Chinese population. Mutat Res 2008; 641: 12-8.
31. David-Beabes GL, Lunn RM, London SJ. No association between the XPD (Lys751G1n) polymorphism or the XRCC3 (Thr241Met) polymorphism and lung cancer risk. Cancer Epidemiol Biomarkers Prev 2001; 10: 911-2.
32. Harms C, Salama SA, Sierra-Torres CH, et al. Polymorphisms in DNA repair genes, chromosome aberrations, and lung cancer. Environ Mol Mutagen 2004; 44: 74-82.
33. Chang JS, Wrensch MR, Hansen HM, et al. Nucleotide excision repair genes and risk of lung cancer among San Francisco Bay Area Latinos and African Americans. Int J Cancer 2008; 123: 2095-104.
34. Spitz MR, Wu X, Wang Y, et al. Modulation of nucleotide excision repair capacity by XPD polymorphisms in lung cancer patients. Cancer Res 2001; 61: 1354-7.
35. Zhou W, Liu G, Miller DP, et al. Gene-environment interaction for the ERCC2 polymorphisms and cumulative cigarette smoking exposure in lung cancer. Cancer Res 2002; 62: 1377-81.
36. Wu X, Zhao H, Wei Q, et al. XPA polymorphism associated with reduced lung cancer risk and a modulating effect on nucleotide excision repair capacity. Carcinogenesis 2003; 24: 505-9.
37. Butkiewicz D, Rusin M, Enewold L, et al. Genetic polymorphisms in DNA repair genes and risk of lung cancer. Carcinogenesis 2001; 22: 593-7.
38. De Ruyck K, Szaumkessel M, De Rudder I, et al. Polymorphisms in base-excision repair and nucleotide-excision repair genes in relation to lung cancer risk. Mutat Res 2007; 631: 101-10.
39. Hou SM, Fält S, Angelini S, et al. The XPD variant alleles are associated with increased aromatic DNA adduct level and lung cancer risk. Carcinogenesis 2002; 23: 599-603.
40. López-Cima MF, Gonzalez-Arriaga P, Garcia-Castro L, et al. Polymorphisms in XPC, XPD, XRCC1, and XRCC3 DNA repair genes and lung cancer risk in a population of northern Spain. BMC Cancer 2007; 7: 162.
41. Matullo G, Dunning A, Guarrera S, et al. DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis 2006; 27: 997-1007.
42. Misra RR, Ratnasinghe D, Tangrea JA, et al. Polymorphisms in the DNA repair genes XPD, XRCC1, XRCC3, and APE/ref-1, and the risk of lung cancer among male smokers in Finland. Cancer Lett 2003; 191: 171-8.
43. Popanda O, Schattenberg T, Phong CT, et al. Specific combinations of DNA repair gene variants and increased risk for non-small cell lung cancer. Carcinogenesis 2004; 25: 2433-41.
44. Raaschou-Nielsen O, Sorensen M, Overvad K, et al. Polymorphisms in nucleotide excision repair genes, smoking and intake of fruit and vegetables in relation to lung cancer. Lung Cancer 2008; 59: 171-9.
45. Zienolddiny S, Campa D, Lind H, et al. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis 2006; 27: 560-7.
46. Wu X, Fan W, Xu S, et al. Sensitization to the cytotoxicity of cisplatin by transfection with nucleotide excision repair gene xeroderma pigmentosun group A antisense RNA in human lung adenocarcinomacells. Clin Cancer Res 2003; 9: 5874-9.
47. Butkiewicz D, Rusin M, Harris CC, et al. Identification of four single nucleotide polymorphisms in DNA repair genes: XPA and XPB (ERCC3) in Polish population. Hum Mutat 2000; 15: 577-8.
48. Akiri G, Nahari D, Finkelstein Y, et al. Regulation of vascular endothelial growth factor (VEGF) expression is mediated by internal initiation of translation and alternative initiation of transcription. Oncogene 1998; 17: 227-36.
49. Miller KL, Karagas MR, Kraft P, et al. XPA, haplotypes, and risk of basal and squamous cell carcinoma. Carcinogenesis 2006; 27: 1670-5.
50. Yokoi M, Masutani C, Maekawa T, et al. The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J Biol Chem 2000; 275: 9870-5.
51. D'Errico M, Parlanti E, Teson M, et al. New functions of XPC in the protection of human skin cells from oxidative damage. EMBO J 2006; 25: 4305-15.
52. Hollander MC, Philburn RT, Patterson AD, et al. Deletion of XPC leads to lung tumors in mice and is associated with early events in human lung carcinogenesis. Proc Nat Acad Sci USA 2005; 102: 13200-5.
53. Zhu Y, Yang H, Chen Q, et al. Modulation of DNA damage/DNA repair capacity by XPC polymorphisms. DNA Repair (Amst). 2008; 7: 141-8.
54. Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res 1998; 58: 604-8.
55. Wolfe KJ, Wickliffe JK, Hill CE, et al. Single nucleotide polymorphisms of the DNA repair gene XPD/ERCC2 alter mRNA expression. Pharmacogenet Genomics 2007; 17: 897-905.
56. Laine JP, Mocquet V, Bonfanti M, et al. Common XPD (ERCC2) polymorphisms have no measurable effect on nucleotide excision repair and basal transcription. DNA Repair 2007; 6: 1264-70.
57. Yin J, Vogel U, Ma Y, et al. Polymorphism of the DNA repair gene ERCC2 Lys751Gln and risk of lung cancer in a northeastern Chinese population. Cancer Genet Cytogenet 2006; 169: 27-32.
58. Matullo G, Palli D, Peluso M, et al. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis 2001; 22: 1437-45.
59. Scagliotti GV, Longo M, Novello S. Nonsmall cell lung cancer in never smokers. Current Opin Oncol 2009; 21: 99-104.
60. Machius M, Henry L, Palnitkar M, et al. Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus. PNAS 1999; 96: 11717-22.
61. Friedberg E. DNA damage and repair. Nature 2003; 421: 436-40.
62. Khan SG, Muniz-Medina V, Shahlavi T, et al. The human XPC DNA repair gene: arrangement, splice site information content and influence of a single nucleotide polymorphism in a splice acceptor site on alternative splicing and function. Nucleic Acids Res 2002; 30: 3624-31.
63. Vodicka P, Kumar R, Stetina R, et al. Genetic polymorphisms in DNA repair genes and possible links with DNA repair rates, chromosomal aberrations and single-strand breaks in DNA. Carcinogenesis 2004; 25: 757-63.
64. Duell E, Wiencke J, Cheng T, et al. Polymorphisms in the DNA repair genes XRCC1 and ERCC2 and biomarkers of DNA damage in human blood mononuclear cells. Carcinogenesis 2000; 21: 965-71.
65. Qiao Y, Spitz MR, Guo Z, et al. Rapid assessment of repair of ultraviolet DNA damage with a modified host-cell reactivation assay using a luciferase reporter gene and correlation with polymorphisms of DNA repair genes in normal human lymphocytes. Mutat Res 2002; 509: 165-74.
66. Hemminki K, Xu G, Angelini S, et al. XPD exon 10 and 23 polymorphisms and DNA repair in human skin in situ. Carcinogenesis 2001; 22: 1185-8.
67. Tang D, Cho S, Rundle A, et al. Polymorphisms in the DNA repair enzyme XPD are associated with increased levels of PAH-DNA adducts in a case-control study of breast cancer. Breast Cancer Res Treat 2002; 75: 159-66.
68. Palli D, Russo A, Masala G, et al. DNA adduct levels and DNA repair polymorphisms in traffic-exposed workers and a general population sample. Int J Cancer 2001; 94: 121-7.
69. Au WW, Salama SA, Sierra-Torres CH. Functional characterization of polymorphisms in DNA repair genes using cytogenetic challenge assays. Environ Health Perspect 2003; 111: 1843-50.
70. Lunn RM, Helzlsouer KJ, Parshad R, et al. XPD polymorphisms: effects on DNA repair proficiency. Carcinogenesis 2000; 21: 551-5.
71. Vogel U, Hedayati M, Dybdahl M, et al. Polymorphisms of the DNA repair gene XPD: correlations with risk of basal cell carcinoma revisited. Carcinogenesis 2001; 22: 899-904.
72. Topinka J, Hertz-Picciotto I, Dostal M, et al. The DNA repair gene XPD/ERCC2 polymorphisms Arg156Arg (exon 6) and Lys751Gln (exon 23) are closely associated. Toxicol Lett 2007; 172: 85-9.
73. King CR, Yu J, Freimuth RR, et al. Interethnic variability of ERCC2 polymorphisms. Pharmacogenomics J 2005; 5: 54-9.
74. Munafo MR, Flint J. Meta-analysis of genetic association studies. Trends Genetics 2004; 20: 439-44.
75. Minelli C, Thompson JP, Abrams KR, et al. The quality of meta-analyses of genetic association studies: a review with recommendations. Am J Epidemiol 2009; 170: 1333-43.
76. Francisco G, Menezes PR, Eluf-Neto J, et al. XPC polymorphisms play a role in tissue-specific carcinogenesis: a meta-analysis. Eur J Hum Genet 2008; 16: 724-34.
77. Zhang D, Chen C, Fu X, et al. A meta-analysis of DNA repair gene XPC polymorphisms and cancer risk. J Hum Genet. 2008; 53: 18-33.
78. Qiu L, Wang Z, Shi X. Associations between XPC polymorphisms and risk of cancers: A meta-analysis. Eur J Cancer 2008; 44: 2241-53.
79. Wang F, Chang D, Hu FL, et al. DNA repair gene XPD polymorphisms and cancer risk: a meta-analysis based on 56 case-control studies. Cancer Epidemiol Biomarkers Prev 2008; 17: 507-17.
80. Kiyohara C, Yoshimasu K. Genetic polymorphisms in the nucleotide excision repair pathway and lung cancer risk: a meta-analysis. Int J Med Sci 2007; 4: 59-71.
81. Attia J, Thakkinstian A, D'Este C. Meta-analyses of molecular association studies: methodologic lessons for genetic epidemiology. J Clin epidemiol 2003; 56: 297-303.
10.1007/s11670-011-0079-2
2011-01-17;Accepted2011-03-17
This work was partly supported by the grants from the National Eleventh-Five-Year Key Task Project of China(No.2006BA102A01), the National “863” High Tech R & D Program of China(No.2006AA02A401) and China-Sweden International Scientific and Technological Cooperative Project (No.09ZCZDSF04100).
*Contributed equally to this work
**Corresponding author
E-mail: zhouqh1016@yahoo.com.cn
©Chinese Anti-Cancer Association and Springer-Veriag Berlin Heldeidelberg 2011
 Chinese Journal of Cancer Research2011年2期
Chinese Journal of Cancer Research2011年2期
- Chinese Journal of Cancer Research的其它文章
- Effects of an Engineered Anti-HER2 Antibody chA21 on Invasion of Human Ovarian Carcinoma Cell In Vitro
- Reduction of Plasma MicroRNA-21 is Associated with Chemotherapeutic Response in Patients with Non-small Cell Lung Cancer
- The Use of CT Perfusion to Determine Microvessel Density in Lung Cancer: Comparison with FDG-PET and Pathology
- Immediate Versus Delayed Treatment with EGFR Tyrosine Kinase Inhibitors after First-line Therapy in Advanced Non-small-cell Lung Cancer
- Clinical Impact of t(14;18) in Diffuse Large B-cell Lymphoma
- Over-expression of Metastasis-associated in Colon Cancer-1 (MACC1) Associates with Better Prognosis of Gastric Cancer Patients
