Genome-wide and candidate gene association studies identify BnPAP17 as conferring the utilization of organic phosphorus in oilseed rape
Ping Xu ,Hao Li ,Haiyuan Li ,Ge Zhao ,Shengjie Dai ,Xiaoyu Cui ,Zhenning Liu,Lei Shi,Xiaohua Wang#
1 College of Agriculture and Forestry Science,Linyi University,Linyi 27600,China
2 National Key Laboratory of Crop Genetic Improvement/Microelement Research Centre/Key Laboratory of Arable Land Conservation (Middle and Lower Reaches of Yangtze River),Ministry of Agriculture and Rural Affairs/Huazhong Agricultural University,Wuhan 430070,China
Abstract Phosphorus (P) is essential for living plants,and P deficiency is one of the key factors limiting the yield in rapeseed production worldwide.As the most important organ for plants,root morphology traits (RMTs) play a key role in P absorption.To investigate the genetic variability of RMT under low P availability,we dissected the genetic structure of RMTs by genome-wide association studies (GWAS),linkage mapping and candidate gene association studies (CGAS).A total of 52 suggestive loci were associated with RMTs under P stress conditions in 405 oilseed rape accessions.The purple acid phosphatase gene BnPAP17 was found to control the lateral root number (LRN) and root dry weight (RDW) under low P stress.The expression of BnPAP17 was increased in shoot tissue in P-efficient cultivars compared to root tissue and P-inefficient cultivars in response to low P stress.Moreover,the haplotype of BnPAP17Hap3 was detected for the selective breeding of P efficiency in oilseed rape.Over-expression of the BnPAP17Hap3 could promote the shoot and root growth with enhanced tolerance to low P stress and organic phosphorus (Po) utilization in oilseed rape.Collectively,these findings increase our understanding of the mechanisms underlying BnPAP17-mediated low P stress tolerance in oilseed rape.
Keywords: genome-wide association studies (GWAS),root morphology traits (RMTs),organic phosphorus (Po),oilseed rape,BnPAP17
1.lntroduction
Phosphorus (P) is of vital importance for plant growth,and it is involved in many key metabolic pathways.Since 80% of the P content in the soil can be fixed from inorganic phosphates (Pi) to organic phosphorus (Po) which cannot be used by plants,the yields of plants are often limited (MacDonaldet al.2011).The Po that cannot be directly absorbed and utilized by plants exists in the soil in the forms of phytate,phospholipids,deoxyribonucleic acid,and others (Denget al.2020).In order to maintain plant yields,global demand for phosphate fertilizers is increasing annually (Kochian 2012).Improving the uptake and utilization efficiency of P by crops through genetic technology is thus an important strategy for reducing the application of P fertilizers (Yanet al.2004).Because oilseed rape (Brassica napusL.Genome AACC,2n=38) (Tanet al.2022) serves as a source of food,feed,and fuel,it is one of the most important oil crops (FAO 2013;http://faostat.fao.org/),yet P is one of the major limiting nutrients for oilseed rape productivity (Zanganiet al.2021).Due to the low mobility of soil P,the root morphology traits (RMTs) of plants play a key role in P acquisition (Wanget al.2017a).To adapt to P stress,the root morphology and architecture can be altered,such as by increasing the root/shoot ratio (Wissuwa 2005),root length (Hufnagelet al.2014),root hair length,root hair density (Gahoonia and Nielsen 2004),and lateral root growth (Halinget al.2013),by reduced root diameter (Lynch 2011),and primary root length (Shiet al.2013),as well as by the induction of proteoid and dauciform roots (Shaneet al.2010).
RMTs are quantitative traits controlled by various genes and loci.Many quantitative trait loci (QTLs) related to RMTs have been recently identified in various crops.InArabidopsis thaliana,a QTL controlling low phosphate root,LPR1,was mapped in the 36 kb region on chromosome 1 and detected in a recombinant inbred line with 411 lines (Svistoonoffet al.2007).In rice,the QTL for P uptake,Pup1,was mapped in the 150 kb region on chromosome 12 and detected in an F3near-isogenic line population with 160 lines (Wissuwaet al.2002).In the common bean (Phaseolus vulgaris),Pup4andPup10were mapped on chromosomes 4 and 10,respectively,and detected in F5:7-derived recombinant inbred lines (Yanet al.2004).In maize,one QTL mapped on chromosome 5 was detected in the recombinant inbred lines with 179 lines (Moussaet al.2021).InB.oleracea,significant QTLs associated with shoot-P and phosphorus utilization efficiency (PUE) were located on chromosomes C3 and C7,and they were detected in 74 commercial cultivars and a 90 doubled haploid (DH) population (Hammondet al.2009).InB.napus,nearly 62 significant QTLs related root volume,root surface area,and total root length were detected in an F10recombinant inbred line with 124 lines (Yanget al.2010).Subsequently,QTLs for primary root length have been identified on chromosomes A7 and C6 with a DH population (190 lines) (Shiet al.2013).A total of 60 QTLs were identified from a paper culture system for root and shoot traits (Zhanget al.2016).Furthermore,multiple genes have been identified and cloned in previous studies,such as the P efficiency genesPSTOL1(phosphorus-starvation tolerance 1 in rice),PHR1(phosphorus-starvation tolerance 1),PHT1(phosphate transporters 1),andSPX(a protein with a singleSYG1/Pho81/XPR1domain),which were related to P deficiency signaling in rice,Chlamydomonas reinhardtii,A.thalianaand oilseed rape (Wykoffet al.1999;Martínet al.2000;Morcuendeet al.2007;Seccoet al.2012;Duet al.2017).The results of the fine mapping and cloning of these genetic loci/genes will be helpful in revealing the genetic mechanisms of RMTs and P efficiency.
Limited by bi-parental segregating populations,low resolution and cost-effectiveness,and greater difficulty in QTL mapping,genome-wide association study (GWAS) can be used to identify relatively higher resolution genetic loci underlying traits because the natural populations take full advantage of ancient recombination events,which shortens the research time (Zhanget al.2014,2023;Wanget al.2017a).Limited by the large number of false positive sites in GWAS,QTL analysis and candidate gene association study (CGAS),based on single nucleotide polymorphisms (SNPs) related to differentially expressed genes (DEGs) that diverge,are used to analyze the contributions of loci to target traits and verify gene functions (Sunduset al.2020a;Tanet al.2022).CGAS has been successfully performed in many crops for identifying complex trait functional genes,includingHd1,Hd3a,andEhd1in the regulation of flowering time in cultivated rice (Takahashiet al.2009),BnaFAD2-A5andBnaFAE1-A8in the regulation of oleic acid production inB.napus(Sunduset al.2020a),as well asBnFATAandBnFAD3in the regulation of ω-3 fatty acid production inB.napus(Sunduset al.2020b).BnPAP17(Purple acid phosphatase 17) is involved in root development in response to low P stress inB.napus(Wanget al.2017a).Moreover,many recent studies have used association studies to detect genetic variations in RMTs and nutrient efficiency in crops using re-sequencing,RDAseq and SNP arrays.Various significant markers were successfully identified,including a total of 40 significant SNP detected for symbiotic nitrogen fixation related traits in bean (Kamfwaet al.2015),six peak SNPs for root efficiency detected in rice under P deficiency (Moriet al.2016),74 significant SNPs associated with P efficiency generated in 192 soybean accessions (Zhanget al.2014),285 peak SNPs associated with RMTs and P efficiency in 405 rapeseed accessions (Wanget al.2017a),and 247 and 166 peak SNPs associated with Ca and Mg concentrations detected in rapeseed leaves (Alcocket al.2017).Further,455 SNP markers were associated with leaf nitrate concentration in rapeseed (Alcocket al.2017).Haplotypes comprise genetic markers on the same chromosome that are inherited together without contemporary recombination.Trait-haplotype association analysis has been successfully performed in the resistance toSclerotina stem rottrait in rapeseed (Weiet al.2015) and P efficiency traits in soybean (Zhanget al.2014).These reports emphasized the potential for association studies in identifying multiple gene functions.
Under the low Pi condition,purple acid phosphatase (PAP),which contains a binuclear metal ion center APase protein,plays an important role in activating the Po around the rhizosphere of plants and promoting the utilization and recycling of phosphorus in plants (Mehraet al.2017;Denget al.2020).As an important APase,PAP can promote the utilization of Po by plants through secretion and activation,which can improve the ability of plants to withstand phosphorus deficiency stress.TheAtPAP12,AtPAP26andAtPAP10genes were reported to be related to the activity of secreted APases and the utilization of Po inArabidopsis(Hurleyet al.2010;Wanget al.2011;Ghahremaniet al.2019).GmPAP21andGmPAP1were associated with APase in soybean roots in response to Pi deficiency (Wuet al.2018).NtPAP12could enhance Pi deficiency resistance through root development in tobacco (Kaidaet al.2009).OsPAP21bwas involved in the cell wall biosynthesis in roots and in improving the utilization rate of Po in rice (Mehraet al.2017).
In this study,the phosphorus-efficient candidate geneBnPAP17was identified through GWAS,QTL and CGAS.Compared with P-inefficient cultivars,the relative expression ofBnPAP17was increased in shoot tissues compared to root tissue under low P conditions in P-efficient cultivars.The haplotype ofBnPAP17Hap3was selected for P efficiency breeding in oilseed rape.In addition,the over-expression ofBnPAP17Hap3could enhance the tolerance to low Pi stress and Po utilization in oilseed rape.Taken together,the findings of this study increase our understanding of the mechanisms underlying theBnPAP17-mediated low P stress resistance response in oilseed rape.
2.Materials and methods
2.1.Plant materials and phenotype evaluation
A total of 405 genetically-diverse rapeseed cultivars were collected worldwide (Appendix A).The cultivars in the panel were grown in a ‘pouch and wick’ hydroponicbased high-throughput phenotyping (HTP) system (Zhanget al.2016) with modified Hoagland solution that was low in P (5 μmol L-1P) in 2014-2015.For each cultivar,16 seeds were sown across eight independent replicates from four different tanks.After sterilization with 70% (v/v)ethanol and NaOCl (2.5% active chlorine),the seeds were grown on top of anchor paper in contact with the modified Hoagland solution without P (where the KH2PO4was replaced with KCl),which was changed every three days.A photo of the cultivar was taken on the 14th day.Root Reader 2D software was used for analyzing the root photos.The primary root length (PRL),lateral root length (LRL),lateral root number (LRN),total root length (TRL),lateral root density (LRD),mean lateral root length (MLRL) and root angle were measured.The shoot and root tissue samples were dried at 80°C and the shoot dry weight (SDW) and root dry weight (RDW) were determined (Shiet al.2013;Wanget al.2017a).The seeds of oilseed rape Westar 10 and over-expression cultivarBnPAP17-OE were germinated and grown in modified Hoagland solution with organic P (where the KH2PO4was replaced with ATP) at low P (0.005 mmol L-1P) and normal P (0.625 mmol L-1P) conditions.
Pearson correlation analysis was conducted to evaluate the significance of relationships between RMTs using GenStat15th (VSN International,Oxford,UK) software.The RMTs were used to allocate the sources of variation and estimate individual means by the restricted maximum likelihood (REML) model,and differences were determined using Studentt-test with significance at the 1% probability level (Wanget al.2017a).
2.2.Differential gene expression in root and shoot tissues at high and low P levels
Total RNA of the P-efficient cultivar ‘Eyou changjia’ (with leaves and root tissues separated) was extracted from three independent biological samples using the Omega Plant RNA Kit (Omega Engineering Inc.,Norwalk,USA).The TopHat v2.0 on a HiSeq 2500 platform (Novo Gene Bioinformatics Institute,Beijing,China) was used for the RNA sequencing.The following criteria were used for the DEGs detected in root and shoot tissues with low and normal P levels:P<0.01,false discovery rate (FDR)<0.01,and |log2Reads per kilobase of exon model per million mapped reads (FPKM)_normal P(NP)/(FPKM)_low P(LP)|>2 (Weiet al.2015).
2.3.Genotyping by the Brassica 60K SNP array and re-sequencing of the association panel
DNA was extracted from young oilseed rape leaf tissues using a modified cetyltrimethyl ammonium bromide method with an adjusted concentration of 50 ng μL-1(Xuet al.2014).TheBrassica60K Illumina Infinium SNP array and Illumina NovaSeq 6000 (Illumina Inc.,San Diego,USA) were employed to screen the panel and SNP data using the v4.1 draft of ‘Darmor-bzh’ as the reference genome (Chalhoubet al.2014).The SNPs meeting the criteria of calling rate>0.80,minor allele frequency (MAF)>0.05 and one blasted (50 bp probe ofBrassica60K SNP array and 150 bp align reads of Illumina NovaSeq 6000) hit on the genome were included for further analysis (Wanget al.2017a).The SNPs located in the DEGs between the low and normal P levels were included for further analysis.
2.4.Population structure,relative kinship and Linkage disequilibrium (LD) analysis of the association panel
Based on the polymorphic SNPs of 405 rapeseed cultivars,the population genetic structure was estimated using STRUCTURE 2.3.4 software (Evannoet al.2005).The criteria of K from 1 to 10,100,000 burn-in period and 100,000 Bayesian Markov Chain Monte Carlo (MCMC) replication number were used to determine the true K value (subgroup number) (Evannoet al.2005).The CLUMPP,PowerMarker 3.25,SPAGeDi,DNASP6 and TASSEL 4.0 software were used for Q matrix,gene diversity,relative kinship matrix,Tajima’s D,and Linkage disequilibrium (LD) decay calculation,respectively (Alcocket al.2017;Wanget al.2017a).
2.5.GWAS and CGAS of oilseed rape RMTs
The Q matrix and kinship matrix were used to show the population structure and relative kinship access of the association panel,respectively.The general linear model (GLM) and mixed linear model (MLM) were used to determine the associations between markers and traits.Unlike the MLM model,the GLM model includes naive and population structure models without controlling for the kinship of the association panel,which could lead to highly spurious associations caused by genotype-phenotype covariance.TheP-value between the genotype and phenotype was calculated using TASSEL 4.0 software.According to the Bonferroni correction for multiple tests,the significant SNPs (1/number of all SNPs) related to traits were determined.An R script was used for quantile-quantile and Manhattan plots (Weiet al.2015;Wanget al.2017a;Xuet al.2022).The LOD scores of QTLs related to LRN and RDW under LP were drawn using Origin 8 software (https://ea-origin.en.softonic.com/).The expression heat maps of P-inefficient and -efficient cultivar root and shoot tissues were drawn using MeV4.9.0 software (https://sourceforge.net/projects/mevtm4/files/mev-tm4/).
2.6.The quantitative real-time RT-PCR
After extraction of the total RNA from leaves of P-inefficient and -efficient cultivars in the natural population using the RNAsimple Kit (DP419,TIANGEN,China),the reversed RNA (followed by Invitrogen™ SuperScript™,ThermoFisher,USA) was conducted in the quantitative real-time RT-PCR using SYBR®Green I (Invitrogen,ThermoFisher).The relative expression levels of genes were quantified using the 2-ΔΔCtmethod (Xuet al.2022) with specific primers 5´-ACACTGCTCCGAGTCTTCAGAA-3´ and 5´-GCGTCGACTACGAATGATCG-3´ ofBnPAP17-A05 (with reference gene ofBnACTIN7) (Yanget al.2014).
2.7.Haplotype and selective sweep analysis of BnPAP17
The mutations of favorable alleles could lead to different phenotypes being dependent on the gene expression and sequence mutations.Since they are located in the sequence ofBnPAP17-A05,the marker haplotypes were inherited together without contemporary recombination.Based on the criteria ofr2>0.4 and cultivars of panel number>3,the haplotype blocks with access to the association population were identified using TASSEL5 software.Based on the criteria of gene diversity<0.2 (calculated by PowerMarker V3.25 software) and Tajima’D<-1.0 (calculated by DnaSP6 software),the selective sweep of SNPs in the sequence ofBnPAP17were determined (Wanget al.2017b).The box-plots of haplotype blocks related to RMTs under low P levels in the rapeseed association population cultivars were drawn using OriginPro 8.0 Software (Wanget al.2017a;Xuet al.2022).
2.8.Vector construction for transformation and transformation in oilseed rape
After amplification of the open reading frame (ORF) ofBnPAP17,the amplified PCR product was cloned into the over-expression vector driven by the double 35S promoter (pMDC83) to verify the correct sequence of 2X35S::BnPAP17.The hypocotyl of oilseed rape was infected byAgrobacterium tumefacienswhich transferred the recombinant vector.The callus was induced in a medium with 1 mg L-12,4-D and 0.3 mg L-1kinetin (KT).The adventitious buds were induced in a medium with 2 mg L-1trans-zeatin (TZ) and 0.1 mg L-1indole-3-acetic acid (IAA).Subsequently,the rooting and transplanting of the adventitious buds formed the regenerated plants.
2.9.GUS staining of BnPAP17
The 2 kb genomic DNA fragment of the 5´ upstream region ofBnPAP17Hap3was amplified and cloned into the pBI121 vector.Then,transgenicArabidopsiswith theBnPAP17Hap3promoter-GUS fusion was constructed.Subsequently,transgenicArabidopsisat 10 and 25 d after the seedling stage were incubated at 37°C in 5-bromo-4-chloro-3-indolyl-b-glucuronic acid solution for 24 h and then cleaned with 75% (v/v) ethanol.Lastly,the transgenicArabidopsiswere observed with an Olympus IX-70 microscope equipped with Nomarski Optics (Liet al.2015).
3.Results
3.1.Phenotypic variations of RMTs in the B.napus association panel under low P conditions
To evaluate the phenotypic variation of RMTs in the association population (Appendix A),10 traits were measured for plants cultured in the greenhouse under low P levels.Extensive phenotypic variations were observed in SDW,RDW,R/S ratio,PRL,LRL,TRL,LRD,LRN,MLRL,and root angle of the association panel of rapeseed under P deficiency in the paper culture experiments (PC) (Table 1).The Shapiro-Wilk normality test indicated that the variations in RMTs in the current population fit a normal distribution (P<0.05;Fig.1).Continuous and wide phenotypic variations were observed for the RMTs among the accessions,ranging from 7.18% in MLRL to 31.33% in RDW,with anaverage variation of 21.41% (Fig.1;Table 1),suggesting that the RMTs were controlled by multiple genes at LP.Correlation analysis of each pair of RMTs showed that LRL (P<0.01,R2=0.9734),LRN (P<0.01,R2=0.7284),and PRL (P<0.05,R2=0.6621) were significantly positively correlated with TRL (Appendix B).
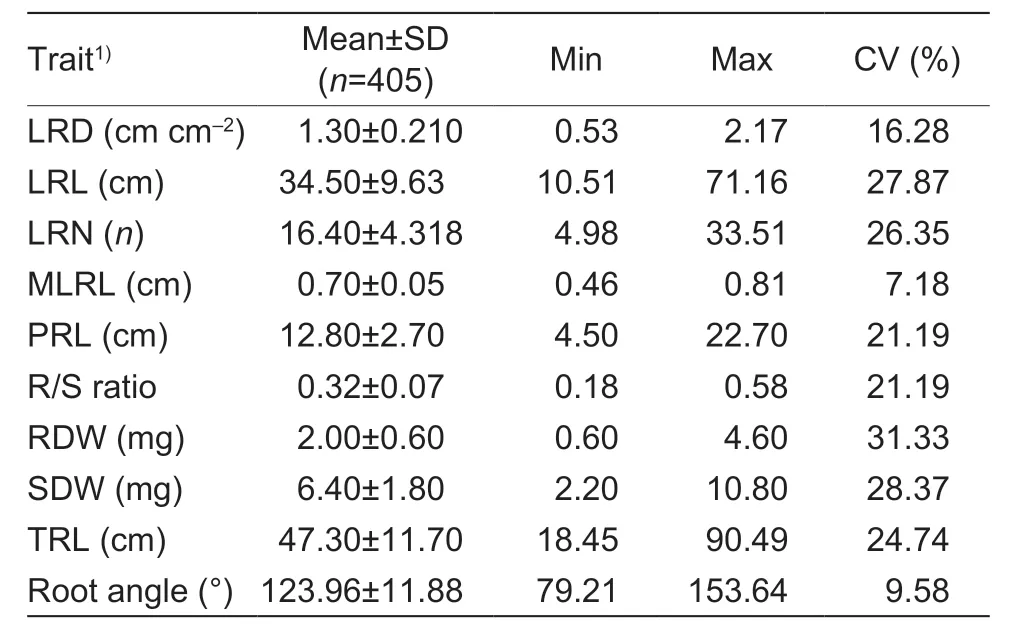
Table 1 Phenotypic variations of LRD,LRL,LRN,MLRL,PRL,R/S ratio,RDW,SDW,TRL and root angle under low phosphorus in the paper culture experiment in the association panel of oilseed rape
3.2.Genomic variations of the candidate genes responding to low P stress
To characterize the genomic variation in the cultivatedB.napusgene pool,the 405 cultivars in the association panel (Appendix A) were genotyped using theBrassica60K Illumina Infinium SNP array.After transcriptome sequencing of the allotetraploidB.napuscultivar ‘Eyouchangjia’ under conditions of contrasting P availability,6,091 DEGs located on 19 chromosomes with a density of 4 Mb-1were identified (Fig.2-A;Table 2;Appendix C).After further exclusion,13,340 SNPs showed high polymorphism and genotypic quality (Appendix C).These 13,340 SNPs were distributed among the 19 chromosomes ofB.napuswith a density of 21 Mb-1(Fig.2-B;Table 2;Appendix D).The DEGs density varied dramatically between the A and C subgenomes (Table 2;Appendix D).The density of DEGs in the A sub-genome was 14 Mb-1,which was approximately twice that found in the C sub-genome (6 Mb-1).The DEGs number on each chromosome ofB.napusranged from 193 on C2 to 608 on A3.The SNP number on each chromosome ofB.napusranged from 365 on C5 to 1,504 on C4 (Table 2;Appendix D).

Fig.2 Densities of differentially expressed genes (DEGs) and candidate markers,population structure and relative kinship of the association panel of oilseed rape.A,the density and number of DEGs of the panel.B,the density and number of candidate markers of the panel.C,LnP(D) and ΔK for the population structure of the 405 rapeseed accessions.D,relative kinship of the panel.E,population structure of the panel (405 rapeseed accessions) when K is equal to 2.

Table 2 Summary of the differentially expressed genes (DEGs) and single nucleotide polymorphism (SNP) numbers,DEGs and SNP densities,and LD decay in the association panel of oilseed rape
3.3.Population structure,relative kinship,and LD decay of the association panel
Since population structure plays a key role in affecting the authenticity of QTL mapping,the population structure was calculated using STRUCTURE 2.3.4 software for GWAS.The population structure was limited by the continuous increase in K values and the inflexion point of the LnP(K) value was not obvious (Fig.3-C-green dotted line).However,the Evanno’s ΔKvalue showed an obvious spike at K=2 (Fig.3-C-red dotted line),meaning that the population could be divided into Group 1 and Group 2 (Fig.3-E),and the numbers of cultivars in these groups were 66 and 339,respectively (Appendix A).In addition,the relative kinship of the association panel,evaluated using the SPAGeDi software,played a critical role in the GWAS.A total of 79% of the pairwise relative kinship coefficient values were 0 (with an average of -0.186).A total of 96% of the pairwise kinship coefficient values were <0.2 (Fig.3-D).These results showed that the cultivars in the association panel were distantly related.Based on 100 kb slide windows,the allele frequency squared correlations (r2) were investigated in the LD extent.Based on setting the cut-offr2to 0.2,the distance of LD decay of the association panel ranged from 125 to 8,250 kb (with an average of 1,100 kb) (Table 2).The LD decay on the A sub-genome was 250 kb,which was approximately one-quarter of that found in the C sub-genome (1,065 kb) (Table 2).
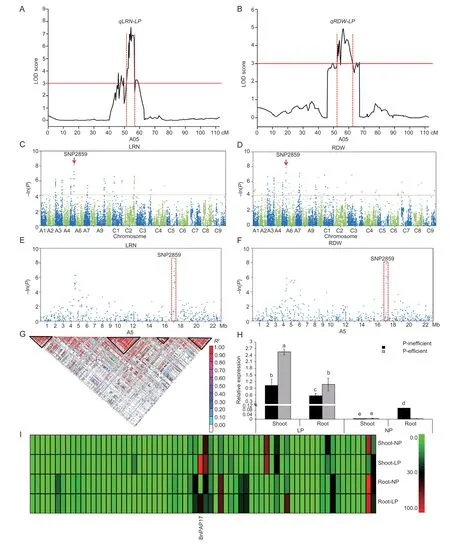
Fig.3 The candidate gene of BnPAP17 relationships to lateral root number (LRN) and root dry weight (RDW) as detected by transcriptome-wide association studies,linkage map analysis and transcriptome analysis.A, QTL related to LRN under low phosphorus on linkage group A5 of the BnTN-DH (Brassica Tapidor-Ningyou 7 DH) population.B,QTL related to RDW under low phosphorus on linkage group A5 of the BnTN-DH population.C,the significant single nucleotide polymorphism (SNP) (SNP2859) related to LRN was detected in the 405 accessions of oilseed rape by genome-wide association studies (GWAS).D,the peak SNP (SNP2859) related to RDW was detected in the association panel of oilseed rape by GWAS.E, the significant SNP (SNP2859) related to LRN was detected in by single chromosome association analysis.F, the peak SNP (SNP2859) related to RDW was detected in the association panel of oilseed rape by single chromosome association analysis.G,linkage disequilibrium (LD) heatmap surrounding the peak SNP2859 on the A5 chromosome (the third inverted triangle from the left).H, relative expression of BnPAP17 in shoot and root tissues of P-inefficient and -efficient cultivars under low and normal phosphorus by qRT-PCR.Bars mean SD (n=9).Different letters represent significance between tissues or plants at P<0.05.I, gene expression of LD blocks of peak SNP2859 in root and shoot tissues through transcriptome sequencing.
3.4.Genome wide association studies of RMTs in oilseed rape
Considering the false positives in genotype-phenotype associations,four common models for association analysis (i.e.,the naive model (GLM),Q model (GLM (Q)),K model (MLM (K)),and MLM model (Q+K)) were compared using a quantile-quantile (QQ) plot (Appendices E and F),and the best model for each trait in the different trials was selected.A total of 93 significant SNPs related to RMTs were scanned in the LP condition in paper culture (PC) withP<7.50×10-5(Appendix G).Under LP conditions,three SNPs for LRD were detected using the Q+K model;seven SNPs for TRL,eight SNPs for LRL,five SNPs for MLRL,four SNPs for SDW,five SNPs for root angle and three SNPs for the R/S ratio were detected by the Q model (Appendices E and G).Altogether,these 93 loci explained 53.7,52.4,47.3,40.9,37.1,33.2,27.8,19.5,and 16.6% of the total phenotypic variances of RDW,LRL,TRL,SDW,root angle,PRL,MLRL,R/S ratio,and LRD,respectively (Table 3;Appendix G).Thirteen of the 93 significant SNPs were co-localized with the position for the same traits in a previous association study (Wanget al.2017a),including one SNP on A2 for LRD,one SNP on C3 for MLRL,one SNP on A2 for PRL,one SNP on A5 for RS-ratio,one SNP on A3 for TRL,two SNPs on A3,and C7 for LRL,two SNPs on A3 and A7 for RDW,and four SNPs on A5,A9,C1,and C2 for LRN detected at LP (Table 4).

Table 3 The number of loci and total phenotypic variance of the same root morphology trait related single nucleotide polymorphism (SNP) as detected by candidate gene association studies (CGAS) in the association panel of oilseed rape

Table 4 The co-locating single nucleotide polymorphism (SNP) for root morphology traits as detected by genome-wide association studies (GWAS) and andidate gene association studies (CGAS) in this study and previous studies (Wang et al.(2017a)) in the association panel of oilseed rape
Many of the RMTs had relationships between them,for instance,TRL was the PRL plus LRL.Considering the phenotype,the correlation analysis of each pair of RMTs in the association panel ofB.napusis shown in Fig.2.For the genome,eight SNPs demonstrated multi-effects and co-location for more than two RMTs detected by GWAS and CGAS (Table 5).The number of RMTs related to the co-located SNP markers ranged from two (LRN and RDWrelated to SNP2583 on chromosome A5) to five (PRL,TRL,LRD,LRN,and RDW related to SNP1138 on chromosome A3) (Fig.4;Table 5).The expression of candidate genes with those SNP intervals are shown in Appendix H.

Table 5 BnPAP17Hap3 acts as a positive regulator of shoot and root growth under low organophosphorus stress1)
3.5.The identification of BnPAP17
A total of 93 SNPs associated with 10 RMTs were identified by GWAS from the 405 rapeseed cultivars.The significant SNP2859 was found to be multi-effective and co-locating with LRN and RDW under low P conditions in the oilseed rape association panel through GWAS (Fig.3-C-F) and previous QTLs (Zhanget al.2016) in theBnTN-DH (BrassicaTapidor-Ningyou 7 DH) population (Fig.3-A and B).In total,77 candidate genes (FDR<0.01;P<0.01) were within the LD decay region confidence interval (Fig.3-G;Table 2) of the peak significant SNP2859 for the LRN and RDW traits on chromosome A5 (Appendix I).Two DEGs (BnaA05g22460DandBnPAP17) were identified between the two P levels in the same tissue based on theP<0.01,FDR<0.01,and |log2(FPKM_NP/FPKM_LP)|>2 criteria in the ‘Eyou changjia’ cultivar according to the transcriptome analyses (Fig.3-I;Appendix I).Compared with normal P levels,the relative expression ofBnPAP17was significantly increased in shoot and root tissues in P-inefficient and -efficient oilseed rape under low P stress (Fig.3-H;Appendix J).
3.6.The BnPAP17Hap3 contributes to shoot and root growth
To identify the natural allelic variation inBnPAP17associated with shoot and root growth,33 polymorphic SNPs in the translated region,coding sequencing region and 1,500 kb upstream ofBnPAP17were detected in the association panel by re-sequencing (Fig.4-A and B;Appendix K).Seven of the 33 SNPs in the promoter sequencing ofBnPAP17formed five haplotypes (Hap1,Hap2,Hap3,Hap4 and Hap5) (Fig.4-C).The LRD,RDW,PRL,RS ratio,LRL,SDW,LRN,TRL,MLRL and Root angle of Hap3 (n=13) were significantly higher than those of the Hap4 group (n=4) (Fig.4-E-N).At the same time,the gene diversity (0.051) and Tajima’s D (-1.700) of Hap3 were significantly lower than those of Hap4 (with gene diversity=1;Tajima’s D=0.740).The main haplotype type,Hap5 (n=179),showed significantly higher shoot and root traits but lower gene diversity (0.242) and Tajima’s D (-1.126) than those in Hap4.The G+C content of Hap3 (0.694) was higher than that of Hap4 (0.576) (Fig.4-D).
3.7.BnPAP17Hap3 positively regulates shoot and root growth under low Po stress
To investigate the role ofBnPAP17Hap3in regulating oilseed rape growth,the full-length cDNA ofBnPAP17Hap3under the control of a constitutive 35S promoter (35::BnPAP17Hap3) was transformed into oilseed rape cultivar “Westar 10”.Compared with the control line (Westar 10),the over-expressingBnPAP17Hap3line (BnPAP17-OE) showed significantly higher values of RDW,SDW,TRL,TSA,RV,PRL and LRN under the normal Pi (0.625 mmol L-1P) solution condition.However,theBnPAP17-OE lines showed lower RDW and SDW than the Westar 10 lines in low Pi (0.005 mmol L-1P) stress (Fig.5;Table 5).Under the low Po stress,theBnPAP17-OE lines showed significantly higher values of RDW,SDW,TRL,TSA,RV,PRL and LRN than Westar 10 (Fig.5;Table 5).Compared with Westar 10,the relativeexpression ofBnPAP17was significantly up-regulated in the over-expression rapeseed lines (Appendix L).
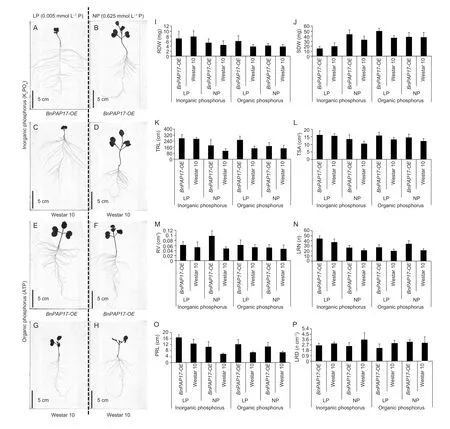
Fig.5 Phenotype of BnPAP17 over-expression in rapeseed under low (LP) and normal (NP) inorganic and organic P conditions.A and B,phenotypes of the BnPAP17 over-expression cultivar under LP and NP,respectively.C and D, phenotypes of cultivar Westar 10 (CK) under LP and NP,respectively.E and F, phenotypes of the BnPAP17 over-expression cultivar under LP and NP,respectively.G and H, phenotypes of cultivar Westar 10 (CK) under LP and NP,respectively.I-P, root dry weight (RDW), shoot dry weight (SDW),total root length (TRL),total surfer area (TSA), root volume (RV),lateral root number (LRN), primary root length (PRL),and lateral root density (LRD) of the BnPAP17 over-expression and CK cultivars under normal/low and inorganic/organic P conditions,respectively.Bars mean SD (n=9).
To better understand the specific expression ofBnPAP17,theBnPAP17Hap3promoter-GUS fusion was constructed inArabidopsis.After detecting the GUS signal of transgenicArabidopsis(10 and 25 d after germination) under low Po stress,theBnPAP17expression was stronger in young leaves,hypocotyl,primary root (without root tips),and leaf veins than in other tissues of the 10 d transgenicArabidopsisplants.Moreover,the expression ofBnPAP17was stronger in leaf veins,flower bud,primary root and the lateral root vascular system (without root tips) than in other tissues of the 25 d transgenicArabidopsisplants (Fig.6;Appendix M).Therefore,these data verified thatBnPAP17could confer the utilization of Po through shoot and root growth of oilseed rape.

Fig.6 Tissue-specific expression of BnPAP17 in Arabidopsis.A,the β-glucuronidase (GUS) signal was stronger in younger leaves than old leaves and root tissues,without root tissue at the top of the root (15 days after germination).B, the GUS signal was stronger in veins than in leaf of Arabidopsis(15 days after germination).C, the GUS signal was stronger in the axial column in root tissues without cortex and root tips(15 days after germination).D,after 20 days,GUS was detected in buds and leaves of Arabidopsis.
4.Discussion
4.1.High-throughput root morphology identification can promote breeding of P-efficient rapeseed
Ideal root architecture and RMTs contribute to phosphorus efficiency breeding,which has prompted crop geneticists to identify low P-related root morphology and architecture in plants.Under low P stress,the plant develops a series of physiological mechanisms related to root morphology and architecture for increasing the root surface contact with P,thereby activating the Po to enhance the absorption capacity for phosphorus (Wissuwa 2005;Shiet al.2013;Hufnagelet al.2014;Xuet al.2022).Limited by cost-effectiveness and a limited number of traits detected in the field in previous studies,the ‘pouch and wick’ hydroponic-based HTP system in the greenhouse (10 RMTs were detected) was used in this study.After planting in the middle of the upper edge of each germination paper,a total of 192 plants were grown in each tank.By adjusting each image to the same scale,the HTP system could reduce the time required to perform such an evaluation and preclude complications due to the variable sizes of the images of different plants through RR2D automatic calculation software (Wanget al.2017a).In addition,phenotypic data obtained for the RMTs of different batches enabled us to determine the length,weight,and angle that provided the most reliable representation of the RMT phenotype.The phenotypic variation of RMTs is shown in Table 1.Our statistical analyses revealed that the LRL (P<0.01,R2=0.9734),LRN (P<0.01,R2=0.7284),and PRL (P<0.05,R2=0.6621) were positively correlated with TRL (Appendix B).The correlations between TRL and other traits (LRL,LRN,and PRL) suggest that accessions with shorter total root length tend to be accompanied by a more compact root architecture.In addition,positive correlations we also observed between shoot (P<0.1,R2=0.4200) and root (P<0.1,R2=0.5906) biomass traits and LRN traits (Appendix B).These results are informative for breeders trying to adapt RMTs to achieve the ideal root architecture and high P efficiency.
4.2.Novel genes were detected by genome-wide and candidate gene association studies
Identifying the QTLs related to RMTs is another strategy for revealing the genetic pattern of P efficiency and guiding P efficiency breeding.Many QTLs related to RMTs have been identified in various plants in recent years (Wissuwaet al.2002;Yanet al.2004;Svistoonoffet al.2007;Hammondet al.2009;Shiet al.2013;Zhanget al.2016;Wanget al.2017a;Xuet al.2022).Based on the fine mapping and cloning of QTLs,multiple P efficiency genes were detected in previous studies (Wykoffet al.1999;Martínet al.2000;Seccoet al.2012).However,limited by the low resolution and cost-effectiveness of bi-parental segregating populations of traditional quantitative trait mapping,and the large number of false association loci of GWAS,CGAS was used for the RMTs ofB.napusgrowing in a solid substrate under P stress conditions during the seedling stage in this study.To the best of our knowledge,6,091 DEGs (Appendix C) were identified through transcriptome sequencing of ‘Eyou changjia’ under conditions of contrasting P availability.After further exclusion,13,340 high polymorphism SNPs (Appendix D) related to the DEGs were detected.In total,52 suggestive association loci for RMTs under P stress conditions were identified.Compared with recent studies on RMTs in rapeseed (Wanget al.2017a),a total of 28 new SNPs and 24 previously identified significant SNPs were detected in the present study (Appendix G),based on theP-values and trait numbers between the studies in 2017 and those presented here.The SNP number and p-value were 30,976 and 3.23×10-5in the previous studies,whereas there were 13,340 (Wanget al.2017a) and 7.50×10-5revealed in this study (Appendix G),respectively.The same 13 significant SNPs related to eight traits (LRD,LRL,LRN,MLRL,PRL,RDW,TRL,and R/S ratio) were detected in the previous study (Wanget al.2017a) and in this study (Table 4).However,the 28 new SNPs (Appendix G) were related to the ten traits in this study,but not in previous studies.
4.3.Selective sweep analysis in the excellent haplotype of BnPAP17
Genetic linkage and pleiotropy can cause one marker to be associated with multiple traits.For picking the important SNPs out of dozens or hundreds of significant SNPs,the co-located SNPs and favorable alleles for more than two RMTs exhibit cumulative effects.In this study,we identified eight co-located SNP loci on the A3,A5,A7,C1,C2,and C7 chromosomes that were associated with more than two RMTs (PRL,TRL,LRD,LRN,RDW,MLRL,LRL,and SDW) under low P conditions (Appendix N).Since the favorable allele mutations lead to gene sequence mutations or gene expression changes featuring different phenotypes,theBnPAP17was found to be sensitive to low P stress in shoot tissues by transcriptome sequencing (Fig.3;Appendix J).As selective breeding plays a key role in oilseed rape breeding (Wanget al.2017b),the different haplotype types of theBnPAP17sequence led to different phenotype types (Fig.4).Phosphorus efficiency breeding related toBnPAP17was re-assessed by Tajima’s D and the genetic diversity of the different haplotype types.Finally,theBnPAP17Hap3andBnPAP17Hap5were selected by unconscious artificial selection with lower Tajima’s D and gene diversity values (Fig.4-D).
4.4.BnPAP17 conferred the utilization of Po for shoot and root growth in oilseed rape
AsB.napusandA.thalianaare cruciferous plants and close relatives,the available knowledge of genes inA.thalianawas applied to the same traits inB.napususing comparative genomics (Hammondet al.2009).The biochemical and functions of the plant PAP members involved in Po utilization and the response to Pi deficiency are known inArabidopsis,soybean and rice (Hurleyet al.2010;Wanget al.2011;Wuet al.2018;Denget al.2020).After over-expression,BnPAP17could enhance the shoot and root growth and Po utilization in oilseed rape (Fig.5),which is similar to the roles ofAtPAP12,AtPAP26,AtPAP10,GmPAP21andOsPAP21binArabidopsis,soybean and rice.The GUS signals of BnPAP17 were detected strongly in the tissues (leaves veins,flower buds and root vascular system without root tips) of the youngArabidopsisplant (Fig.6),indicating thatBnPAP17conferred the utilization of Po during shoot and root growth in oilseed rape.Overall,the findings of this study confirmed thatBnPAP17mediates the shoot and root growth by enhancing the utilization of Po in response to low P tolerance in oilseed rape.
5.Conclusion
This study investigated the function ofBnPAP17in regulating the shoot and root growth by enhancing tolerance to low P stress and Po utilization in oilseed rape.Therefore,the results indicate thatBnPAP17may be useful for enhancing plant tolerance to low P stress.
Acknowledgements
The authors thank Martin R.Broadley from Nottingham University,UK for offering the HTP system and the greenhouse.This work was financially supported by the National Natural Science Foundation of China (32201868 and 32001575).
Declaration of competing of interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2023.05.002
 Journal of Integrative Agriculture2024年4期
Journal of Integrative Agriculture2024年4期
- Journal of Integrative Agriculture的其它文章
- Invasion of fall armyworm led to the succession of maize pests in Southwest China
- Impacts of agri-food e-commerce on traditional wholesale industry: Evidence from China
- Transcriptomic and metabolomic analysis provides insights into lignin biosynthesis and accumulation and differences in lodging resistance in hybrid wheat
- Responses of growth performance,antioxidant function,small intestinal morphology and mRNA expression of jejunal tight junction protein to dietary iron in yellow-feathered broilers
- Genome-wide identification of the CONSTANS-LIKE (COL) family and mechanism of fruit senescence regulation by PpCOL8 in sand pear (Pyrus pyrifolia)
- ldentification of S-RNase genotype and analysis of its origin and evolutionary patterns in Malus plants
