Coinfection of SARS-CoV-2 Omicron variant and other respiratory pathogens in children
Wei Li·Bing-Han Wang·Bao-Hai Chen·Yi Sun·Lin Li·Wen-Qing Xiang·Ahmed Faisal Ali·Lin-Xuan Su·Hai-Yan Mao ·Hong-Qiang Shen·Qiang Shu
Since the outbreak of coronavirus disease 2019 (COVID-19), the entire world has been affected by the epidemic [1,2].On November 9, 2021, a variant of the novel coronavirus B.1.1.529 was detected for the first time from case samples in South Africa.The Omicron strain spread and mutated significantly fast.This variant has the characteristics of escape immunity, which can induce relatively extreme disease and reduce the neutralization of antibodies in vaccinators, and is more likely to be reinfected [3, 4].The clinical presentation of severe acute respiratory syndrome coronavirus 2 (SARSCoV-2) in children has mild symptoms and a lower risk of hospitalization and death.The most common symptoms are fever, cough, headache, diarrhea, and sore throat [5].Coinfection with bacteria, fungi, influenza virus, human parainfluenza virus (HPIV), respiratory syncytial virus (RSV),adenoviruses (ADV), human metapneumovirus (HMPV),human rhinovirus (HRV), and so on has been described as a factor associated with more severe clinical symptoms and outcomes in children with SARS-CoV-2 [6–8].
Our study aimed to investigate the influence of changes in prevention and control strategies on the infection of SARSCoV-2 and other respiratory pathogens, as well as their coinfection patterns and clinical symptoms in children.
Throat swabs were collected from pediatric patients in our hospital from December 1, 2022, to December 31, 2022.The inclusion criteria were as follows: children who were diagnosed with acute respiratory tract infection and were under 14 years old [9].Information, including demographic data, case history, symptoms, and clinical results for each patient, was also collected.
SARS-CoV-2 infection was confirmed by RNA or antigen detection.For RNA detection, throat swabs were collected and detected using a commercial reverse transcription-polymerase chain reaction kit (Guangzhou Daan Gene, China and Hunan Sansure Biotech, China) with amplification targeting theORF1a/bandNgenes.For antigen detection,throat swabs were collected and detected using a commercial colloid gold assay kit (Nanjing Vazyme, China) targeting the SARS-CoV-2 N antigen.Other respiratory pathogen infections were confirmed by 11 pathogen nucleic acid assays or antigen detection from 17,450 throat swabs.To detect 11 pathogens’ nucleic acids, throat swabs were obtained and detected using a commercial multiplex reverse transcriptionpolymerase chain reaction assay (Health Biomed, Ningbo,China).Eleven common respiratory pathogens are listed,including influenza A virus (IAV), influenza B virus (IBV),HPIV, RSV, ADV, HMPV, HRV, human bocavirus (HBOV),human coronavirus (HCOV), chlamydia (CH), andMycoplasma pneumonia(MP).For antigen detection, throat swabs were stored in a buffer solution (KaiBiLi, Hangzhou, China).Five common respiratory pathogens were screened (including RSV, ADV, IAV, IBV, and MP).
The whole-genome sequences were edited using Geneious Prime (https:// www.genei ous.com/).Additional sequences were downloaded via the Global Initiative of Sharing All Influenza Data (https:// www.gisaid.org/).The full-length genome sequence of Wuhan-Hu-1 (NC_045512) as the reference strain was downloaded from GenBank.Multiple sequence alignment was performed using MUSCLE [10].The phylogenetic tree was constructed using the neighborjoining method.One thousand bootstrap replicates were run to assess the reliability of the phylogenetic trees.The clinical characteristics were described for all patients and according to whether they were coinfected with SARS-CoV-2 and other respiratory pathogens.Continuous variables that were normally distributed or skewed were presented as the mean(standard deviation) or median (interquartile range, IQR),respectively, while the categorical variables were described by the number and percentage.Depending on whether the continuous variables were normally distributed, Student’sttest and Wilcoxon signed-rank test were conducted to compare the inter-group differences.Student’sttest is used to infer the probability of difference usingtdistribution theory to compare whether the difference between two averages is significant.It is mainly used for normally distributed data with small sample content (e.g.,n< 30) and an unknown population standard deviation.In the Wilcoxon signedrank test, the rank of the absolute value of the difference between the observed value and the center position of the null hypothesis is added separately according to different signs as its test statistic.It is suitable for pairwise comparison inttests, but the difference in pairwise data is not required to obey a normal distribution; only a symmetrical distribution is needed.Additionally, the Chi-square test is a method widely used hypothesis test, which is the deviation degree between the actual observed value and the theoretical inferred value of the statistical sample.The Chi-square value indicates the degree of inconsistency, which decreases as the deviation decreases.All analyses were performed using R 4.2.2.Two-sidedPvalues are presented throughout, and aPvalue < 0.05 denoted statistical significance.
Whole-genome targeted sequencing technology was used to obtain SARS-CoV-2 genome assemblies from ten randomly selected SARS-CoV-2 patients [median (IQR) age:3.1 (10.4) years, female: 30.0%] who were admitted to the hospital in December 2022.As shown in Supplementary Fig.1, the results revealed that ten genomes were clustered into two major sublineages: Omicron variant BA.5.2 (3/10,30%) and BF.7.14 (7/10, 70%).
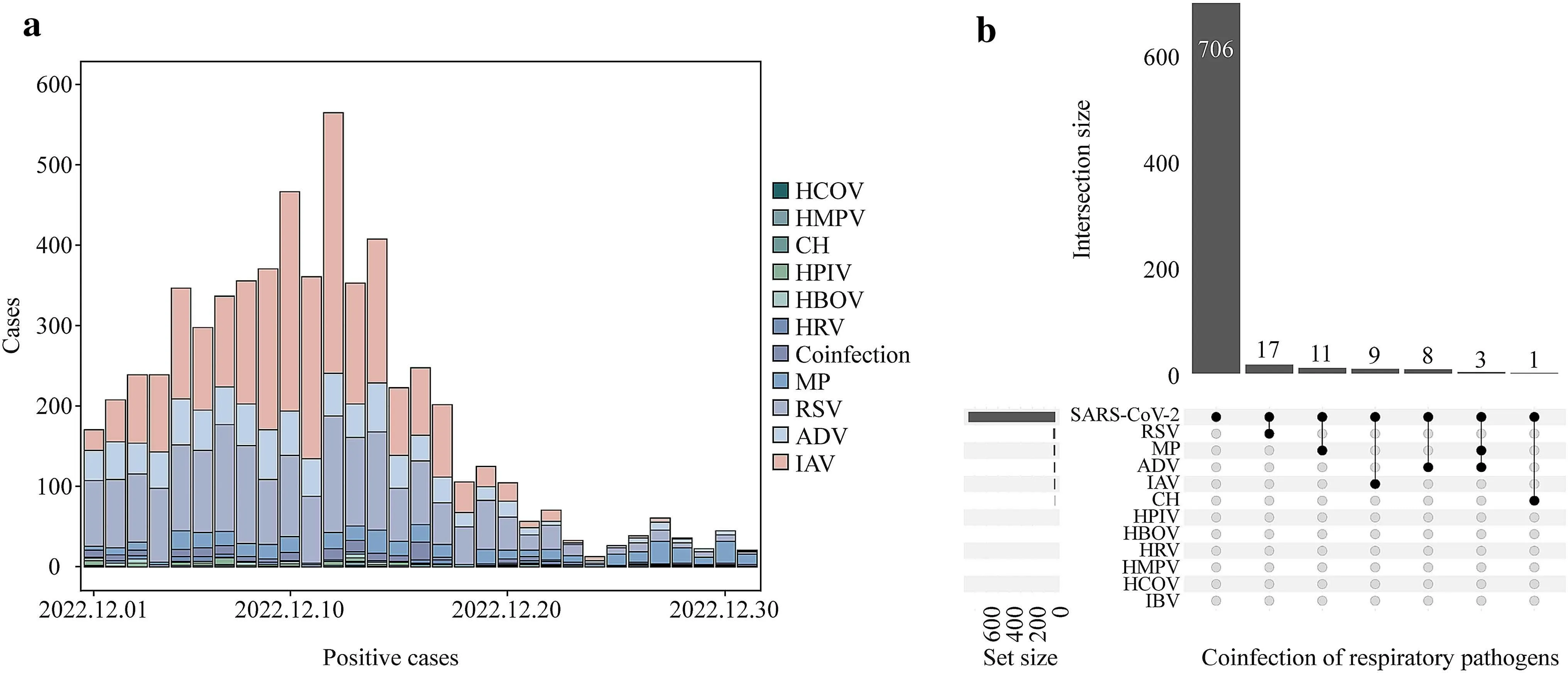
Fig.1 Positive cases of 11 respiratory pathogens and coinfection pattern in December 2022.a Positive cases of HCOV human coronavirus; b intersection size of coinfection respiratory pathogens.HMPV human metapneumovirus, CH chlamydia, HPIV human parainfluenza virus, HBOV human bocavirus, HRV human rhinovirus, MP Mycoplasma pneumonia, RSV respiratory syncytial virus, ADV adenoviruses, IAV influenza A virus, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2, IBV influenza B virus
Seventeen thousand four hundred and fifty individuals[median (IQR) age: 4.0 (5.0) years, female: 47.5%] were tested for 11 respiratory pathogens, including MP, ADV,IVA, IVB, RSV, CH, HCOV, HMPV, HRV, HBOV, and HPIV, with the positive detection rates of them were 3.8%,5.5%, 14.5%, 0.1%, 11.37%, 0.3%, 0.4%, 1.8%, 4.0%, 2.8%,and 3.6%, respectively.Among them, 172 (1.0%) patients were coinfected with two or more viruses.The most typical coinfection pattern was ADV-MP (n= 43, 25.0%), followed by IVA-IVB (n= 18, 10.5%), RSV-MP (n= 15, 8.7%), RSVHBOV (n= 13, 7.6%), and RSV-HPIV (n= 11, 6.4%).The majority of the positive detections of the above-mentioned 11 pathogens occurred in early and middle December, with a rising and then falling pattern.After December 20, there were fewer than 100 positive detections per day.Specifically,the positive detection of IAV was mostly from December 5 to 14; the positive detection of ADV and RSV was mostly concentrated in the early and middle of December; the positive detection of all viruses except MP showed a downward trend in the second half of the month; and the detection of MP was largely steady throughout the month (Fig.1 a).
In the study period, 755 patients [median (IQR) age: 5.0(7.2) years, female: 43.0%] were tested for other respiratory pathogens among positive SARS-CoV-2 patients, and 49 (6.5%) of them were positive for 11 respiratory pathogen nucleic acids or five respiratory pathogen antigen assays.These patients had a median age of 5 years (IQR: 7.2 years),and 43.0% of them were female.The positive rates for males(6.7%) and females (6.2%) were not significantly different(P= 0.860).Forty-six (93.9%) patients with coinfection had only two viruses, and three (6.1%) had three viruses(SARS-CoV-2, MP and ADV).The coinfection of SARSCoV-2 and RSV (n= 17, 34.7%) was the most common coinfection pattern, followed by MP (n= 11, 22.4%), IAV(n= 9, 18.4%), ADV (n= 8, 16.3%), MP-ADV (n= 3, 6.1%),and CH (n= 1, 2.0%).The specific coinfection patterns are shown in Fig.1 b.
According to age and gender, we matched the patients with SARS-CoV-2 coinfected with other viruses and the patients only infected with SARS-CoV-2 and compared their symptoms.The median (IQR) age of the 98 patients included was 4.5 (5.8) years, and 40.8% of them were female(Table 1).Patients who were coinfected were more likely to have cough (coinfected vs.non-coinfected: 85.7% vs.59.2%,P= 0.007), runny nose (46.9% vs.20.4%,P= 0.010), and less pharyngeal hyperemia (93.9% vs.77.6%,P= 0.043).No statistically significant difference was detected in the symptoms of thermal peak, expectation, sore throat, arthralgia or myalgia, and vomiting between the two patient groups.
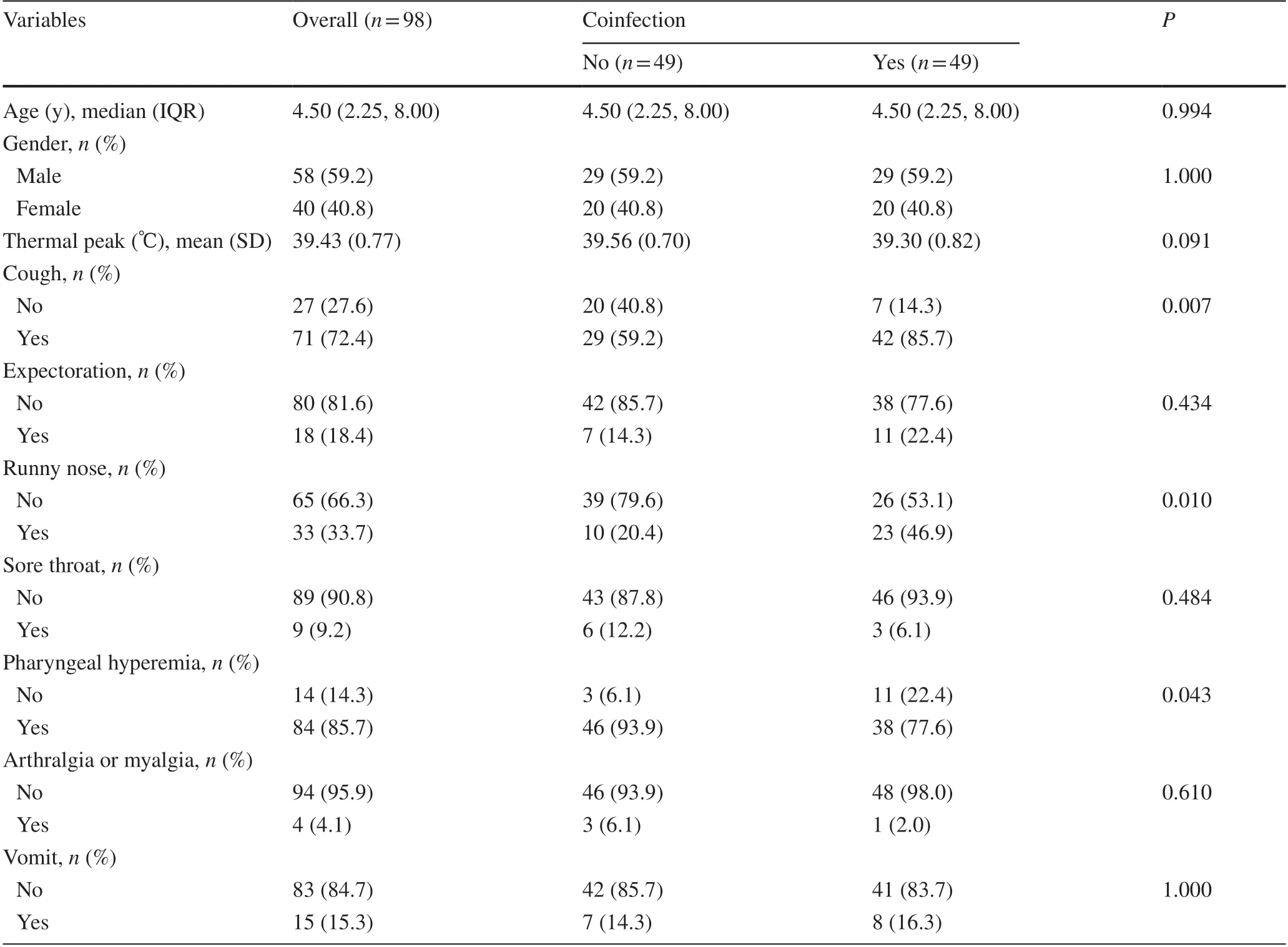
Table 1 Clinical characteristics of the children overall and by whether they were coinfected with SARS-CoV-2 and other respiratory pathogens
In this study, we retrospectively evaluated the overall prevalence of SARS-CoV-2 and other respiratory pathogens among children with acute respiratory infections during the Omicron epidemic.Omicron has demonstrated an increased transmissibility relative to Delta and immune escape capability [11, 12].This study found that 11 other respiratory pathogens showed a high positive number in the first half of the month and a sharp decline in the second half of the month, such as IAV, ADV, MP, and RSV.Previous studies have also revealed that the prevalence of SARS-CoV-2 affects the transmission of other respiratory pathogens.For example, public health interventions and continuous use of face masks reduced the spread of influenza virus [13, 14].
Currently, the Omicron variant has spread into more than 100 sublineages, including the already-dominant BA.1,BA.2, BA.2.12.1, BA.4, BA.5, BQ.1, XBB, and CH.1.1[15–18].Lu et al.analyzed 369 viral genomes from diagnosed COVID-19 patients and found that BA.5.2, BA.5.3,and BA.2.75 were the most common sublineages in the autumn and winter of 2022 [3].In our study, we found that the BF.7.14 and BA.5.2 sublineages were the main predominant SARS-CoV-2 Omicron variants.Perhaps, due to only ten children enrolled in our research for whole-genome sequencing, neither the XBB and CH.1.1 subtypes nor any other subtypes were detected.In a subsequent study, we intend to detect a greater sample size to conduct a survey of the epidemic subtype of SARS-CoV-2.
RSV and MP were the most commonly identified virus and bacterium in children infected with SARS-CoV-2 in the previous studies [19, 20], while we additionally determined the coinfection pattern between SARS-CoV-2 and other respiratory pathogens (viruses and mycoplasma).After being infected with SARS-CoV-2, the body's immunity is weakened, and its susceptibility to other respiratory viruses is increased, which may also be one of the reasons for coinfection.The rate of coinfection was 6.5%, and the most prevalent coinfection pattern in our research period was SARS-CoV-2 and RSV coinfection (n= 17, 34.7%).Similar to adults, coinfection with bacteria, fungi, and respiratory viruses has been described as a factor associated with more severe clinical outcomes in children with SARSCoV-2.Although all patients in our trial were outpatients,our results revealed that patients who were coinfected had higher rates of cough and runny nose and a lower rate of pharyngeal hyperemia, which indicated that children coinfected with SARS-CoV-2 and other respiratory pathogens may experience more serious respiratory symptoms in the outpatient department.The coinfection sample size was not large enough.In future studies, we plan to increase the number of samples to confirm the characteristics of coinfections between Omicron and other respiratory pathogens.
In conclusion, the primary epidemic strains among children in this study were BF7.14 and BA.5.2.During the same time span, infections with other respiratory pathogens decreased.Outpatient children coinfected with SARS-CoV-2 and other respiratory viruses showed more serious respiratory symptoms.Coinfection with SARS-CoV-2 is a phenomenon worthy of attention, and vaccination is the most important and effective means to prevent acute respiratory infectious diseases.
Supplementary InformationThe online version contains supplementary material available at https:// doi.org/ 10.1007/ s12519- 023- 00744-4.
AcknowledgementsThe authors acknowledge the financial support from all the funders.
Author contributionsLW, WBH, CBH, and SY contributed equally to this study.LW, MHY, SHQ, and SQ conceptualized and designed the clinical study.WBH, CBH, SLX, SY, LL, XWQ, and AAF carried out the data curation and formal analysis.LW wrote the manuscript with critical input from SQ.All authors approved the final version of the manuscript.
FundingThis study was funded by the Science and Technology Project in Zhejiang Province (LGC21H200004).
Data availabilityThe data that support the findings of this study are not openly available due to clinical data and are available from the corresponding author upon reasonable request.
Declarations
Ethical approvalThis study was approved by the Medical Ethics Committee of Children’s Hospital, Zhejiang University School of Medicine(No.2021-IRB-182).All of the reported investigations were performed in accordance with the principles of the Declaration of Helsinki as revised in 2000.
Conflict of interestNo financial or non-financial benefits have been received or will be received from any party related directly or indirectly to the subject of this article.Qiang Shu is the Chief Editor forWorld Journal of Pediatrics.The paper was handled by the other Editor and has undergone rigorous peer-review process.Qiang Shu was not involved in the journal’s review of, or decisions related to, this manuscript.The authors have no conflict of interest to declare.
 World Journal of Pediatrics2024年1期
World Journal of Pediatrics2024年1期
- World Journal of Pediatrics的其它文章
- Editors
- Information for Readers
- Instructions for Authors
- Current status of Mycoplasma pneumoniae infection in China
- PACS gene family-related neurological diseases: limited genotypes and diverse phenotypes
- Neighborhood predictors of short sleep duration and bedtime irregularity among children in the United States: results from the 2019–2020 National Survey of Children’s Health
