Leaf Morphology Genes SRL1 and RENL1 Co-Regulate Cellulose Synthesis and Affect Rice Drought Tolerance
Liu Dan#Zhao Huibo#Wang Zi’an,Xu Jing,Liu YitingWang Jiajia,Chen MinminLiu XiongZhang Zhihai,Cen JiangsuZhuLiHu JiangRen DeyongGao ZhenyuDong GuojunZhang QiangShen LanLi Qing,Qian QianHu SongpingZhang Guangheng
Research Paper
Leaf Morphology GenesandCo-Regulate Cellulose Synthesis and Affect Rice Drought Tolerance
Liu Dan1,2, #, Zhao Huibo1,3, #, Wang Zi’an1, 2,Xu Jing1,Liu Yiting1,2, Wang Jiajia1,Chen Minmin1, Liu Xiong1, Zhang Zhihai1,Cen Jiangsu1, ZhuLi1, 3, Hu Jiang1, Ren Deyong1, Gao Zhenyu1, Dong Guojun1, Zhang Qiang1, Shen Lan1, Li Qing1,Qian Qian1, 3, Hu Songping2, Zhang Guangheng1,3
(; Qian Qian Academician Workstation, National Nanfan Research Institute (Sanya), Chinese Academy of Agricultural Sciences, Sanya 572024, China; These authors contributed equally to this work)
The morphological development of rice (L) leaves is closely related to plant architecture, physiological activities, and resistance. However, it is unclear whether there is a co-regulatory relationship between the morphological development of leaves and adaptation to drought environment. In this study, a drought-sensitive, roll-enhanced, and narrow-leaf mutant () was induced from a semi-rolled leaf mutant () by ethyl methane sulfonate (EMS), which was obtained from Nipponbare (NPB) through EMS. Map-based cloning and functional validation showed thatencodes a cellulose synthase, allelic to/. Themutation resulted in reduced vascular bundles, vesicular cells, cellulose, and hemicellulose contents in cell walls, diminishing the water-holding capacity of leaves. In addition, the root system of themutant was poorly developed and its ability to scavenge reactive oxygen species (ROS) was decreased, leading to an increase in ROS after drought stress. Meanwhile, genetic results showed thatandsynergistically regulated cell wall components. Our results revealed a theoretical basis for further elucidating the molecular regulation mechanism of cellulose on rice drought tolerance, and provided a new genetic resource for enhancing the synergistic regulation network of plant type and stress resistance, thereby realizing simultaneous improvement of multiple traits in rice.
cellulose; cell wall; drought tolerance; leaf morphology; rice
As the main plant organ involved in capturing light energy, exchanging gases, and synthesizing organic matter through photosynthesis in rice (Braybrook and Kuhlemeier, 2010), the morphology of leaves has a close relationship with plant drought resistance. It is also one of the important agronomic traits in rice architecture. Ideal leaf morphology can reduce water loss and intercept solar radiation by the canopy. Therefore, it is significant to search for the ideal leaf morphology of rice to enhance its adaptability to environmental stress (Kadioglu and Terzi, 2007).
Cellulose plays a crucial role in plant morphology and root development. Plant cell walls mainly consist of cellulose, hemicellulose, and pectin, forming intricate networks within cells. Cellulose microfibers create a cellulose-hemicellulose network through hemicellulose chains and embed themselves in the pectin matrix (Carpita and Gibeaut, 1993). The cell wall acts as a barrier between the protoplasts of rice tissue cells and the environment. Besides maintaining cell morphology, protecting protoplasts, and enhancing tissue mechanical strength, it is closely related to water metabolism, transpiration, nutrient transport, and hormone secretion, playing a crucial role in effective resistance to biotic and abiotic stresses in rice. Thegene affects water transport in rice leaves by influencing the components of the secondary cell wall (Fang et al, 2012). Cellulose is one of the main components of plant cell walls, and its content directly affects the structure of plant cell walls and the development of plant tissue morphology. The CESA and CSL gene families play key functions in cellulose synthesis, responsible for synthesizing cellulose and the β-1,4-glycosidic bond of most hemicelluloses, respectively (Lerouxel et al, 2006). CSL gene family includes six gene families, CSLA, CSLB, CSLC, CSLD, CSLE, and CSLG. Among them, the CSLD gene families are highly homologous with CESA (Richmond and Somerville, 2000). However, the root system of the plant affects water and inorganic salt absorption, and is closely related to plant drought tolerance. Somemutants display serious phenotypic defects. For instance,has been cloned in rice, and its loss-of-function may lead to abnormal root hair development (Kim et al, 2007).primarily affects the cell wall of the root hair apex, and the loss of this gene can result in root hair defects (Wang et al, 2001). The mutanthas deformed root hairs (Bernal et al, 2008). Inknockout plants, both the stem and root are significantly reduced (Bernal et al, 2007). Recent studies have shown that cell walls also play an important role in plant responses to abiotic stress. Chen et al (2005) isolated an allele of,, whose mutant shows strong tolerance to drought, salt, and mannitol osmotic stresses. Zhu J H et al (2010) reported a mutant of, which encodes a cellulase-like synthase, AtCSLD5. The loss-of-function of this gene causes subtle defects in the cell walls, leading to plants accumulating high levels of reactive oxygen species (ROS) under stress, greatly increasing their sensitivity to drought and osmotic stresses.
Stomatal and cuticular waxy layers are the main tissues in response to drought stress in leaves. Under drought stress, leaf stomatal conductance and pore size decrease (Bertolino et al, 2019), the accumulation of the waxy layer in the leaf epidermis increases (Jenks et al, 2001), and photosynthetic capacity diminishes. These behaviors are often regulated by various genes, such as, which reduces rice drought tolerance by affecting leaf epidermis development and stomatal closure (Huang et al, 2019). The overexpression ofandcan decrease stomatal density in leaves and enhance the drought tolerance of rice (Yoo et al, 2010; Li X M et al, 2017). Meanwhile, the overexpression ofcan promote stomatal closure and increase drought resistance of rice (Dey et al, 2016). Bothandregulate drought resistance of rice by affecting drought-induced wax accumulation in the leaf cuticle (Zhu and Xiong, 2013, Xue et al, 2017). The transformation ofinto rice can improve the photosynthetic capacity and drought resistance of rice leaves (Karaba et al, 2007).
In recent years, an increasing number of genes related to leaf morphology and drought resistance have been identified and cloned in rice. However, it is not clear whether there is a co-regulatory relationship between leaf morphology development and adaptation to a drought environment. In this study, a new short-stalked and narrow-leaf-rolling rice mutant,(), was obtained through ethyl methane sulfonate (EMS) mutagenesis of, a previously cloned half-leaf-rolling gene. Based on these results, map-based cloning, functional analysis, and drought tolerance evaluation of the target genewere carried out to preliminarily clarify the roles ofandin the coordinated regulation of leaf morphogenesis and drought resistance in rice. The results provided a theoretical basis for further improving the molecular mechanism of droughttolerance, germplasm innovation, and molecular design breeding of drought-resistant rice.
RESULTS
Phenotypic characterization of mutant renl1
We used EMS to induce mutagenesis in the semi-rolled leaf mutant, and successfully identified a short-stalked and narrow-rolled leaf mutant,. To elucidate the molecular mechanism that determines rice roll-enhanced and narrow leaf formation, we conducted a comprehensive characterization of. At the seedling stage, themutant exhibited a significantly reduced plant height and a narrower, more curled, and shorter leaf phenotype compared with, (Fig. 1-A to -D). Specifically, the plant height ofwas 62.1 cm, which was 26.8% and 23.7% lower than that ofthe wild type Nipponbare (NPB) and, respectively (Fig. 1-E). The flag leaf length was 22.5 cm, shortened by 13.7 and 13.4 cm relative to NPB and, respectively (Fig. 1-F). The flag leaf width was 0.76 cm, decreasing by 0.49 and 0.56 cm relative to NPB and, respectively (Fig. 1-G). The measurement of the flag leaf crimp indicated that the mutantsandhad significantly higher flag leaf crimp than NPB, reaching 35.6% and 39.2%, respectively (Fig. 1-H). Simultaneously, the analysis of chlorophyll a and chlorophyll b contents in the leaves revealed a significant reduction in(Fig. 1-I). These results indicated thatseriously affected the development of leaf and plant type. Additionally, we observed that the loss-of-function ofhad a significant impact on the panicle and grain traits of rice, notably affecting rice yield (Fig. S1).

Fig. 1. Phenotypic characterization of Nipponbare (NPB) and mutants(and).
A–D, Phenotypes of tillering plant (A, scale bars are 10 cm), local flag leaf (B, scale bar is 1 cm), blade cross section (C, scale bar is 1 cm), and flag leaf morphology (D, scale bar is 10 cm).
E–I, Comparisons of plant height (E), flag leaf length (F), flag leaf width (G), flag leaf crimp (H), and chlorophyll content (I) between wild type (NPB) and mutants (and). Data are Mean ± SD (= 3), and the lowercase letters above the bars represent the significant differences at the 0.05 level by one-way analysis of variance.
RENL1 affects histological structure of leaves
To investigate the formation mechanism of narrow and rolled leaves in, we examined the cross sections of flag leaves using stereomicroscopy and frozen sectioning methods. The results demonstrated that the mutanthad significantly more bulliform cells (BCs) than NPB (Fig. 2-A and -E). In, however, the number of BCs decreased significantly, and their size and shape were highly irregular (Fig. 2-B). The average number of large and small vascular bundles inwas 6.5 and 27.3, respectively, which were significantly lower than those in NPB and(Fig. 2-C and -D).had 3.3 BCs less than NPB and 6.7 BCs less than. The numbers of BCs in NPB andwere apparently higher than those in(Fig. 2-E). Previous studies have shown that leaf curling inis due to an increase in the amount of adaxial BCs (Xiang et al, 2012), which aligns with our observations. In summary, the narrowing and curling of’s leaves are due to a reduction in the formation of BCs and vascular bundles.
RENL1 affects rice root development
The root system is the main organ of a plant for absorbing water and mineral nutrition from the external environment, playing a crucial role in coping with water stress (Ghosh and Xu, 2014). Measurements of the rice root system under drought stress showed that the development of the root system, including its diameter and depth, is positively correlated with the plant’s ability to acquire water (Yoshida and Hasegawa, 1982). Statistical analysis of root traits in 5-day-old rice seedlings showed that the length of main roots and the number of roots in theandmutants were significantly lower than those in NPB, and the length of main roots and the number of roots inwere significantly lower than those in(Fig. 3-Ato -C). On the other hand, the main root diameters inwere not significantly different from those in NPB, but tended to decrease, while the main root diameters inwere significantly lower than those in NPB (Fig. 3-D). According to the results, the total root length ofand NPB was not significantly different, whereas the total root length inwas significantly lower than that in NPB (Fig. 3-E). The total root surface area directly affects the water absorption capacity of the roots. Our results showed that the total root surface area of NPB andwere not significantly different, while that ofdecreased by 112.9 and 84.5 mm2compared with NPB and, respectively (Fig. 3-F). These results suggested thatmay affect the development of rice roots, which can reduce the water absorption capacity of rice plants.
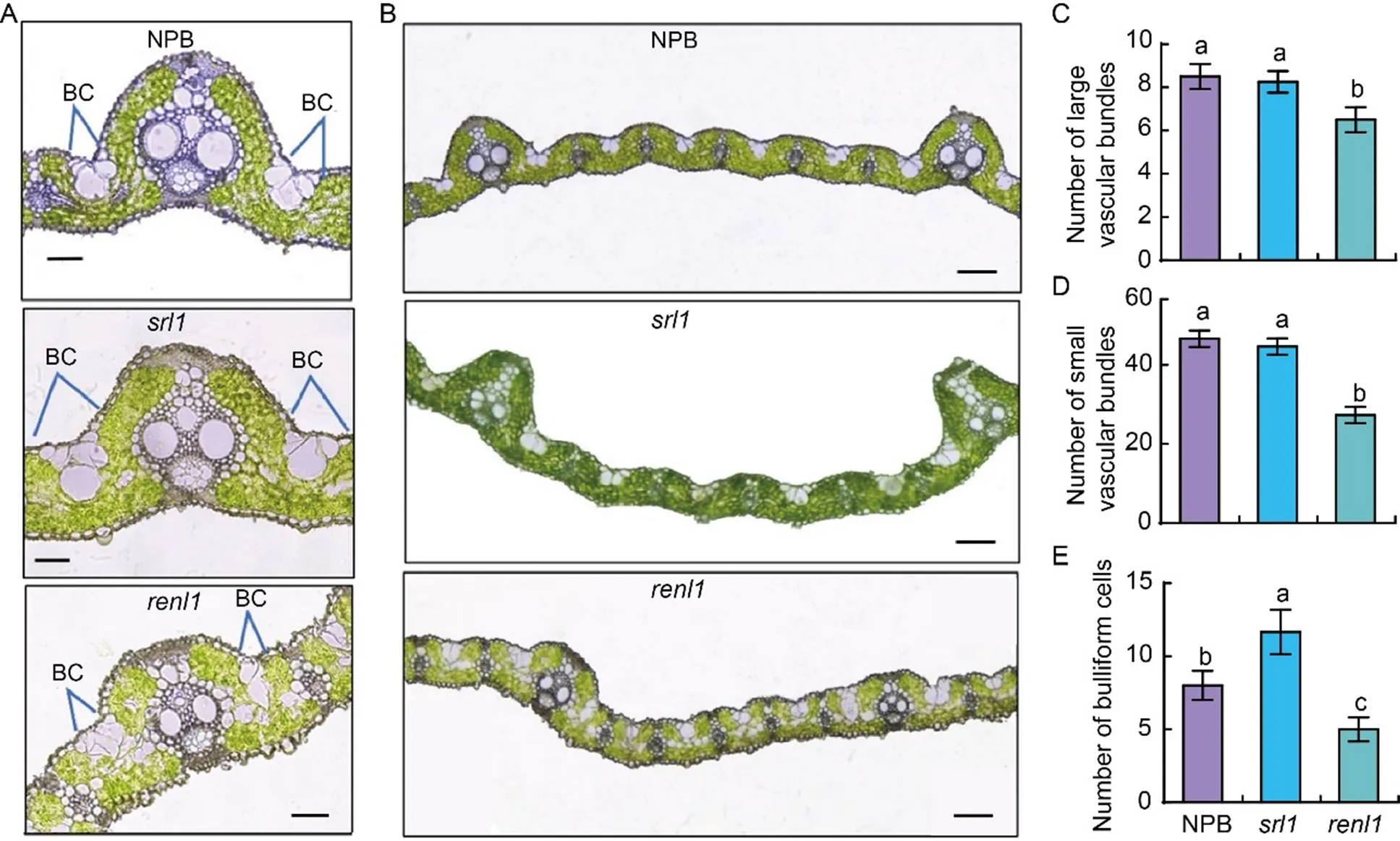
Fig. 2. Leaf cytological morphology analysis.
A and B, Frozen sections of main veins (A, scale bars are 50μm), and large and small veins of Nipponbare (NPB),, and(B, scale bars are 100μm). BC, Bulliform form.
C–E, Numbers of large (C) and small (D) vascular bundles, and bulliform cells (E). Data are Mean ± SD (= 3). The lowercase letters above the bars represent the significant differences at the 0.05 level by one-way analysis of variance.
Map-based cloning of RENL1
We used leaf width as the primary phenotype to map the genes involved in the regulation of narrow rolled leaves in the mutant. We crossedwith a semi- rolled leaf mutantand identified the phenotype of the F1hybrids. The segregation ratio of the F2generation was based on the difference in leaf width. The gene was mapped to a physical distance of 12.1 kb on chromosome 12 between markers C-2 and C-7 through map-based cloning (Fig. 4-A). Within this interval, there are five open reading frames (ORF). The full length of the interval was sequenced and compared with the genome of wild type to confirm the gene as, with a single-base substitution from G to A occurring at position 1775, causing the amino acid changed from serine to l-phenylalanine (Fig. 4-B). The mutation ofmay be responsible for the narrow leaf and dwarf phenotype of
To confirm thatis, we disruptedinusing CRISPR-Cas9 genome editing technology and analyzed the resulting phenotypes (Fig. 4-C). The knockout lines (_cas9)exhibited a narrow leaf and dwarf phenotype similar to(Fig. 4-D to -F). The results indicated thatis the target gene involved in regulating leaf width and leaf rolling in,and it is also an allele of.
RENL1 affects development of leaf epidermal structure and stomata
encodes a cellulase-like protein of the glycosyltransferase family, and this protein plays a significant role in the development of the plant cell walls.encodes a glycosylphosphatidylinositol- anchored protein, which anchors to the outer surface of the plasma membrane. Previous studies of theallelehave speculated that it functions primarily in extracellular environments such as the cell walls(Li W Q et al, 2017). Because the components of the plant cell walls have specific autofluorescent properties, we can observe autofluorescence in plant leaf cells under a fluorescence microscope. Through this phenomenon, we can clearly distinguish the size and structure of rice leaf tissue. We sectioned the test materials by hand and observed it under a fluorescence microscope. Fig. 5-A showed the leaf tissue structure under blue fluorescence with a light-emitting diode(LED) wavelength of 470 nm. Compared with NPB, the percentage of mesophyll cells inanddecreased by 5.3% and 14.4%, respectively. The percentage of mesophyll cells inalso decreased significantly compared with that inand, reaching 12.1% and 9.1%, respectively. The leaf structure was observed under green fluorescence with an LED wavelength of 530 nm (Fig. 5-B), and the green highlighted portion indicated by arrows was the mechanical tissue of the leaf and the thick-walled tissue. In, the green light intensity indicated by the arrow was significantly weaker than that in the NPB and. In, however, the mechanical tissue emitted a weaker green light, suggesting that the mechanical tissue and the thick-walled tissue of theandhad serious defects. These observations suggested that the loss-of-function ofmay lead to severe damage to leaves and abnormal stomatal development.

Fig. 3.affects rice root development.
A, Phenotypes of seedling root. Scale bar is 2 cm.
B–F, Length of main root (B), the number of roots per plant (C), diameter of main root (D), total root length per plant (E), and total root surface area per plant (F) in Nipponbare (NPB) and mutants (and). Data are Mean ± SD (= 3). The lowercase letters above the bars represent the significant differences at the 0.05 level by one-way analysis of variance.
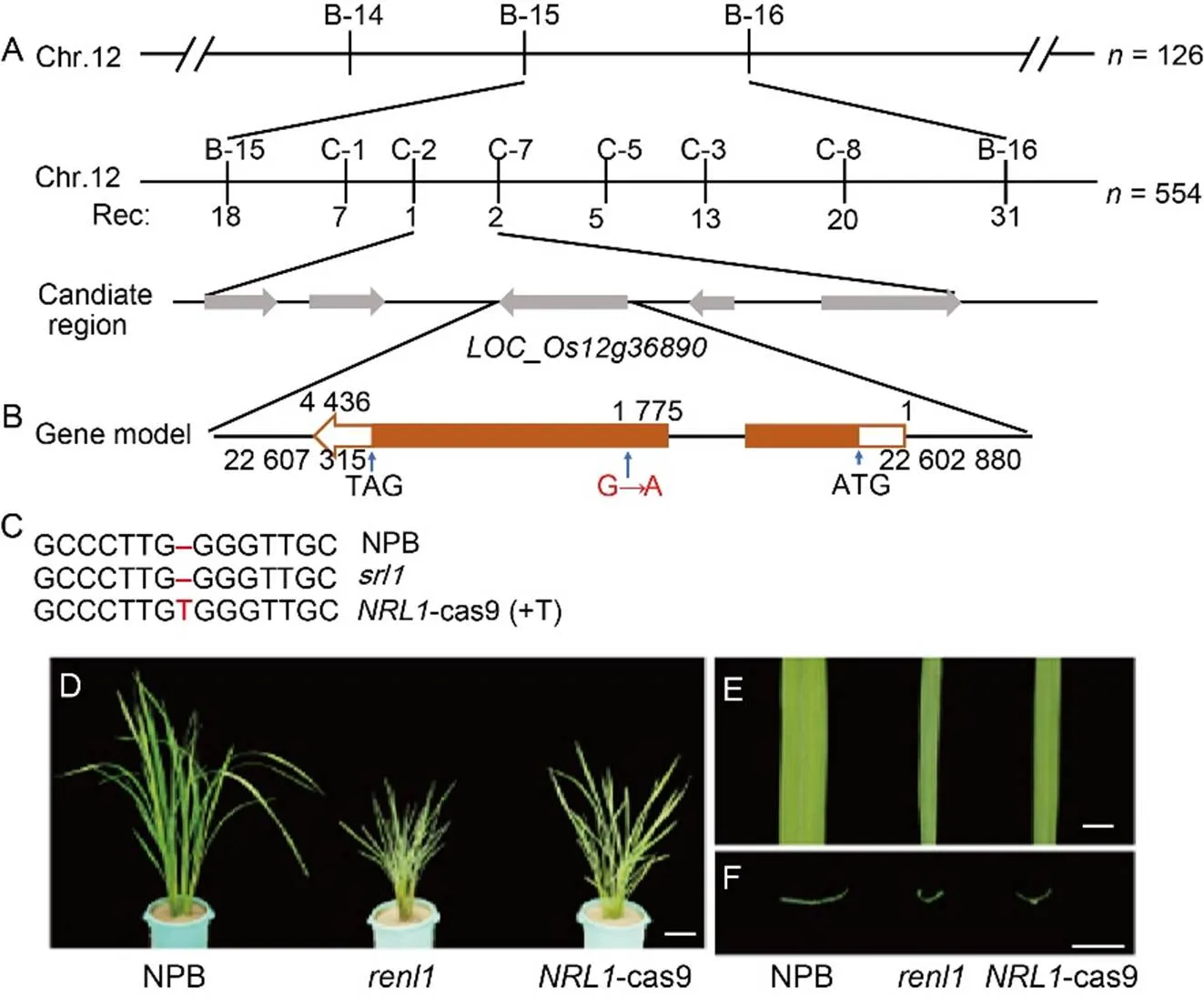
Fig. 4. Map-based cloning of.
A,gene was mapped to an interval between molecular markers B-15 and B-16 on chromosome (Chr.) 12 and further delimited to a 12.7 kb genomic region between markers C-2 and C-7. The numbers below the markers indicate the number of recombinants (Rec).
B, Structure ofgene, a G→A base substitution occurred at position 1 775 of the open reading fragment.
C, Gene knockout site detection. ‘T’ in red represents the site knocked by CRISPR/Cas9.
D, Phenotypes of Nipponbare (NPB),, and-cas9 at the tillering stage. Scale bar is 10 cm.
E, Flag leaf type of NPB,, and-cas9. Scale bar is 1 cm.
F, Cross section of flag leaves of NPB,, and-cas9. Scale bar is 1 cm.
In order to better understand the changes of mutant leaves, we examined the flag leaves, particularly the abaxial sides of rice leaves, in mutants,, and, andNPB using a scanning electron microscope. Clearly, the arrangement of silicon cells in NPB was smooth and compact, while in the three mutants, that along the longitudinal vein was more sparse and irregular (Fig. 5-C). Additionally, the papillary structures and vesicular processes on the wild type NPB leaf epidermis were regular and well- organized. However, in all the three mutants, these papillary structures exhibited morphological changes and were disordered (Fig. 5-D). Inand, most of the papillary structures were developmentally defective, withshowing more severe changes. Moreover, we observed a significant difference in stomatal morphology between the wild type NPB and the mutants (Fig. 5-E). In NPB, claw-like protrusions around the stomata formed clusters, with their tips pointing towards the center of the stomata. The proximity of these protrusions influences stomatal opening (Shen et al, 2005). However, in the other three mutants, the claw-like structures did not develop properly. Stomatal width measurements revealed a significant reduction in stomatal opening in the three mutants compared with NPB(Fig. 5-F). The stomatal width ofwas notably smaller than those ofand(Fig. 5-F). However, there were no significant differences in stomatal length among the four plant materials (Fig. 5-G). Furthermore, we assessed the stomatal density in the three mutants by applying nail polish to the leaves at the tillering stage under a microscope. The results showed that the stomatal density in all the three mutants was lower than that in NPB, withhaving a significantly lower density than(Fig. 5-H).
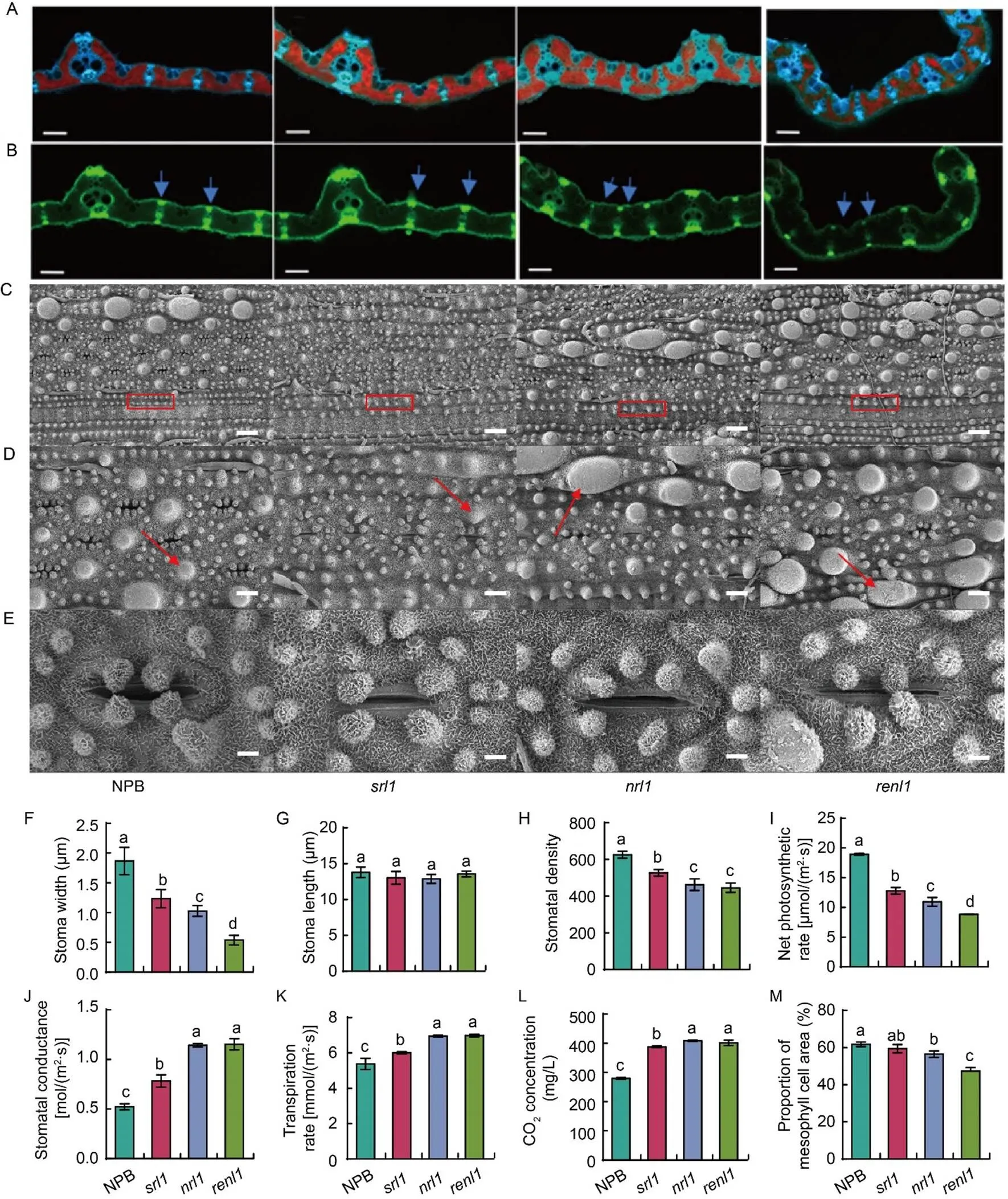
Fig. 5. Morphology, physiology, and scanning electron microscope observation of rice leaves in wild type (Nipponbare, NPB) and mutants (,, and).
A and B, Flag leaf fluorescence sectionunder blue fluorescence with a light-emitting diode (LED) wavelength of 470 nm (A) andunder green fluorescence with an LED wavelength of 530 nm (B). Scale bars are 100 μm.
C–E, Scanning electron microscope observation of abaxial epidermis. Cork-silica cell pairs are represented by red boxes (C), papillae and nodular papillae are indicated with arrows (D), and stomata on the abaxial epidermis (E). Bars in C, D, and E are 20 μm, 10 μm, and 2 μm, respectively.
F–M, Blade stomatal width (F), blade stomatal length (G), leaf epidermal stomatal density (H), net photosynthetic rate (I), stomatal conductance (J), transpiration rate (K), intercellular CO2concentration (L), and proportion of mesophyll cells to leaf cells (M) in wild type and mutants. Data are Mean ± SD (= 3). The lowercase letters above the bars represent the significant differences at the 0.05 level by one-way analysis of variance.
Leaves serve as the primary sites for photosynthesis and gas exchange with the external environment. Damage to the leaves can affect rice plant photosynthesis and water retention. Results from photosynthetic rate measurements and related parameters indicated that the net photosynthetic rate of the three mutants was significantly lower than that of NPB (Fig. 5-I), while stomatal conductance (Fig. 5-J), transpiration rate (Fig. 5-K), and intercellular CO2concentration (Fig. 5-L) were significantly higher in the mutants compared with the wild type. Notably,exhibited a significantly lower net photosynthetic rate thanand, with higher stomatal conductance, transpiration rate, and intercellular CO2concentration compared with(Fig. 5-I to -L). These results suggested thatandhad significant effects on leaf photosynthetic rate, stomatal conductance, transpiration rate, and intercellular CO2concentration. Additionally, the decreased photosynthetic rate inandmay be associated with a significant reduction in mesophyll cells (Fig. 5-M).
Effects of RENL1 and SRL1 on cell wall development
The development of the cell walls may have been altered, as evidenced by observations using a fluorescence microscope and a scanning electron microscopy. We subsequently measured the contents of cell wall-related components and found that the cellulose contents in the leaves of,, andwere significantly lower than that in NPB (Fig. 6-A). Additionally, the leaf hemicellulose contents inandwere significantly lower than that in NPB (Fig. 6-B). We further examined the relative expression levels of rice tillering-stage-related cellulose synthase genes (OsCSLD family genes, OsCESA family genes, and) and the lignin synthesis gene(Fig. 6-C and -D), which revealed down-regulation ofand, inand.was significantly upregulated in all the three mutants, and the classical kinesin genewas significantly up-regulated inand.,,,,, andexhibited a significant down-regulation trend in all the three mutants. These results clarified thatandplay a crucial role in the pathway of cellulose synthesis in the rice cell walls.
Expression levels ofandat the rice plant’s tillering stage were significantly down-regulated in all the three mutants (Fig. 6-E). However, in- overexpressing lines, whiledisplayed significant up-regulation, the expression level ofdid not change significantly (Fig. 6-F). These results suggested a potential relationship between these two genes. However, it is worth noting that yeast two-hybrid testing demonstrated thatanddo not interact directly, leading us to speculate that their interaction may be indirect.
Impact of RENL1 and SRL1 on rice leaf water-holding capacity
Leaves serve as the primary organs for gas exchange andwater transpiration between plants and the environment. Previous studies have shown thatexhibited defects in leaf epidermis integrity and stomatal morphology, resulting in a significant decrease in net photosynthetic rate, and increases in transpiration rate and stomatal conductance. These changes may adversely affect rice leaf water-holding capacity, subsequently reducing drought tolerance. When we measured and analyzed relative values, we found that the leaf water contents in,,anddid not significantly differ from NPB (Fig. 6-G). However, the relative water content in,, andwas significantly lower than that of NPB, withexhibiting the lowest relative water content(Fig. 6-H). Results fromwater loss rate measurements of flag leaves at the heading stage revealed that all the three mutants lost water faster than NPB, withdisplaying the highest water loss rate (Fig. 6-I). These findings collectively indicate that the water-holding capacity of the three mutants was significantly lower than that of NPB, andexhibited the poorest water-holding capacity.
RENL1 and SRL1 co-influence rice drought tolerance
To ascertain the relationship between,, and drought tolerance of rice, we conducted simulated drought experiments on two-week-old rice seedlings using an 18% polyethylene glycol (PEG) solution (Fig. 7-A). After 1 d of treatment, most leaves in the four rice seedlings showed a drooping phenotype. After a week, when most leaves had withered, we re-watered them and calculated the survival rate. The results revealed that most NPB seedlings resumed growth, with a survival rate of 88% after rewatering. In contrast, the,, andmutants had significantly lower survival rates than NPB (Fig. 7-B), withhaving the lowest survival rate and nearly all plants perishing. We also measured the expression levels of drought-tolerant genes beforeand after 18% PEG treatment (sampling at 24 h) (Fig. 7-C and -D)., a drought-and low-temperature-induced gene, was significantly down-regulated in the three mutants after PEG treatment. In contrast, the expression levels ofandwere significantly up-regulated inand, while the expression level ofsignificantly decreased compared with NPB after treatment (Fig. 7-D). These findings collectively indicated decreased drought tolerance in the three mutants.

Fig. 6. Measurements of physiological indices and relative expression levels of genes in rice leaves.
A and B, Cellulose (A) and hemicellulose (B) contents of rice leaves in wild type (Nipponbare, NPB) and mutants (,, and).
C and D, qRT-PCR analysis of cell wall cellulose-related synthetic genes (C for OsCSLD family and D for OsCESA family).
E and F, Analysis of the expression levels ofandin mature leaves (E) and in NPB and-overexpressing lines (F).
G–I, Leaf water content (G), relative water content (H), and water loss rate in leaf (I) in NPB,,, and.
Data are Mean ± SD (= 3). The lowercase letters above the bars represent the significant differences at the 0.05 level by one-way analysis of variance.
We further compared the expression levels ofandin the four rice materials at the seedling stage. The results showed that the expression levels of both genes in the,, andmutants were significantly down-regulated compared with NPB (Fig. 7-E). We also measured the expression levels ofandin NPB before and after 18% PEG treatment, revealing an impact on their expression (Fig. 7-F).
However, it is essential to note that the PEG- simulated drought stress may not fully represent rice growth under actual drought conditions. To support our conclusions, we conducted experiments on real soil drought. When we withheld water from four-week-old rice seedlings for two weeks, most mutant leaves withered. After one week of rewatering, most wild type plants resumed growth, whereas most of the three mutants perished compared with the wild type (Fig. S2-A and -B). Statistically, all three mutants had significantly lower survival rates than NPB, withshowing the lowest survival rate and the highest mortality (Fig. S2-C). In summary, the loss of function of SRL1 and RENL1, affecting the epidermal structure and stomatal morphology of rice leaves, leads to faster water loss and reduced drought tolerance. In, the combined mutations ofandexacerbated this effect, further reducing rice’s drought tolerance.
Effects of drought stress on antioxidant properties of rice
Under drought stress conditions, a substantial accumulation of ROS occurs in the cell walls(Tenhaken, 2014). To counteract this, the activities of superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), and other antioxidant enzymes increased significantly to scavenge the excess ROS. Two-week-old rice seedlings were subjected to 18% PEG treatment for 6, 12, and 24 h, respectively. Afterward, their leaves were sampled, and the contents of H2O2and MDA, along with the activities of SOD, POD, and CAT, were measured. The results revealed that the H2O2contents inandwere significantly higher than those in NPB andat 6 h, reaching 3.83 and 3.21 μmol/g, respectively. After 24 h of treatment, the H2O2contents in all the three mutants surpassed that in NPB, withshowing the most significant increase (Fig. 8-A). The MDA content inexceeded that in NPB throughout the treatment period, butandreached 7.58 and 7.83 nmol/g after 24 h of treatment, respectively, significantly higher than that in NPB (Fig. 8-B). In contrast, the SOD activity in NPB significantly increased at the beginning of the treatment, while the three mutants displayed lower SOD activity (Fig. 8-C). The POD and CAT activities in NPB remained higher than those in the three mutants as the treatment duration extended (Fig. 8-D and -E). Furthermore, we observed that themutant exhibited the highest increase in H2O2and MDA content and the lowest SOD, POD, and CAT activities throughout the treatment period. These findings indicated that the antioxidant enzyme systems in the three mutants were compromised, preventing them from adequately responding to drought stress, scavenging ROS, mitigating oxidative and membrane damage, and ultimately affecting the drought tolerance of rice. Notably, the extent of damage to the antioxidant enzyme system inwas significantly greater than that in the other three rice materials, contributing to its lower drought tolerance.
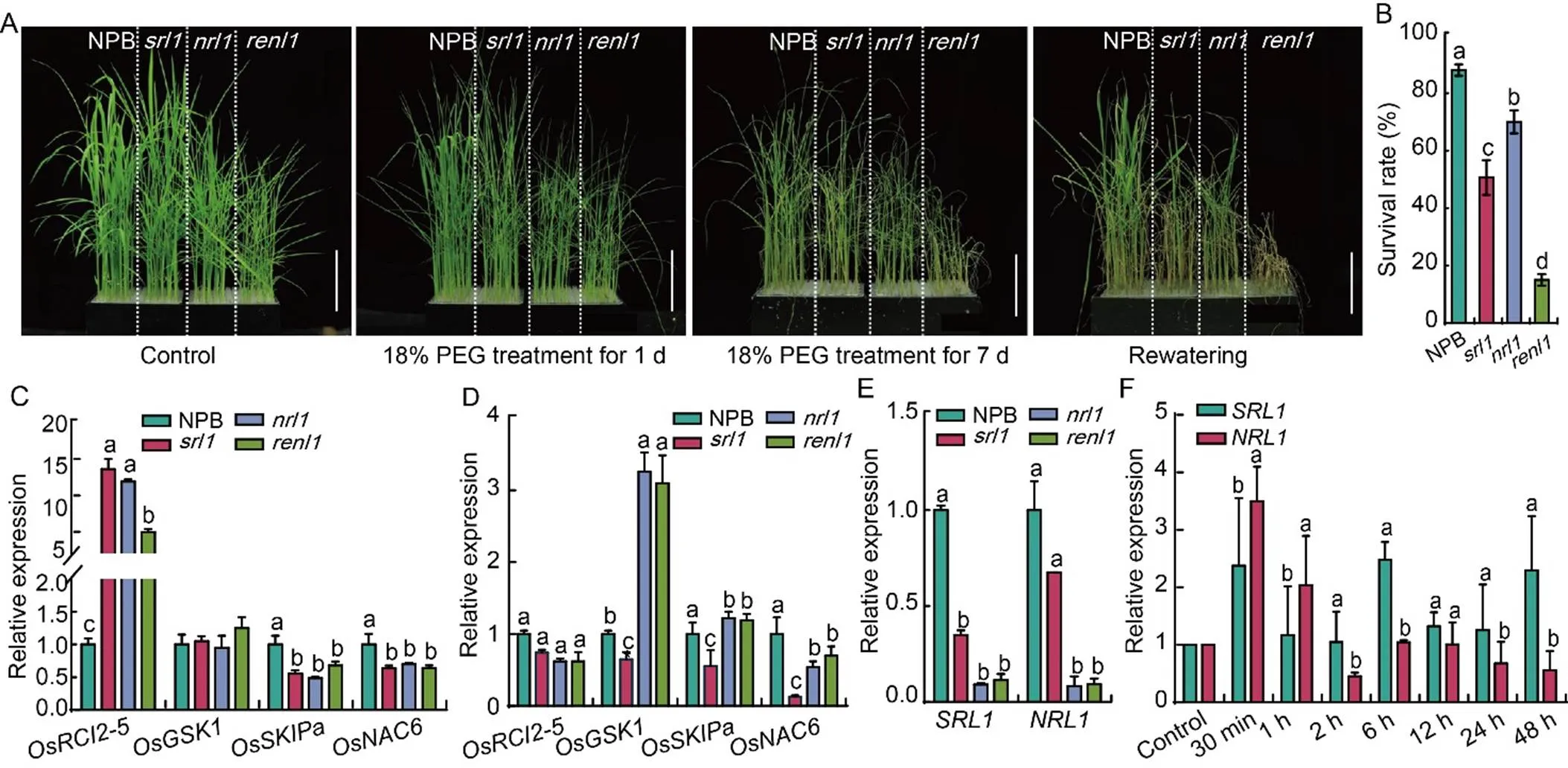
Fig. 7.andaffect drought tolerance of rice.
A, Drought tolerance of wild type (Nipponbare, NPB) and mutants (,, and) under 18% polyethylene glycol (PEG) stress treatment. Scale bars are 10 cm.
B, Survival rates of NPB,,, andafter rewatering.
C and D, Relative expression levels of drought tolerant genes in NPB,,, andbefore (C) and after (D) 18% PEG stress for uninterrupted 48 h.
E, Relative expression ofandgenes in NPB,,, andat the seedling stage.
F, Relative expression of drought-induced genes after 18% PEG treatment for different times.
In B to F, data are Mean ± SD (= 3). The lowercase letters above the bars represent the significant differences at the 0.05 level by one-way analysis of variance.

Fig. 8. Drought stress on antioxidant enzyme activities of rice leaves in wild type (Nipponbare, NPB) and mutants (,, and).
A–E, Changes of H2O2content (A), malonaldehyde (MDA) content (B), superoxide (SOD) activity (C), peroxidase (POD) activity (D), and catalase (CAT) activity (E) in NPB,,, andunder drought stress.
Data are Mean ± SD (= 3). The lowercase letters above the bars represent the significant differences at the 0.05 level by one-way analysis of variance.
DISCUSSION
The scarcity of water resources and the growing prevalence of drought have emerged as critical factors, leading to reduced rice yields (Todaka et al, 2015). The exploitation of water-saving and drought-resistant rice germplasm resources and variety selection can enhance land use efficiency and rice production capacity, carrying substantial implications for addressing food security issues in our country. The relationship between plant morphology and stress resistance in rice is well- established, although its genetic regulatory mechanisms remain elusive. The cell wall, composed primarily of polysaccharides, plays a pivotal role in plant adaptation and survival. Cellulose, synthesized at the plasma membrane, along with pectin and hemicellulose assembled in the Golgi apparatus, constitutes the major structural components of the cell wall (McFarlane et al, 2014). While the primary cell wall offers flexibility in maintaining cell shape, the secondary cell wall, apart from providing mechanical support and protection, plays a vital role in plant morphogenesis and resistance to dehydration stress. In this study, we identified the narrow and rolled leaf gene, which is also an allele of. The functions ofhave been extensively documented. It induces changes in arabinoxylan structure, cellulose content, and high galacturonic acid content in the primary cell, leading to structural defects in the primary cell wall in(Li et al, 2009). Hu et al (2010) found thatmutants exhibit a leaf-rolling phenotype primarily due to a reduction in the size and number of vesicular cells on the adaxial axis. Furthermore, Zhu L et al (2010) demonstrated the significant role ofin regulating the width and curl of rice leaves. In, severe defects were observed in pollen formation, anther dehiscence, stomatal development, and various tissues. It was also established that theOsCSLD4 proteinplays a crucial role in regulating cell proliferation during the mitotic phase of cell mitosis (Yoshikawa et al, 2013). While most of these studies have focused on the effects ofon rice growth, development, and morphology, the impact of cell wall defects and cellulose content on abiotic stress has been less explored. In addition to highlighting the significance ofin leaf development, our study revealed thatinfluenced drought tolerance in rice by affecting cellulose content.
The synthesis pathway of the rice cell wall is intricate and subject to regulation by various factors, including the cytoskeleton, GPI-anchored proteins, hormones, phosphatidylinositol, and the supply of ribonucleotides (Zhong and Ye, 2007). Studies have demonstrated that changes in cell wall-related components can impact drought tolerance in rice. For instance, mutations in the galacturonase gene,, lead to thickening of root and leaf cell walls, thus improving the plant’s drought resistance (Zhang et al, 2021). Conversely, overexpression of the E3 ubiquitin ligase genereduces rice’s drought tolerance by affecting genes related to leaf roll and cell wall development (Ning et al, 2011). The synthesis of the cell wall, abscisic acid response, drought response, and the significant upregulation of SOD-related genes collectively contribute to enhanced drought tolerance in(Liang et al, 2018). Our study revealed that the loss-of-function ofandprofoundly affected the expression of cellulose synthesis-related genes. The decrease in cellulose content in the cell wall resulted in defects in leaf structure and stomatal morphology. These defects, on one hand, led to reduced photosynthesis and accelerated water loss, substantially diminishing the water-holding capacity of rice leaves, thereby reducing the plant’s drought tolerance. It is worth noting that the recently identified allele of,, is anchored to the outer surface of the plasma membrane through a secretory transport pathway and plays a crucial role in extracellular fluids such as the cell wall. Loss offunction also results in an altered cell wall composition and changes in the expression of genes and proteins involved in cell wall synthesis, further leading to defects in rice leaf epidermis and increased susceptibility to water deficit (Li W Q et al, 2017). The smallerBCs inandmay be attributed to rapid water loss. Additionally,also impacted root development, as the analysis of root traits indicated weaker root development in these mutants compared with the wild type. This may affect the mutants’ ability to absorb water, contributing to their decreased drought tolerance. Therefore, the reduced drought tolerance ofandmay be associated with slow root water uptake and rapid leaf water loss.also affected the ability to scavenge ROS under drought stress. The production of ROS is one of the earliest plant cell responses to various stresses. ROS can trigger the inactivation of cell- damaging enzymes, biofilm breakdown, and changes in gene expression through protein oxidation (Finkel and Holbrook, 2000). Under drought stress conditions, the activities of various peroxidases in rice plants maintain a balance between ROS accumulation and a clear system. In general, higher ROS scavenging capacitycorresponds to greater drought resistance, and lower MDA accumulation, signifying stronger drought resistance.
encodes a plasma membrane-bound glycosylphosphatidylinositol-anchored protein that negatively regulates the formation of vesicular cells, thereby influencing leaf rolling (Xiang et al, 2012). In eukaryotes, glycosylphosphatidylinositol anchoring is typically an alternative for attaching proteins to the plasma membrane (Udenfriend and Kodukula, 1995). Whileis not directly involved in cellulose synthesis, our yeast two-hybrid experiments with these two genes did not reveal a direct interaction betweenand. Nevertheless, our results suggested thatplays a significant role in the cellulose synthesis pathway. Similar to, the loss offunction results in reduced cellulose content in rice, affecting the expression of genes related to cellulose and hemicellulose synthesis. The mechanistic similarities betweenandin diminishing drought tolerance in rice indicate a correlation between these two genes. Genetic analysis has indicated thatis situated downstream of, a conjecture confirmed by the expression analysis ofin- over expression lines.
With the global population increasing and water resources becoming scarcer, the cultivation of drought-resistant rice has become a crucial research focus. Previous studies on the molecular mechanisms of stress tolerance in rice have primarily centered on transcription factors and their molecular mechanisms, such as bZIP(Maruyama et al, 2014), MYB/MYC(El-Kereamy et al, 2012), and WRKY (Song et al, 2010). The identification of drought-tolerant rice germplasm resources, the isolation and cloning of key genes, and the discovery of excellent haplotypes for adaptive responses to drought stress have all been major limiting factors in breeding new drought- resistant rice varieties. Considering the role ofin regulating leaf type and its impact on plant drought tolerance, as well as the significance of cellulose content in enhancing drought tolerance in rice, this provided an important research direction for breeding drought-resistant rice varieties and studying the mechanisms of rice drought resistance in the future.
METHODS
Rice materials and growth conditions
Themutant, with a semi-rolled leaf, was obtained from a population ofL. ssp.cultivar NPB through mutagenesis using EMS. Subsequently, themutant, characterized by an extremely rolled leaf, was derived fromvia EMS mutagenesis. These mutants, along with the wild type NPB, were cultivated at two locations: the Fuyang District experimental base of the China National Rice Research Institute in Hangzhou (30°04′ N, 119°55′ E), Zhejiang Province, China, and the South Breeding Base of China National Rice Research Institute in Lingshui (18°30′ N, 110°01′ E), Hainan Province, China, under natural conditions. Transgenic materials were grown in a designated field for transgenic rice at the Fuyang District experimental base of the China National Rice Research Institute. Standard field management practices were applied to all field materials, including disease and pest controls. To better understand the function of, we crossedwith NPB to isolate a single mutant of the puregene from the F2population.
Phenotypic characterization and chlorophyll content measurement
At the heading stage, six replicates of flag leaves from NPB,,,andmaterials were randomly selected for measuring leaf width (w), leaf natural width (n), and leaf length.The leaf curl ratio (LRI, %) = (w –n)/w × 100%. After maturity, plant height and various yield and grain characteristics, including panicle length, the number of primary branches per panicle, the number of secondary branches per panicle, the number of grains per panicle, seed-setting rate, 1000-grain weight, grain length, and grain width were measured following the rice standard evaluation system (http://www.Knowledge bank.irri.org/ses/).
At the heading stage, flag leavesfromNPB,,andwere randomly collected in triplicate, and the contents of chlorophyll a and chlorophyll b were determined using a spectrophotometer (Wellburn, 1994). Leaf samples were cut into 0.5 cm pieces and soaked in 80% acetone in the dark for more than 24 h. Optical density (OD) values at 663, 645, and 470 nm were measured using a DU800 photometer (Beckman Coulter, CA, USA).
Frozen section of leaf tissue
Leaves from NPB,,, andplants were sampled at the tillering stage, and the middle sections (approximately 0.1–0.3 cm) of the leaves were excised. These samples were placed in OCT (optimum cutting temperature compound) cryopreservation agent and frozen in a -20 ºC refrigerator for 1 h. Subsequently, the samples were sliced into frozen sections using a frozen slicer (Leica CM1950, Wetzlar, Germany). Observations were made using a microscope (Nikon Eclipse 90i, Tokyo, Japan) at 200× (10 × 20) and 400× (10 × 40) magnifications.
Map-based cloning of RENL1
To map thegene, a mapping population was created by crossingwith, an allelic mutant of thegene. The narrow rolled leaf mutants from the F2generation were used for mapping gene. A total of 232 simple sequence repeat markers distributed across 12 rice chromosomes were employed to map. The initial mapping narrowed down theposition to a physical distance of 12.1 kb between the markers C-2 and C-7. The 12.1 kb genomic DNA was amplified by PCR and sequenced using SeqMan software (DNASTAR, Inc, MA, USA), with PCR products analyzed via 4% agarose gel electrophoresis.
Electron microscope scanning and fluorescent slice
At the tillering stage, leaves from NPB,,, andwere collected separately, and cut into 0.5 cm × 0.5 cm segments. These segments were immersed in a 2.5% glutaraldehyde solution for over 4 h. Afterward, the leaves were washed three times with phosphate buffer for 15 min each time and fixed in a phosphate buffer at 4 ºC. Subsequent steps included dehydration with gradient ethanol (30%, 50%, 70%, 80%, 90%, and 95%), incubation in an isoamyl alcohol acetate (1:1) mixture and coating with gold-palladium. The samples were observed using a vertical scanning electron microscope (Hitachi TM-1000, Tokyo, Japan).
For the observation of leaf tissue structure, sections were prepared from the middle of fresh rice flag leaves, and fluorescence microscopy was conducted at a 200-fold field of view (10 × 20) using a fluorescence microscope (NIKON EGLIPSE 90i, Tokyo, Japan). The fluorescence wavelengths used were 470 nm and 530 nm (Deng et al, 2020).
Measurement of stomatal number and photosynthetic parameters
To assess the stomatal density of leaves, the epidermis of the middle leaves in NPB,,,andplants at the heading stage was coated with transparent nail polish. Stomatal numbers were determined at a 400× (10 × 40) field of view using an optical microscopy (Nikon, Tokyo, Japan) with five replicates for each sample.
The photosynthetic rate, stomatal conductance, transpiration rate, and intercellular CO2concentration of flag leaves were measured using a Li-6400 portable photosynthetic analyzer (LI-COR, Lincoln, NE, USA) between 10:00 am and 12:00 pm on a sunny day, with five replicates for each sample.
RNA isolation and qRT-PCR analysis
Total RNA was extracted using Axygen’s Miniprep kit (Axygen Scientific Inc, CA, USA). Reverse synthesis of cDNA was carried out using ReverTra Ace quantitative PCR RT Master Mix reverse transcription kit (Toyobo, Osaka, Japan). qRT-PCR was conducted using 2× SYBR premix extaqtm (Takara, Kyoto, Japan) and Bio-Rad CFX PCR instruments (Bio-Rad Laboratories, CA, USA). The PCR amplification was performed in 40 cycles, including 95 ºC denaturation for 60 s, 95 ºC denaturation for 15 s, and 60 ºC extension for 60 s. Information on primers used is presented in Table S1.
Vector construction and plant transformation
To construct the overexpression plasmid, the cDNA fragment ofwas amplified and integrated into the pCAMBIA1300S vector to create the p35S::plasmid.
Theknockout plasmid pC1300-2×35S::Cas9 was built using the rice polygene knockout system from Wang Kejian Laboratory, China National Rice Research Institute. The CRISPR target ofwas linked to SK-gRNA vector, and the positive intermediate vector SK-gRNA-was selected. The final expression vector pC1300-2×35S::CAS9-was constructed through restriction endonuclease ligation.
All constructed plasmids were validated through sequencing. The plasmids were introduced intocompetent EHA105 through electroporation, with the overexpression vector transformed into NPB, and the pC1300-2×35S:: CAS9-knockout vector transformed intomutant.
Detection of leaf water content, relative water content, and water loss rate
At the heading stage, the flag leaves of NPB,,andwere collected. The fresh weight (f) of leaves of each group was measured, and the leaves were soaked in distilled water for 70 min. After reaching a constant weight, the saturated fresh weight (t) was determined. The leaves were then oven-dried at 70 ºC for 12 h, and the dry weight (d) of the leaves was measured. Leaf water content was calculated as (f–d)/f× 100%, and leaf relative water content was calculated as (f–d)/(t–d) × 100% (Barrs and Weatherley, 1962). Six flag leaves were randomly selected from each group, with three replicates. The water loss rate of leaveswas measured by randomly six flag leaves from different plants and conducting three repetitions. The initial weight of the freshly removed leaves was recorded, and the leaves were then placed in an incubator with a constant temperature (28 ºC) and humidity (approximately 60%). The leaf weight of each group was measured every 30 min, and measurements were stopped when the change in leaf weight was not significant.
Drought treatment
NPB,,, andplants were grown in a 96-well hydroponic cassette (25 cm × 7 cm × 6 cm) in a thermostatic greenhouse with a temperature of 28 ºC and humidity maintained at 70%–80%. Three replicates were used for each treatment group. The plants in the hydroponic cassette were cultivated in rice nutrient solution (Beijing Kuleber Technology Co., Ltd, Beijing, China) and exposed to 18% PEG for two weeks. After 24 h of treatment, the relative expression levels of,, and related drought-induced genes were analyzed. Following 7 d of rewatering treatment, the seedlings that did not recover were considered to have not survived, and the survival rate was calculated as the percentage of surviving seedlings out of the total treated seedlings. The plants were subjected to a two-week drought period when they reached four weeks of age, followed by one week of rewatering, and the survival rate was determined.
Analysis of root characteristics at the seedling stage
NPB,,, andplants were cultivated in a hydroponic box within a controlled-temperature greenhouse. Five-day-old rice seedlings were scanned and photographed using a scanner(Shanghai Zhongjing Technology Co., Ltd, Shanghai, China). Root length, main root diameter, and root surface area were analyzed using ImageJ image processing software (National Institutes of Health, MA, USA).
Measurement of contents of H2O2 and MDA, and activities of related antioxidant enzymes
H2O2and MDA contents, and SOD, POD, CAT enzyme activity test kits were purchased from Suzhou Grace Biotechnology Co., Ltd (Suzhou, China). The two-week-old hydroponic seedlings were treated with 18% PEG, and about 0.1 g of leaf samples was collected at 6 h, 12 h, and 24 h. Each sample was immediately frozen in liquid nitrogen, and this process was repeated three times, following the kit instructions.
Statistical analysis
All statistical analysis was performed using a one-way ANOVA test with a significant difference via IBM SPSS.
Acknowledgements
This study was supported by the Nanfan Special Project of Chinese Academy of Agricultural Sciences (Grant No. ZDXM2315), the National Natural Science Foundation of China (Grant Nos. 32372125, 31861143006, and 32188102), Special Support Program of Chinese Academy of Agricultural Sciences (Grant NO. NKYCLJ-C-2021-015), Specific Research Fund of the Innovation Platform for Academicians of Hainan Province, and 2023 College Student Innovation and Entrepreneurship Project of Jiangxi Agricultural University, China (Grant No. S202310410095).
SUPPLEMENTAL DATA
The following materials are available in the online version of thisarticle at http://www.sciencedirect.com/journal/rice-science; http://www.ricescience.org.
Fig. S1.affects yield traits of rice.
Fig. S2. Soil drought tolerance of mutants.
Table S1. Oligonucleotides used in this study.
Barrs H D, Weatherley P E. 1962. A re-examination of the relative turgidity technique for estimating water deficits in leaves., 15(3): 413–428.
Bernal A J, Jensen J K, Harholt J, Sørensen S, Moller I, Blaukopf C, Johansen B, de Lotto R, Pauly M, Scheller H V, Willats W G T. 2007. Disruption ofresults in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis., 52(5): 791–802.
Bernal A J, Yoo C M, Mutwil M, Jensen J K, Hou G C, Blaukopf C, Sørensen I, Blancaflor E B, Scheller H V, Willats W G T. 2008. Functional analysis of the cellulose synthase-like genes,, andin tip-growing Arabidopsis cells., 148(3): 1238–1253.
Bertolino L T, Caine R S, Gray J E. 2019. Impact of stomatal density and morphology on water-use efficiency in a changing world., 10: 225.
Braybrook S A, Kuhlemeier C. 2010. How a plant builds leaves., 22(4): 1006–1018.
Carpita N C, Gibeaut D M. 1993. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth., 3(1): 1–30.
Chen Z Z, Hong X H, Zhang H R, Wang Y Q, Li X, Zhu J K, Gong Z Z. 2005. Disruption of the cellulose synthase gene,, enhances drought and osmotic stress tolerance in Arabidopsis., 43(2): 273–283.
Deng Q W, Liu Z X, Zhang Q, Gao Y, Jiang Y J, Zheng Y X, Hu W. 2020. Fluorescence microscopic observation and free-hand section techniques for rice leaves., 33(1): 24–27. (in Chinese with English abstract)
Dey A, Samanta M K, Gayen S, Maiti M K. 2016. The sucrose non-fermenting 1-related kinase 2 geneimproves drought tolerance and grain yield in rice by modulating cellular osmotic potential, stomatal closure and stress-responsive gene expression., 16: 158.
El-Kereamy A, Bi Y M, Ranathunge K, Beatty P H, Good A G, Rothstein S J. 2012. The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism., 7(12): e52030.
Fang L K, Zhao F M, Cong Y F, Sang X C, Du Q, Wang D Z, Li Y F, Ling Y H, Yang Z L, He G H. 2012. Rolling-leaf14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves., 10(5): 524–532.
Finkel T, Holbrook N J. 2000. Oxidants, oxidative stress and the biology of ageing., 408: 239–247.
Ghosh D, Xu J. 2014. Abiotic stress responses in plant roots: A proteomics perspective., 5: 6.
Hu J, Zhu L, Zeng D L, Gao Z Y, Guo L B, Fang Y X, Zhang G H, Dong G J, Yan M X, Liu J, Qian Q. 2010. Identification and characterization of, a novel gene regulating leaf morphology and plant architecture in rice., 73(3): 283–292.
Huang L C, Chen L, Wang L, Yang Y L, Rao Y C, Ren D Y, Dai L P, Gao Y H, Zou W W, Lu X L, Zhang G H, Zhu L, Hu J, Chen G, Shen L, Dong G J, Gao Z Y, Guo L B, Qian Q, Zeng D L. 2019. A Nck-associated protein 1-like protein affects drought sensitivity by its involvement in leaf epidermal development and stomatal closure in rice., 98(5): 884–897.
Jenks M A, Andersen L, Teusink R S, Williams M H. 2001. Leaf cuticular waxes of potted rose cultivars as affected by plant development, drought and paclobutrazol treatments., 112(1): 62–70.
Kadioglu A, Terzi R. 2007. A dehydration avoidance mechanism: Leaf rolling., 73(4): 290–302.
Karaba A, Dixit S, Greco R, Aharoni A, Trijatmiko K R, Marsch- Martinez N, Krishnan A, Nataraja K N, Udayakumar M, Pereira A. 2007. Improvement of water use efficiency in rice by expression of, androught and salt tolerance gene., 104(39): 15270–15275.
Kim C M, Park S H, Je B I, Park S H, Park S J, Piao H L, Eun M Y, Dolan L, Han C D. 2007., a cellulose synthase-like D1 gene, is required for root hair morphogenesis in rice., 143(3): 1220–1230.
Lerouxel O, Cavalier D M, Liepman A H, Keegstra K. 2006. Biosynthesis of plant cell wall polysaccharides: A complex process., 9(6): 621–630.
Li M, Xiong G Y, Li R, Cui J J, Tang D, Zhang B C, Pauly M, Cheng Z K, Zhou Y H. 2009. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth., 60(6): 1055–1069.
Li W Q, Zhang M J, Gan P F, Qiao L, Yang S Q, Miao H, Wang G F, Zhang M M, Liu W T, Li H F, Shi C H, Chen K M. 2017./modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice., 92(5): 904–923.
Li X M, Han H P, Chen M, Yang W, Liu L, Li N, Ding X H, Chu Z H. 2017. Overexpression of, which encodes a novel cysteine-rich peptide, enhances drought tolerance and increases ABA concentration in rice., 93(1/2): 21–34.
Liang J Y, Guo S Y, Sun B, Liu Q, Chen X H, Peng H F, Zhang Z M, Xie Q J. 2018. Constitutive expression ofconfers the rice response to drought stress and abscisic acid., 11(1): 59.
Maruyama K, Urano K, Yoshiwara K, Morishita Y, Sakurai N, Suzuki H, Kojima M, Sakakibara H, Shibata D, Saito K, Shinozaki K, Yamaguchi-Shinozaki K. 2014. Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts., 164(4): 1759–1771.
McFarlane H E, Döring A, Persson S. 2014. The cell biology of cellulose synthesis., 65: 69–94.
Nakashima K, Tran L S P, van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K. 2007. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice., 51(4): 617–630.
Ning Y S, Jantasuriyarat C, Zhao Q Z, Zhang H W, Chen S B, Liu J L, Liu L J, Tang S Y, Park C H, Wang X J, Liu X L, Dai L Y, Xie Q,Wang G L. 2011. The SINA E3 ligase OsDIS1 negatively regulates drought response in rice., 157(1): 242–255.
Richmond T A, Somerville C R. 2000. The cellulose synthase superfamily., 124(2): 495–498.
Shen H S, Chen J C, Huang J H, Tang B S. 2005. Microstructure and distribution of silica bodies in rice epidermis., 34(2): 137–140. (in Chinese with English abstract)
Song Y, Ai C R, Jing S J, Yu D Q. 2010. Research progress on functional analysis of ricegenes., 17(1): 60–72.
Tenhaken R. 2014. Cell wall remodeling under abiotic stress., 5: 771.
Todaka D, Shinozaki K, Yamaguchi-Shinozaki K. 2015. Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants., 6: 84.
Udenfriend S, Kodukula K. 1995. How glycosyl-phosphatidylinositol- anchored membrane proteins are made., 64: 563–591.
Wang X, Cnops G, Vanderhaeghen R, de Block S, van Montagu M, van Lijsebettens M. 2001., a cellulose synthase-like gene important for root hair growth in Arabidopsis., 126(2): 575–586.
Wellburn A R. 1994. The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution., 144(3): 307–313.
Xiang J J, Zhang G H, Qian Q, Xue H W. 2012.encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells., 159(4): 1488–1500.
Xue D W, Zhang X Q, Lu X L, Chen G, Chen Z H. 2017. Molecular and evolutionary mechanisms of cuticular wax for plant drought tolerance., 8: 621.
Yoo C Y, Pence H E, Jin J B, Miura K, Gosney M J, Hasegawa P M, Mickelbart M V. 2010. TheGTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of., 22(12): 4128–4141.
Yoshida S, Hasegawa S. 1982. The rice root system: Its development and function.: International Rice Research Institute. Drought Resistance in Crops with Emphasis on Rice. Los Baños, the Philippines: International Rice Research Institute: 97–114.
Yoshikawa T, Eiguchi M, Hibara K I, Ito J I, Nagato Y. 2013. Ricegene encodes cellulose synthase-like D4 and is specifically expressed in M-phase cells to regulate cell proliferation., 64(7): 2049–2061.
Zhang G H, Hou X, Wang L, Xu J, Chen J, Fu X, Shen N W, Nian J Q, Jiang Z Z, Hu J, Zhu L, Rao Y C, Shi Y F, Ren D Y, Dong G J, Gao Z Y, Guo L B, Qian Q, Luan S. 2021.encodes a polygalacturonase that modifies cell wall structure and drought tolerance in rice., 229(2): 890–901.
Zhong R Q, Ye Z H. 2007. Regulation of cell wall biosynthesis., 10(6): 564–572.
Zhu J H, Lee B H, Dellinger M, Cui X P, Zhang C Q, Wu S, Nothnagel E A, Zhu J K. 2010. A cellulose synthase-like protein is required for osmotic stress tolerance in., 63(1): 128–140.
Zhu L, Hu J, Yan M X, Gao Z Y, Liu J, Qian Q, Guo L B. 2010. Rnai and expression analysis of a gene, which controls plant narrow and rolled leaf in rice., 24(5): 873–880. (in Chinese with English abstract)
Zhu X Y, Xiong L Z. 2013. Putative megaenzyme DWA1 plays essential roles in drought resistance by regulating stress-induced wax deposition in rice., 110(44): 17790–17795.
7 August 2023;
31 October 2023
Zhang Guangheng (zhangguangheng@caas.cn); Hu Songping (husongping1969@163.com); Qian qian (qianqian@caas.cn)
Copyright © 2024, China National Rice Research Institute. Hosting by Elsevier B V
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://doi.org/10.1016/j.rsci.2023.10.001
(Managing Editor: Wang Caihong)
- Rice Science的其它文章
- Leaf Morphology Genes SRL1 and RENL1 Co-Regulate Cellulose Synthesis and Affect Plant drought Tolerance
- Effects of Main Nitrogen-Regulating Gene AreA on Growth, Pathogenicity, and Fumonisin Synthesis of Fusarium proliferatum
- Grain Yield, Biomass Accumulation, and Leaf Photosynthetic Characteristics of Rice under Combined Salinity-Drought Stress
- Alternative Splicing of OsCYL4 Controls Drought Resistance via Regulating Water Loss and Reactive Oxygen Species-Scavenging in Rice
- Potential Secretory Transporters and Biosynthetic Precursors of Biological Nitrification Inhibitor 1,9-Decanediol in Rice as Revealed by Transcriptome and Metabolome Analyses
- OsbZIP01 Affects Plant Growth and Development by Regulating OsSD1 in Rice