Automated screening of primary cell-based aptamers for targeting and therapy of pancreatic cancer
Zhukng Guo ,Bijing Jin ,Yile Fng ,Lin Jin ,Song Li ,Yn Deng ,Zhu Chen,Hui Chen,Yunying Zhng,Ri Usmn,Nongyue He,,*
a State Key Laboratory of Digital Medical Engineering,School of Biological Science and Medical Engineering,Southeast University,Nanjing 210096,China
b Hunan Key Laboratory of Biomedical Nanomaterials and Devices,Hunan University of Technology,Zhuzhou 412007,China
c Department of Molecular Biology,Jiangsu Cancer Hospital,Nanjing 210009,China
Keywords: Aptamer Cell-SELEX Automatic instrument Combination therapy Pancreatic cancer
ABSTRACT Although it has been developed for many years,nucleic acid aptamer screening technology still fails to be widely used,a considerable part of it is due to the variability of tumor cell morphology,which leads to the use of immortalized cell lines in the laboratory to screen nucleic acid aptamers for recognition ability of tumor cells in the diseased body.To address this,primary cells that can be stably passaged were isolated and extracted from spontaneous tumors of genetically engineered pancreatic ductal adenocarcinoma model mice in this study.Next,an automated screening instrument for nucleic acid aptamers developed autonomously by our group was used to perform efficient aptamer screening using a limited number of cells,and the obtained nucleic acid aptamers were affinity verified at the cellular level.Finally,to answer the question of the cell growth environment difference on the recognition ability of nucleic acid aptamers,we verified its targeting ability to tumors in vivo on a nude mice xenograft tumor model,and further used a common antitumor drug doxorubicin combined with nucleic acid aptamers to verify the drug loading ability of this aptamer combined with the targeting therapeutic ability.
Pancreatic cancer is one of the common malignant tumors of the digestive tract,and has a "king of cancer" designation in the field of tumor.The five-year survival rate after pancreatic cancer diagnosis is approximately 10%,making it one of the malignant tumors with the worst prognosis [1-3].The clinical symptoms of pancreatic cancer are insidious and atypical,and the diagnostic difficulty is extremely high,about 90% of which are ductal adenocarcinomas originating from the glandular duct epithelium [4,5].Its incidence and mortality rates have risen markedly in recent years.Pancreatic cancer is not diagnosed at an early stage,which also contributes to its high operative mortality rate,while the cure rate is low [6-8].Pancreatic ductal adenocarcinoma of murine origin has many characteristics like human pancreatic cancer,such as pancreatic endothelial cell neoplasia,strong immune reaction.Murine pancreatic ductal adenocarcinoma harbors KRAS and TP53 gene mutations,whereas in a study of human pancreatic adenocarcinoma,it was found that 80% and 70% of patients,respectively,expressed both mutant proteins [9-11].Therefore,this experiment chose to start from pancreatic cells of murine origin and explore the diagnosis and treatment of pancreatic cancer by targeting and treatment research [12-14].
With the development of biomaterial technology,several new targeting tools have been introduced into the oncology clinic,among them,aptamers as a new type of material with wide range applications have gradually attracted the attention of many researchers [15-20].One of these is nucleic acid aptamers that target cell surface proteins for recognition,the chemical nature of which is an oligonucleotide sequence with specific targeting effects [21-24].The technology to screen them is known as cell systematic evolution of organisms by exponential enrichment (Cell-SELEX).Cell-SELEX technology emerges from an oligonucleic acid library containing many different sequences through continuous screening and amplification,ultimately enabling the process of enrichment for high affinity nucleic acid sequences [25-28].The advantage of this technique is that it does not require specific information about the target before the screen is carried out,but rather directly obtains the fragment with the highest affinity for the cell by performing the screen on the whole cell.Thus,the obtained nucleic acid aptamers can be used not only as probes for tumor recognition but also for novel tumor marker discovery [29-32].Though aptamers for nucleic acids have some advantages comparing with antibody,such as lower cost,better batch to batch reproducibility,better thermal stability and stronger binding affinity,they also have problems such as excessively long screening process and high requirement of operator expertise for practical application [33-37].To this end,our group has independently designed an instrument that can automatically carry out the screening of nucleic acid aptamers,by which the 24-h uninterrupted work of the instrument greatly reduces the experimental flow,while circumventing the possible mistakes of manual operation [38-40].We complete the Cell-SELEX of primary pancreatic cells (for details,see Supporting information) and expand our experimental studies based on this instrument.
First,we verified the targeting ability of the nucleic acid aptamers.It is important to note that the KPC primary cells used in this experiment were derived from genetically engineered induced spontaneous pancreatic cancer mice.First,we modified the 5′ 6-FAM fluorescent group on the 5′ end of the nucleic acid aptamer and the random library,then incubated the modified aptamer and library with KPC cells and PSC cells respectively for 30 min and washing three times with binding buffer.The average fluorescence of the two cells was counted and compared using flow cytometry(Fig.1A).It can be seen from the figure that the binding ability of the nucleic acid aptamer obtained in this experiment to KPC cells is much stronger than its binding ability to PSC cells,which further indicates that this aptamer does have targeting ability to KPC cells.
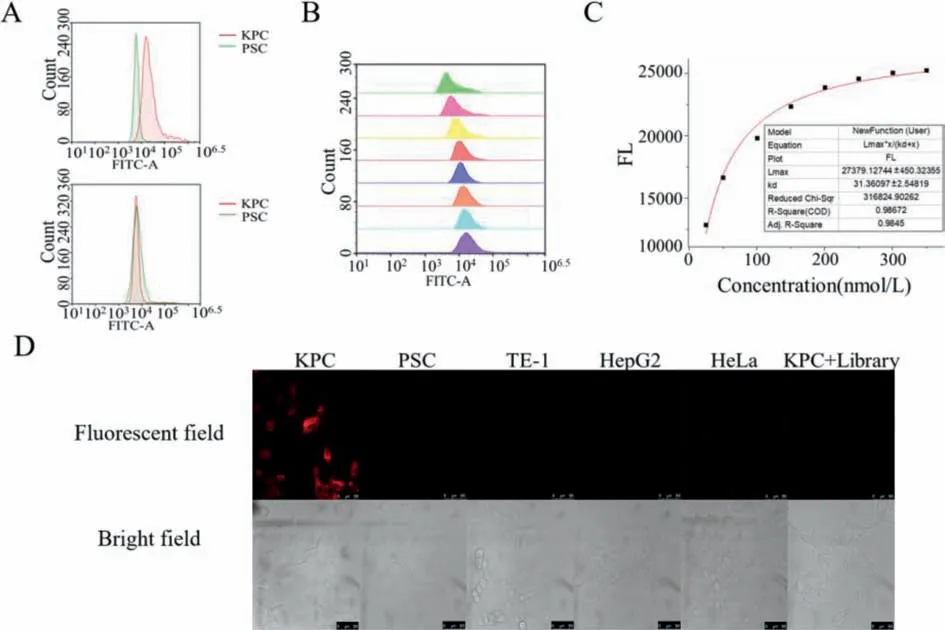
Fig.1.(A) Fluorescence intensity of KPC cells and PSC cells after incubated with aptamer (above) and random library (below).(B) Fluorescence intensity of different concentrations of aptamers with KPC cells.(C) Kd value of the modified aptamer with KPC cells.(D) Fluorescent microscopy images of aptamer incubated with different kind of cells and random library incubate with KPC cells.Scale bar=50 μm.
On this basis,we selected eight different concentrations of fluorescent modified nucleic acid aptamers at 25,50,100,150,200,250,300,and 350 nmol/L to bind KPC cells (Fig.1B).We then calculated theKdvalue of this nucleic acid aptamer with KPC cells,it can be concluded that theKdvalue is about 31.36 with theRsquare of 0.99 (Fig.1C),which proves that this aptamer has strong ability in targeting KPC cells.
Then,nucleic acid aptamer modified with TAMRA fluorescent group at the 5′ end were incubated with different kinds of cells,to avoid nonspecific binding of fluorophores to KPC cells,we also added a set of random libraries modified with TAMRA and incubated with KPC cells.After the incubation was completed,confocal microscope was used to image (Fig.1D).As can be seen in the figure,only the KPC cells incubated with the aptamer appeared fluorescent,so it can be deduced that this nucleic acid aptamer can specifically binds to KPC cells.
After obtaining suitable aptamers,we turned to answer the question of the influence of different cell growth environments on the binding ability of nucleic acid aptamer.We designed aninvivoimaging experiment in nude mice for verification.All animal welfare and experimental procedures involved in this article were reviewed and approved by the Animal Ethics Committee of Southeast University.The nucleic acid aptamer modified with Cy5.5 fluorescent group were resuspended using binding buffer and injected into the tail vein of KPC nude mice and 5-8F nude mice respectively.These two experimental nude mice were photographed with a small animal living body imager (Figs.2A and B).The photographing time was set at 0,5,15,30 min,and 1 and 2 h after injection.After theinvivoimaging,the nude mice were decapitated and dissected,and the stripped organs and tumor tissues were photographed (Fig.2C).It can be seen from the following figure that after treatment by nucleic acid aptamer the KPC nude mice showed significant fluorescence enrichment at the tumor site and lasted for a long time.In addition,the liver and kidney of the two nude mice both showed different levels of fluorescence intensity.However,the fluorescence intensity in tumor tissues of KPC nude mice was obviously higher than that of 5-8F nude mice.This illustrates that our screen obtained aptamers can achieve targeting and enrichment to KPC tumor tissues in nude mice.Therefore,it can be confirmed that the nucleic acid aptamer obtained by primary cells through automatic instrument screening in the experiment also can target the tumor tissue of cell derived xenograft(CDX) model in nude mice.

Fig.2.(A) Distribution of fluorescently labeled aptamers in KPC nude mice over time.(B) Distribution of fluorescently labeled aptamers in 5-8F nude mice over time.(C)Images of anatomical organs and isolated tumor tissues from the nude mice in the groups shown in A,B.
After theinvivotargeting experiment was completed,we turned to exploring the possibility of nucleic acid aptamers for drug loading therapy.To achieve this,we validated the ability of nucleic acid aptamer for combination therapy at cellular level.We treated the KPC cells with doxorubicin hydrochloride (DOX) at concentrations of 0.2,0.4,1 and 2 μg/mL,then observed the results using light microscopy (Fig.3A).KPC cells underwent various degrees of cell death following treatment with various concentrations of DOX,among them,essentially all cells in the 0.2 μg/mL group died after treatment.
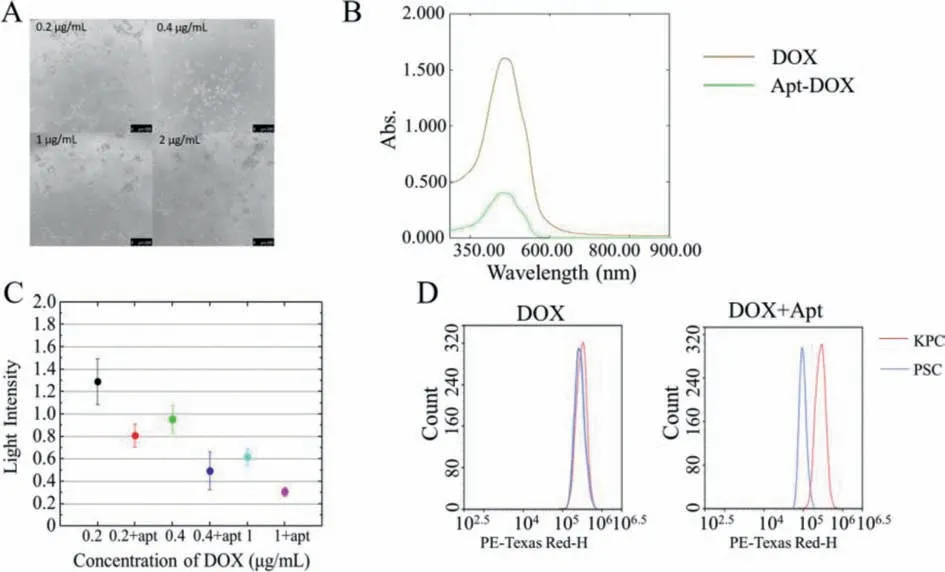
Fig.3.(A) Effect of doxorubicin hydrochloride administration on KPC cells.Scale bar=250 μm.(B) Changing characteristic peaks before and after doxorubicin hydrochloride mixed with aptamer.(C) Absorbance of cells after doxorubicin hydrochloride/doxorubicin hydrochloride+aptamer treatment.The mean ± standard deviation (SD)was calculated for each group of samples (n=8),and a T-test was performed for group design materials.(D) Absorbance of doxorubicin hydrochloride and doxorubicin hydrochloride-aptamer binding to KPC cells and PSC cells.
The loading of DOX into the aptamer was performed through simple mixing at room temperature (Fig.3B).As an anthracycline of drug,DOX have fluorescence properties that become quenched after intercalation to DNA.The decrease in absorbed light in turn demonstrates the occurrence of binding between DOX and the aptamer.We used aptamer and binding buffer to dilute doxorubicin hydrochloride respectively and control the final concentration of the drug to 0.2,0.4 and 1 μg/mL.After incubating them with KPC cells,the absorbance at 570 nm was measured using a microplate reader (Fig.3C).Pearson’s correlation coefficient value between the administered concentration and light intensity was -0.823,and it presented a level of significance ofP<0.01,indicating a significant negative correlation between the administered concentration and light intensity,which in turn confirmed that the administered concentration was significantly positively correlated with KPC cell killing efficacy.In the group with a DOX concentration of 0.2 μg/mL,the mean relative fluorescence intensity was 0.81 in the experimental group with added aptamer and 1.29 in the control group without aptamer,the introduction of aptamer reduced the survival of KPC cells.Similarly,the corresponding data in the remaining two groups were 0.49/0.95 and 0.31/0.62.By analyzing the results of independent samplesT-test for each of the six experimental groups with different administration amounts,each corresponding sample presented a significant difference (P<0.001) on light intensities.It can be concluded that the addition of the nucleic acid aptamer significantly enhanced the cytotoxicity of DOX in KPC cells.
Next,we combined DOX with KPC cells and PSC cells,using aptamer as a variable and observed fluorescence using flow cytometry (Fig.3D).From the fluorescence intensity data,it can be seen that the binding effect of DOX on KPC cells and PSC cells was not selective before the introduction of the aptamer,and the two groups of cells presented same binding ability.However,with the introduction of the aptamer,DOX presented a specific recognition ability for KPC cells,and the fluorescence intensity of the KPC cells group was significantly higher than that of the PSC cells.From this we can conclude that aptamers can indeed enhance the targeted killing effect of doxorubicin hydrochloride on KPC cells.
Finally,we chose to unfold the validation of the therapeutic ability of aptinvivo.To verify whether the aptamer obtained due to the screening of primary cells can be used for the actual treatment of tumorsinvivo,we performed nude mice xenograft tumor experiments with corresponding treatment experiments.First,15 female nude mice weighing between 19 g and 22 g were selected for KPC cell xenograft experiments.When tumors grew to 25-50 mm3,randomly divided the nude mice into three groups with five mice in each group,the three groups were respectively set as: aptamer-DOX (Apt-DOX) group,DOX group and negative control group.
The nude mice were observed and recorded daily during the administration process,nude mice in the DOX group continued to lose weight with the administration,exhibited poor activity status and comparatively slow tumor growth.The nude mice in the Apt-DOX group showed better growth status,increased body weight,and particularly slow tumor growth.The growth status of the nude mice in the negative control group was general,the body weight went down,and the tumor grew rapidly.At the end of administration,all nude mice were uniformly sacrificed by decapitation after gas anesthesia using isoflurane,the tumor tissues were stripped off,and the tumor tissues of all 15 nude mice were collected and sequentially photographed (Fig.4A),from top to bottom three lines,which were 1-5 negative control group,6-10 DOX group,11-15 Apt-DOX group.Throughout the treatment,we simultaneously recorded the body weight change of each nude mouse to characterize their health degree (Fig.4B).
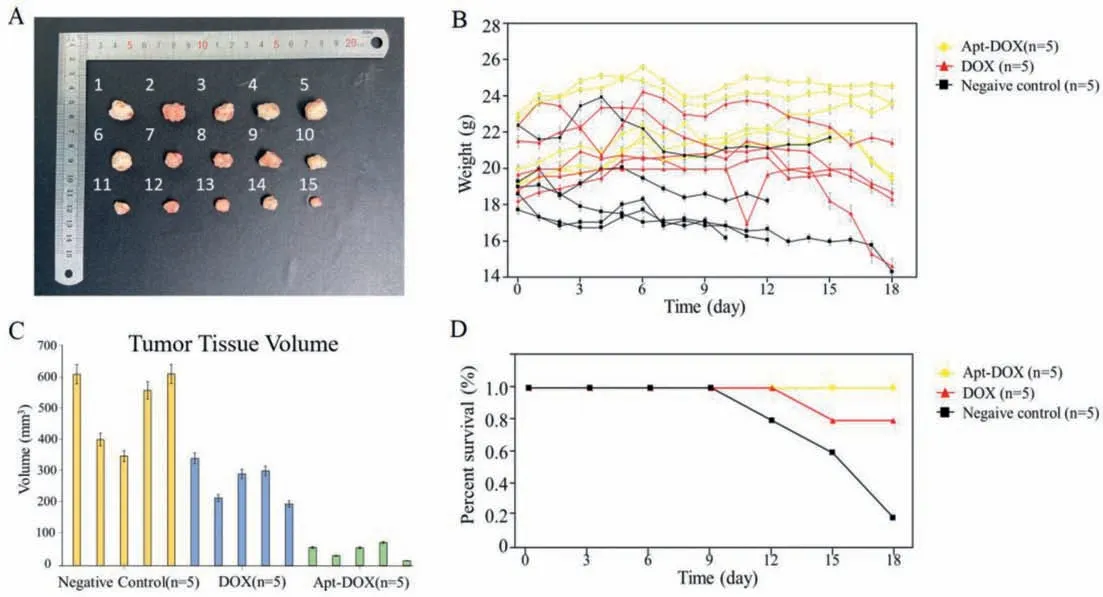
Fig.4.(A) Stripped tumor tissues of the nude mice after treatment,1-5: Negative control group;6-10: DOX group;11-15: Apt-DOX group.(B) Body weight of the mice among three groups during treatment.(C) Volume of the stripped tumor tissues of the mice among three groups.The mean ± SD was calculated for each group of samples(n=5).(D) Survival curves of the mice among three groups during treatment.
After the photographs were finished,the whole tumor tissues were measured and the tumor tissue volume distribution plots among the 3 groups were drawn,and the tumor growth inhibition rate was calculated (Fig.4C).The average tumor volume was 508.1 mm3in the negative control group,270.6 mm3in the DOX group and 47.1 mm3in the Apt-DOX group.The tumor growth inhibition rate of the Apt-DOX group is 90.7% and the DOX group is 46.7%.
From these,it can be concluded that comparing with other groups,the nude mice treated with the aptamer doxorubicin conjugate had a longer survival cycle as well as better survival status(Fig.4D),only nude mice from the Apt-DOX group showed a 100%survival ratio at the end of the treatment.With the introduction of the aptamer,the tumor growth of the nude mice were significantly inhibited and the targeted therapy was highly effective.In turn,it can be stated that the nucleic acid aptamers obtained in this experiment from the primary tumor tissues through the primary cell extraction culture screening process,still have a strong targeting effect on the transplanted tumor tissues grown in different environments in nude mice,possessing the potential forinvivotumor targeted therapy.
In this article,we started from pancreatic cancer primary cells and completed the screening of nucleic acid aptamer using an automated nucleic acid aptamer screening instrument.The specific recognition ability and cytotoxicity verification at the cellular level and tumor level were performed using this aptamer.We then focus on answering the biggest problems encountered in the practical application of Cell-SELEX.Most of the current aptamer screening is based on laboratory passage cells.However,different growth environments in primary cells,laboratory passage cells and tumor cellsinvivowill lead to differences in cell phenotypes.And this difference will produce uncertain interference to the recognition effect of nucleic acid aptamer on cells,in turn,led to the unconvincing ability of nucleic acid aptamer to recognize tumorsinvivo.
Currently,one of the avenues to tackle this problem is theinsitutissue slide based SELEX strategy.The advantage of this approach is that the obtained aptamers have excellent clinical adaptation,but deficiencies remain in the difficulty of target acquisition and high-throughput screening.In response to this challenge,we continued our subsequent experiments.We used primary cells to complete the nucleic acid aptamer screening.Then we used laboratory passaged cells to verify the specific targeting ability and cytotoxicity of the Aptamer-Doxorubicin hydrochloride.Finally,we used nude mice CDX model forinvivotargeting and therapeutic ability verification.
From the results,we have fully validated the recognition ability of the nucleic acid aptamer obtained by primary cells screening against cells of various growth environments,provided new possibilities for practical applications of Cell-SELEX technology.From the results,we fully verified the recognition ability of nucleic acid aptamers obtained from primary cell screening against various growth environment cells.Next,one of our ideas is to continue intensify the research on the automated screening instrument for nucleic acid aptamers,allowing it to adapt to more diverse targets,combining various new SELEX technologies such as thein-situtissue slide based SELEX mentioned above,trying to provide more new ideas for the practical application of Cell-SELEX progress.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by the National Key Research and Development Program of China (Nos.2017YFA0205301 and 2018YFC1602905),National Natural Science Foundation of China (Nos.62071119,62075098,81902153,61527806),and the Open Funding of State Key Laboratory of Oral Diseases (No.SKLOD2022OF05).
Supplementary materials
Supplementary material associated with this article can be found,in the online version,at doi:10.1016/j.cclet.2023.108528.
 Chinese Chemical Letters2024年2期
Chinese Chemical Letters2024年2期
- Chinese Chemical Letters的其它文章
- The 3rd Xihua Chemistry and Biomedicine Forum
- Professor Hualiang Jiang: A tribute to an esteemed visionary chemist and pharmacist
- Recent advances in visible light-mediated chemical transformations of enaminones
- Development of porphyrin-based fluorescent sensors and sensor arrays for saccharide recognition
- Recent advances of versatile reagents as controllable building blocks in organic synthesis
- Synthetic host-guest pairs as novel bioorthogonal tools for pre-targeting☆
