Evaluation of the binding affinity and antioxidant activity ofphlorizin to pepsin and trypsin
Jing Zhng,Ln Tng,Xi Hu,Zhen Zeng,Wen Wu,Fng Geng,Hui Li,Di Wu,*
a Meat Processing Key Laboratory of Sichuan Province, School of Food and Biological Engineering, Chengdu University, Chengdu 610106, China
b School of Chemical Engineering, Sichuan University, Chengdu 610065, China
Keywords:Interaction Phlorizin Pepsin Trypsin Antioxidation
ABSTRACT Phlorizin (PHL) is a natural compound with strong antioxidant properties mainly found in apples.In this paper,the interaction mechanism of PHL with pepsin and trypsin was comparatively evaluated by computer simulation,fluorescence spectra,circular dichroism (CD),and Fourier transform infrared (FT-IR) spectra at a molecular level.Fluorescence spectra showed that PHL quenches the pepsin/trypsin by static quenching.Thermodynamic parameters indicated that PHL binds to pepsin mainly through hydrogen bonds and van der Waals forces,and that of trypsin was electrostatic forces.The ground state complexes PHL and protease have a moderate affinity of 105 L/mol PHL binds more strongly to trypsin than to pepsin.CD and FT-IR spectra results showed that pepsin/trypsin decreased the β-sheet content and slightly changed its secondary structure upon PHL.These experimental results are mutually verified with the predicted computer-aid simulation results.Upon PHL and trypsin binding,the antioxidant capacity of PHL was elevated.Nevertheless,the antioxidant capacity of PHL was decreased after binding to pepsin.This work elucidates the binding of PHL binding mechanisms to pepsin/trypsin and provides useful information for the digestion of PHL to improve the application of PHL in food processing.
1. Introduction
Consumption of fruits is good for health and can reduce the risk of diseases[1].Among them,fruit flavonoids have been shown to be beneficial in enhancing health[2].People are more concerned about the impact of food on their health as their consumption levels increase.The consumption trend has shifted to functional foods with high nutrition and health care functions.The number and types of functional foods containing biologically active compounds have gradually increased.Apples are a common fruit in people’s daily life,which contains a variety of flavonoids.The addition of apple flavonoids in foods and beverages can improve the nutritional value of foods,such as functional gel desserts[3],snack bars[4],and drinking yogurt[5].
Phlorizin (PHL) is one of the dihydrochalcone groups of flavonoids that are homology to medicine and food,which are mainly found in the young leaves,rhizomes,and apple fruits of tissues such as apple andLithocarpuspolystachyusRehd[6].PHL has various health-promoting effects including cardiovascular protection[7],antiviral[8],antioxidant activity[9],and antidiabetic activity[10].Several researches have found that PHL can inhibit the sodium-glucose transporter protein system (SGLT1) on the mucosal of the small intestinal epithelium[11],as well as block intestinal glucose absorption by improving insulin sensitivity and glucose uptake[12].In addition,PHL has a good inhibitory effect on tyrosinase and may prevent some skin diseases[13].
When flavonoids are ingested by organisms,they may interact with digestive enzymes,affecting the digestion and absorption of nutrients[14].The flavonoid compounds antioxidant activity of the compounds may also alter[15].As we all know,antioxidants are closely related to physical health[16-18],and it is important to study the antioxidant activity of flavonoids.Pepsin and trypsin are the main digestive proteases.Pepsin is an aspartic protease[19]and has a molecular mass of 35 kDa[20].It is often used in digestion models to reveal the event of the biological compound on their structural conformation.The interaction of pepsin with baicalin,myricetin,rutin,and other flavonoids have been more widely studied.Among them,the combination of aloe-emodin with pepsin decreases its antioxidant activity[21].Li et al.[22]reported interaction of pepsin with flavonoids affects the microenvironment of proteases.Trypsin is a serine protease[23]with a relative molecular mass of about 24 kDa and three catalytic active sites His-57,Asp-102,and Ser-195[24].It is secreted by the pancreas and is the most abundant protease in nature,converted from inactive trypsinogen.Furthermore,trypsin is a trigger enzyme[25]that stimulates a variety of digestive enzymes in the digestive system and plays a weighty impact on the digestion of food proteins.Ren[26]reported that resveratrol interacted with trypsin and that the antioxidant activity of resveratrol and the catalytic activity of trypsin was reduced after binding.PHL is likely to binding to pepsin and trypsin during digestion,and there is still a lack of research on the digestive part of PHL,so it is very necessary to study the binding of interaction and activity changes of PHL with pepsin and trypsin.
In this paper,the binding of PHL with pepsin and trypsin was analyzed.Computational simulation methods and multi-spectral techniques such as fluorescence spectra,time-resolved fluorescence,CD and FT-IR spectra were employed to detect the binding mechanisms between PHL to pepsin and trypsin.Including quenching types,thermodynamic parameters,binding mechanism,and conformational changes.In addition,the changes in the antioxidant capability of PHL after binding to proteases were investigated by antioxidant activities.
2. Materials and methods
2.1 Materials
PHL (CAS:60-81-1,≥ 98% purity) was supplied from Aladdin Chemical Reagent (Shanghai,China).Pepsin (from porcine,activity:1:3 000) was supplied from Shanghai Macklin Biochemical Co.,Ltd (Shanghai,China).Trypsin (from porcine pancreas,activity:1:2 500) was supplied from Sangon Biotech Co.,Ltd (Shanghai,China).The low content of PHL has a better water solubility.PHL stock solution (1 × 10-3mol/L) was prepared in ultrapure water.Pepsin stock solution (1 × 10-4mol/L) was prepared in disodium hydrogen phosphate-citrate buffer pH 2.0 (0.01 mol/L NaCl).In phosphate buffer pH 7.4 (0.01 mol/L PBS),a trypsin stock solution (1 × 10-4mol/L) was made.The sample stock solution was stored in the dark at 4 °C.2,4,6-Tri(2pyridyl)-s-triazine(TPTZ),2,2’-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) and other chemical reagents were for an analytical grade.
2.2 Molecular docking
Molecular docking simulations were performed with the YASARA program (17.4.17)[27].The structure of PHL was obtained from PubChem (CID: 6072).The native structures of pepsin (PDB ID:3PEP) and trypsin (PDB ID: 2ZQ1) were obtained from the Protein Data Bank database.Protein removes water molecules as well as metal ions,adds hydrogen atoms and performs energy minimization.The system environments of pH 2.0 and pH 7.4 were selected for semi-flexible docking of PHL with pepsin/trypsin,respectively.
2.3 Molecular dynamics (MD) simulation
MD simulation by the AMBER14 force field.The best results of docking of PHL with pepsin and trypsin were used as the initial conformation.A grid box of 68 Å × 68 Å × 68 Å was placed to meet the entire complex system.Add Na or Cl ions and remove water molecules to make the system ionically balanced.All simulations were run in multiple time steps of 1.25 fs and 2.5 fs.Long-range Coulomb interactions were observed with particle mesh Ewald summation and a cutoff of 8 Å.MD simulations in the range of 70 ns were performed at 25 °C.
2.4 Fluorescence spectra
Fluorescence spectra was measured with a Cary Eclipse fluorophotometer (Varian,California,USA).Fluorescence intensities were corrected using previously reported methods[28].Disodium hydrogen phosphate-citrate buffer pH 2.0 or phosphate buffer pH 7.4 were used to dilute the PHL to pepsin/trypsin combination to experimental concentrations.The concentrations of pepsin(3.0 × 10-6mol/L) and trypsin (6.0 × 10-6mol/L) were kept constant,respectively.The PHL concentration was serially increased from 0 to 4.2 × 10-5mol/L at increments of 3.0× 10-6mol/L.Fluorescence spectra in the 300-460 nm region were observed at temperatures of 4,25 and 37 °C.The excitation wavelength of 280 nm and the scanning speed is 240 nm/min.Pepsin and trypsin had 2.5 nm and 5 nm excitation/emission slit widths,respectively.The Stern-Volmer equation was used to calculate the relevant parameters[29]:
whereF0andFare the fluorescence intensities of the protease alone and the protease complexed with PHL,respectively.KSVis the quenching constant and [Q] is the concentration of the quencher PHL.The binding constant (Ka) and the Hill coefficient (n) were determined by a modified Stern–Volmer equation[30]:
The values of enthalpy change (ΔHo) and entropy change (ΔSo)were estimated using the Van’t Hoff equation to fit the values ofKand the associated temperature (T),and the free energy change(ΔGo) was computed using the Gibbs–Helmholtz equation,whereR(8.314 J/(mol·K) is the gas constant[31,32]:
Synchronous fluorescence spectra of PHL with pepsin/trypsin was recorded at 25 °C with scans ranging from 250 nm to 360 nm(Δλ=15 nm) and 280 nm to 420 nm (Δλ=60 nm),respectively.The concentration ratios,slit widths and the scanning speed of the mixture was similar to the fluorescence spectra.
2.5 Time-resolved fluorescence measurements
Time-resolved fluorescence measurements atλex=280 nm andλem=350 nm were accomplished by a Jobin Yvon fluorolog-3 spectrofluorometer (Horiba,Les Ulis,France) at 25 °C.The concentration of pepsin/trypsin was 3.0 × 10-6mol/L and 6.0 × 10-6mol/L,respectively.The fluorescence lifetime data of the system through the tail fitting method and the fluorescence lifetimeτwere obtained by the formula[33]:
whereτis the average fluorescence lifetime,andτiandαiare the delay times and relative amplitudes corresponding to thei-fitting,respectively.
2.6 CD measurements
The CD spectra of pepsin and trypsin solutions were collected with a CD spectrometer (Applied Photophysics,Surrey,UK) at 25 °C.The wavelengths of 200-260 nm.The concentration of pepsin and trypsin were fixed at 1.5 × 10-5mol/L and 2.0 × 10-5mol/L,respectively.The band width was 1 nm and all data were collected under a constant nitrogen atmosphere (3 L/min).
2.7 FT-IR measurements
FT-IR spectral experiments were measurements on the infrared spectrometer (Thermo Fisher Scientific,Sunnyvale,USA).The spectral data of pepsin/trypsin were recorded in 600-4 000 cm-1in the presence and absence of PHL at 25 °C with a resolution of 4 cm-1,respectively.The pepsin concentration was 5.0 × 10-5mol/L and the trypsin concentration was 2.0 × 10-5mol/L.Analysis of data from 1 500-1 700 cm-1.Data processing was performed with Omnic(Thermo Nicolet Corporation,Wisconsin,USA) and PeakFit (Beijing HuanZhongRuiChi Technology Co.,Ltd.,Beijing,China) software.Curve fitting was performed with the Savitzky-Golay function.The area of each peak was used to compute the proportion of secondary structure.
2.8 Assays for antioxidant activities
2.8.1 Determinationoffreeradicalscavengingabilityby ABTSassay
The ABTS scavenged assay was performed referring method[34]with slight modifications.ABTS solution of 7.0 × 10-3mol/L and K2S2O8solution of 2.45 × 10-3mol/L were prepared,respectively.The two solutions were mixture 1:1 to obtain ABTS cation radical working solution,which was standing in the dark for 12-16 h at 25 °C.The sample solutions of different concentrations were taken,and diluted ABTS cation radical solution was added in turn.Reaction under darkness for 6 minutes at 25 °C.The absorbance of the sample solution was determined at 734 nm.Instead of the PHL sample solution,the control group was given distilled water.Calculate the ABTS cation radical scavenging rate of the sample according to the formula[35]:
whereAcontrolandAsamplerepresent the absorbance of the control and samples,respectively.
2.8.2 Determinationofreducingcapacitybyferricion reducingantioxidantpower(FRAP)assay
FRAP analysis was modified from the previously reported protocol.Fresh FRAP working solution was prepared (3.0 × 10-2mol/L acetic acid-sodium acetate buffer (pH 3.6),1.0 × 10-2mol/L TPTZ solution in 4.0 × 10-2mol/L hydrochloric acid,2.0 × 10-2mol/L FeCl3solution mixed in a volume of 10:1:1,respectively) and mix samples of different concentrations well with FRAP working solution.Incubated for 10 min at 25 °C absorbances were determined at 593 nm.Instead of the PHL sample solution,the blank group was given distilled water.The reducing power was calculated with the formula[36]:
whereAblankrepresents the absorbance of the blank,Asamplerepresents the absorbance of the samples.
2.9 Statistical analysis
Data were presented as the mean of the three replicates and their standard deviation values (mean ± SD).GraphPad Prism 9.0 software (GraphPad Software,California,USA) was used for data processing,and SPSS 20.0 software (International Business Machines Corporation,Armonk,USA) was used for one-way analysis of variance (ANOVA) with statistically significant differencesP<0.05).
3. Results and discussion
3.1 Molecular docking studies
Molecular docking can predict the binding force and binding sites between ligands and proteins.Using the Lamarck genetic algorithm[37],the optimal binding energy conformation was chosen for study after 100 docking conformations were created.The possible binding modes of PHL to pepsin are shown in Fig.1(A,B).PHL is surrounded by amino acid residues of Gly-217,Asp-32,Thr-77,Met-289,Pro-292,Ile-73,Ser-35,Asp-215,Gly-34,Tyr-75,Tyr-189,Thr-218.PHL binds to the catalytically active amino acid residues Asp-32 and Asp-215 of pepsin occupying the cavity of the protease[38].This may have a certain influence on the activity of pepsin.In addition,Thr,Ser,Tyr,Ile,and other amino acids are all hydrophobic amino acid residues.Therefore,there may exist van der Waals forces in the interaction.Hydrogen bonds also exist between PHL and pepsin residues Gly-76 (3.22 Å) and Ile-128 (2.98 Å).Interactions involving hydrogen bonds and van der Waals forces contribute to the improved conformational stability.The possible ways of binding PHL to trypsin are displayed in Fig.1C and 1D.The PHL is surrounded by amino acid residues of Gly-219,Trp-215,Asp-189,Tyr-228,Ser-214,Gly-226,Val-213,Gly-216,Ser-190,Val-227,Leu-99,Thr-98,Gln-192,Gln-192,Ser-195.Asn-97 (2.89 Å) was attached to the PHL by hydrogen bonding,which increased the binding affinity of the PHL to trypsin.The active sites of trypsin are Asp-102,His-57,and Ser-195.It can be observed that the PHL binds directly to the Ser-195 of trypsin.This suggests that PHL binding to trypsin may affect the activity of trypsin.Li also found something similar when he studied the procyanidin B3 with trypsin[39].In summary,hydrogen bonds,van der Waals forces and electrostatic forces are mainly involved in the binding of PHL to proteases.The optimal combination of the energy of PHL with trypsin (7.90 kcal/mol) is smaller than that of PHL with pepsin (8.11 kcal/mol).It can be seen from the optimal combination of energy that the binding of PHL to trypsin is more stable.In the presence of both proteases,PHL may preferentially bind to trypsin.Binding patterns predicted by molecular docking help better understand the binding mechanism.
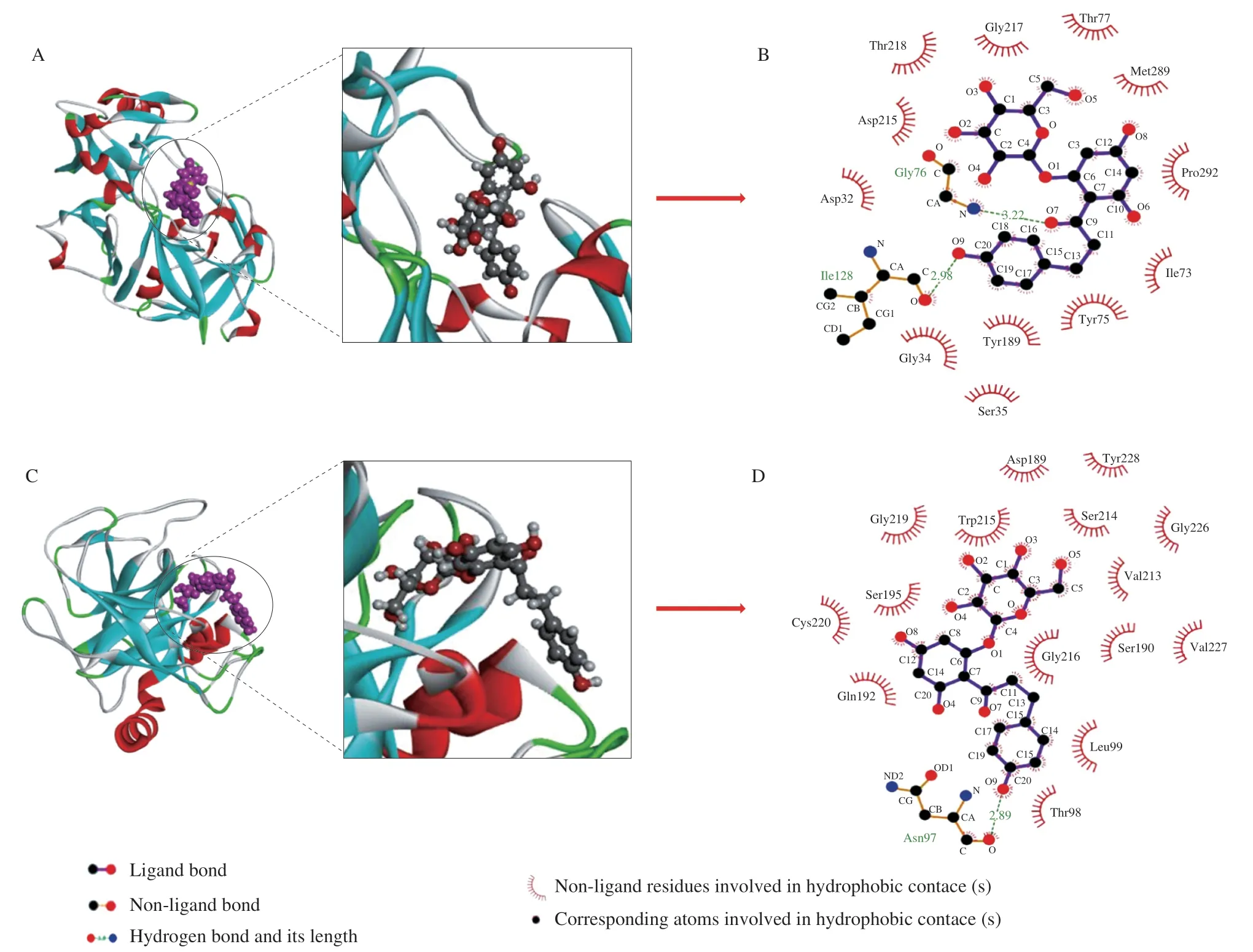
Fig.1 3D and 2D plots of the optimal binding sites for the PHL with pepsin (A,B) and trypsin (C,D).
3.2 MD studies
MD allows the analysis of the conformational changes of PHL with pepsin/trypsin over time and the stability.The results of root mean square deviation (RMSD),radius of gyration (Rg),and root mean square fluctuation (RMSF) is used to analyze the binding of PHL and pepsin/trypsin (Fig.2).RMSD can measure the stability of the system.The RMSD of the PHL and pepsin system fluctuated with time in the simulation of 0-70 ns,suggesting that the secondary structure of the protease may change after PHL binding to pepsin.The average RMSD value of 1.99 Å for free pepsin was lower than the average RMSD value of 2.69 Å after the addition of PHL,which may be due to the addition of PHL resulting in an unconsolidated pepsin conformation.The average RMSD value of the trypsin system after the addition of PHL (1.89 Å) was higher than that of trypsin(1.46 Å),which speculated that PHL will bind to protease effects the stability of the system.In contrast,RMSD changes less after PHL binding to trypsin.PHL is more stable with the trypsin system.Rg could be obtained as the change in protein tightness[40].The average Rg values for the pepsin system without and with PHL were 19.74 Å and 20.42 Å,respectively.The average Rg values for the trypsin system were essentially unchanged (16.39 Å and 16.83 Å).Overall,the bending radius of the proteases remained almost unchanged and PHL did not affect the tightness of the proteins.The RMSF shows the effect on the number of atoms and can understand the elasticity of the protein[41].The RMSF values of pepsin residues/trypsin were slightly lower than those of PHL and pepsin/trypsin system residues (Fig.2E and 2F)).The stability of the system decreased after the binding of PHL to pepsin/trypsin,which corroborates with the findings of the RMSD analysis.
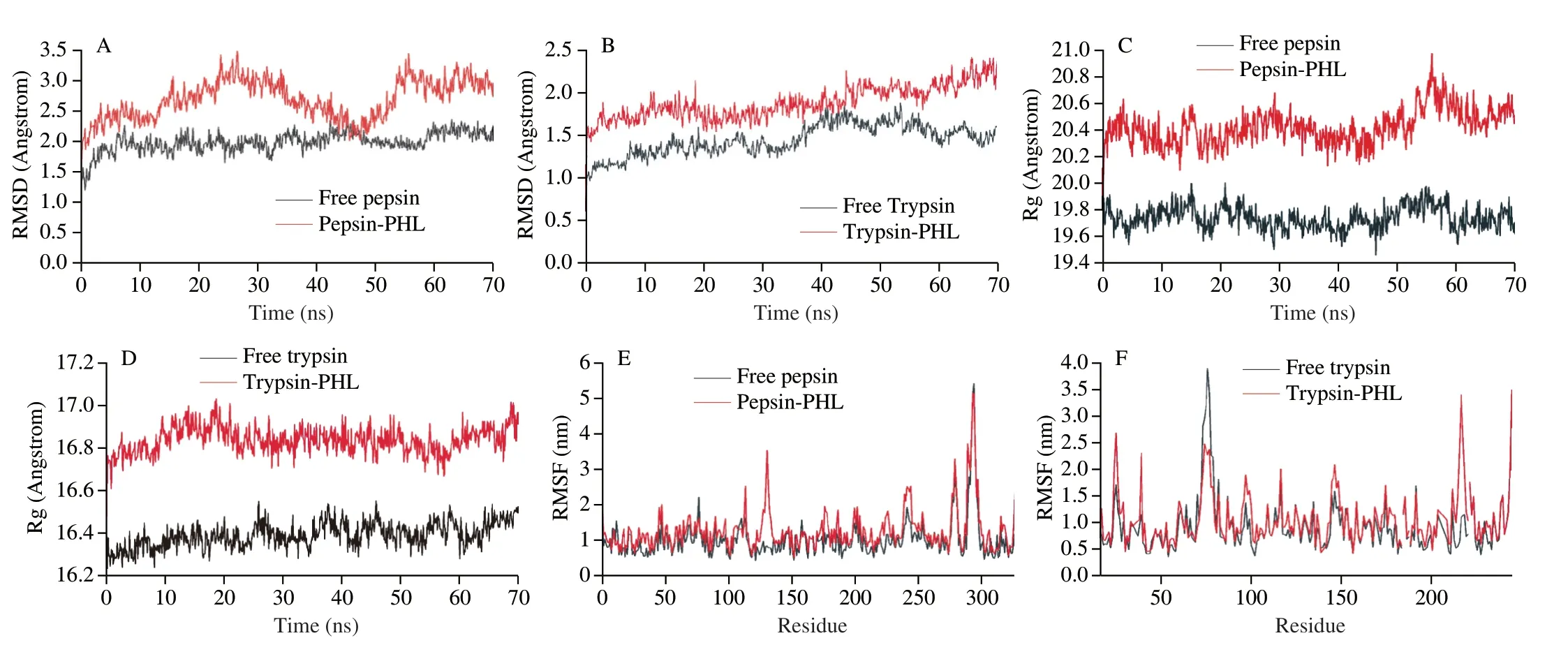
Fig.2 RMSD (A,B),Rg (C,D),and RMSF (E,F) of free pepsin,free trypsin,PHL with the pepsin/trypsin complex system.
MD experiments can also compare free pepsin/trypsin and changes in protease secondary structure content after adding PHL.Structures of both proteases areβ-sheet,which are verified by the FT-IR results (Fig.3).The structural change trends of the two proteases were different after the addition of PHL at 70 ns.Pepsinβ-sheet content reduced,whereas theβ-turn content grew somewhat,while trypsinβ-sheet content declined.It can be speculated that the process of PHL binding to pepsin/trypsin impacts the conformation of the protease to become more unconsolidated.
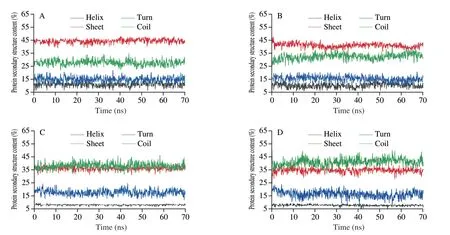
Fig.3 Secondary structural changes of free pepsin/trypsin (A,C) and pepsin/trypsin complex system with PHL (B,D).
3.3 Fluorescence quenching studies
Steady-state fluorescence is a typical technique for studying small molecule protein interaction.This method can provide information alteration in the protease microenvironment.The fluorescent moieties of pepsin and trypsin consist of multiple Try residues and Tyr residues.PHL has no fluorescence and will not interfere with experimental measurements.The fluorescence spectrum at 25 °C is described in Fig.4A and 4B.The pepsin/trypsin has a strong absorption fluorescence peak of around 340 nm.Meanwhile,the fluorescence intensity of both proteases was gradually quenched as the PHL concentration increased.The maximum emission wavelength of pepsin/trypsin had different degrees of red shift after the addition of PHL.It showed PHL might bind with Try or Tyr amino acids of pepsin/trypsin.The red shift distance of trypsin after the binding was greater than pepsin indicating a more pronounced effect of PHL on trypsin.This may influence the digestive effect of PHL.
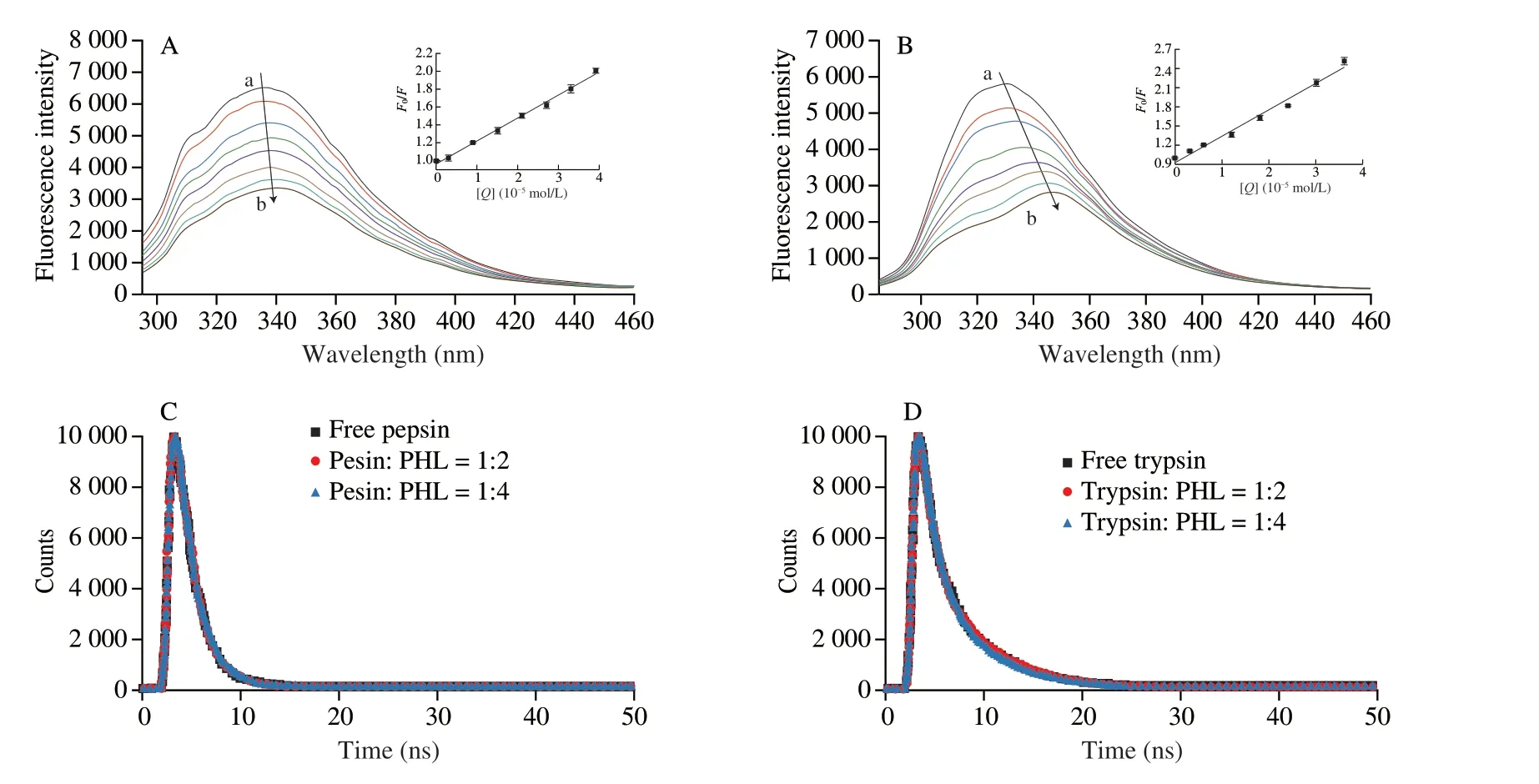
Fig.4 Fluorescence emission spectra of pepsin (3.0 × 10-6 mol/L) bounding of PHL at 25 °C (concentration ratio of pepsin to PHL=1:0,1:1,1:3,1:5,1:7,1:9,1:11,1:13) (A).Fluorescence emission spectra of pepsin (6.0 × 10-6 mol/L) bounding of PHL at 25 °C (concentration ratio of pepsin to PHL=1:0,1:1,1:2,1:3,1:4,1:5,1:6,1:7) (B).Time-resolved fluorescence decay curves of pepsin/trypsin in the presence and absence of PHL (C,D).The inset is the Stern-Volmer plots of interaction for PHL with protease at 25 °C.
Time-resolved fluorescence can effectively judge static and dynamic fluorescence quenching.The fluorescence lifetime of the protein upon static quenching is not disturbed during complex formation[42].The time-resolved fluorescence decay curves of pepsin/trypsin with and without PHL were similar in shape (Fig.4C and 4D).From the decay parameters can be seen that theχ2value is less than 1.3,indicating that the data fit values are good and acceptable[43].The average fluorescence lifetimes of pepsin/trypsin alone were 4.48 ns and 1.98 ns,respectively.This is consistent with previous studies with theτvalues[44,45].Theτvalues after PHL binding with pepsin and trypsin were 4.38 ns and 1.99 ns,respectively.Theτvalues of free protease and PHL remained essentially unchanged,indicating that PHL binding with proteases was static quenching.This further corroborated the consequences of fluorescence titration.
3.4 Binding force and thermodynamic parameter studies
Recording the fluorescence spectra of PHL under three temperature conditions and calculating theKaandnfor PHL and pepsin/trypsin.Thenof both systems is close to 1,indicating that PHL binds to the protease with one active binding site.TheKavalues at 4 °C,25 °C,and 37 °C were (0.115 ± 0.047) × 105L/mol,(0.068 ± 0.125) × 105L/mol,(0.021 ± 0.048) × 105L/mol between PHL with pepsin,and (1.143 ± 0.027) × 105L/mol,(0.465 ± 0.124) × 105L/mol,(0.371 ± 0.175) × 105L/mol between PHL with trypsin,respectively (Table 1).It is shown that the binding affinity of the basal complex formed by PHL with protease was moderate.The binding constant of PHL interacting with trypsin was greater than PHL with pepsin,revealing that PHL was stronger bound to trypsin than to pepsin.Perhaps PHL preferentially binds to trypsin during digestion.
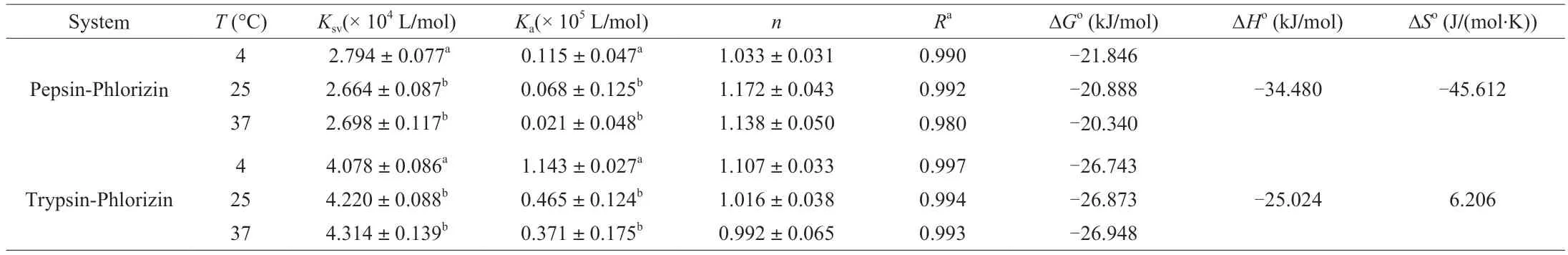
Table 1 Binding parameters and thermodynamic parameters for the PHL with pepsin/trypsin systems at different temperatures.
The type of interaction force between compounds and proteases is related to ΔG°,ΔH°,and ΔS°.Thermodynamic parameters were calculated.The values of ΔG° were negative (Table 1) indicating that the reaction of PHL to pepsin/trypsin is spontaneous exothermic.PHL binds readily to pepsin/trypsin,which is maybe beneficial for the digestion and absorption of PHL.When PHL binds to pepsin,ΔH° <0 and ΔS° <0 (-34.480 kJ/mol and -45.612 J/(mol·K)),depicted that Van der Waals forces and hydrogen bonding are the main binding forces in the binding process.ΔH° <0 and ΔS° >0(-25.024 kJ/mol and 6.206 J/(mol·K)) when PHL bound to trypsin.It can be determined that the force in the interaction of PHL and trypsin is mainly electrostatic.
3.5 Conformational changes
Synchronous fluorescence is associated with protein tyrosine(Tyr) and tryptophan (Trp) residues with a difference of 15 nm and 60 nm in emission and excitation wavelengths[46],respectively.The synchronous fluorescence spectra as Fig.5A−D of the binding ofPHL and pepsin/trypsin.The peak of pepsin/trypsin Tyr remained stable when PHL concentration increased,whereas the peak position of Trp was a blue shift and red shift,respectively.This revealed that Trp residues played a crucial role in PHL-protease interaction.The binding of PHL to pepsin resulted in a reduced polarity and enhanced hydrophobic environment for the Trp,while the environment around Tyr remained intact.The opposite change was observed for trypsin.
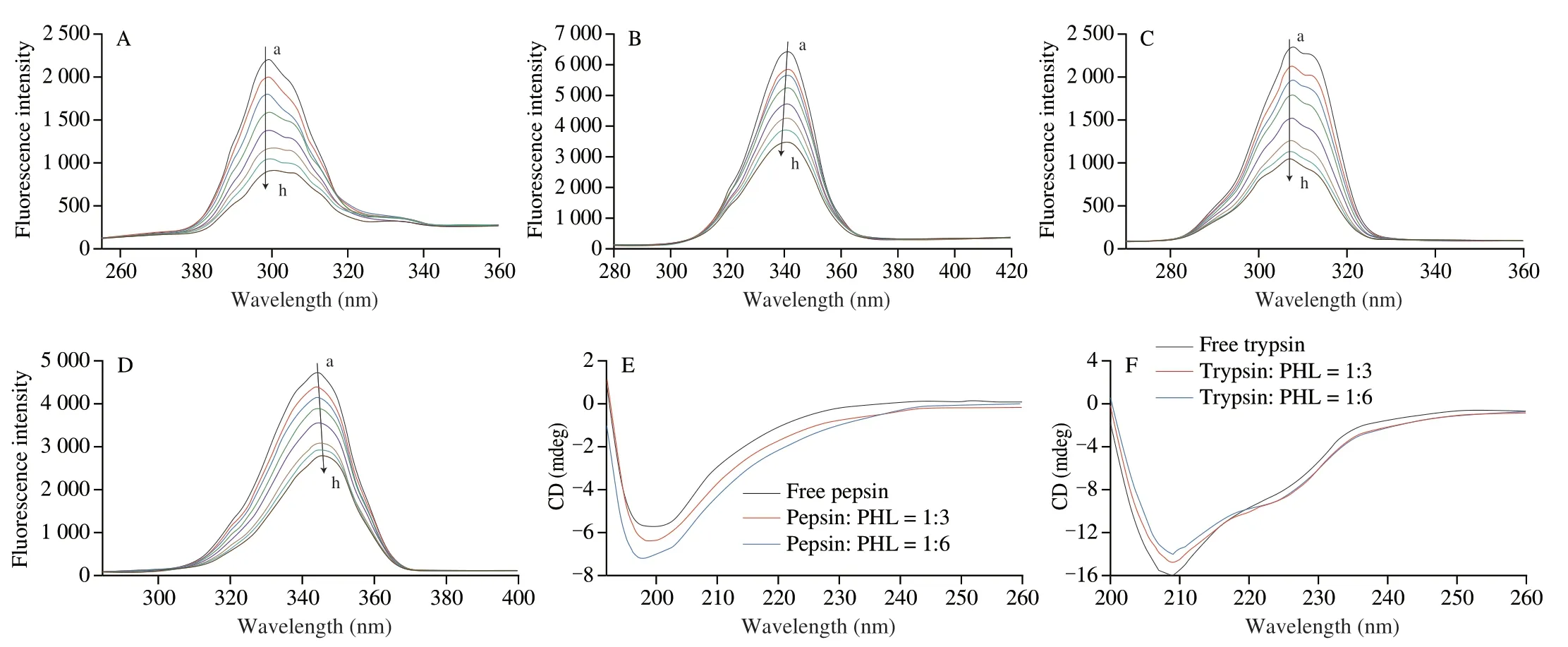
Fig.5 The synchronous fluorescence spectra of different concentrations of PHL with pepsin/trypsin Δλ=15 nm (A,C) and Δλ=60 nm (B,D) at 25 °C,respectively.CD spectra of free pepsin/trypsin and PHL with pepsin/trypsin complex system at 25 °C (E,F).
CD spectra is a sensitive and fast tool for analyzing protein secondary structure changes.The CD spectra of PHL with pepsin/trypsin are demonstrated in Fig.5E and 5F. Free pepsin has a negative band at 200 nm,which is a characteristic peak of random curl.Indicated that there are manyβ-sheets and random coils in the structure of pepsin[47].The band peak shifted slightly after PHL was bound to the protease,which indicated that the secondary structure of the protease changes after PHL was bound to pepsin.Free trypsin has a peak at 209 nm.The intensity of the peak changed slightly but the position of the peak does not change after the addition of PHL,presumably that the structure of trypsin is still dominated byβ-sheets[48].Simultaneous fluorescence measurements have also led to such conclusions.
The conformational changes of the protease after PHL binding to pepsin/trypsin can also be assessed by FT-IR.The amide I band(1 500-1 600 cm-1) of the protein is mainly C=O stretching and the amide II bands (1 600-1 700 cm-1) are mainly C-N stretching and N-H bending[49].There are two absorption peaks for pepsin/trypsin with and without PHL (Fig.6A and B).The positions of the main peaks of the protease amide bands were shifted after the addition of PHL.It is suggested that the PHL interacts with proteases.Fig.6C−F showed the fitted spectra of the protease amide I band (1 600-1 700 cm-1).The content of the secondary structure was estimated by analyzing the peak area.After the addition of PHL,theβ-sheet content of pepsin decreased (2.58%) less than that of trypsin (6.27%).The comparative analysis found that PHL had an efficient effect on trypsin.
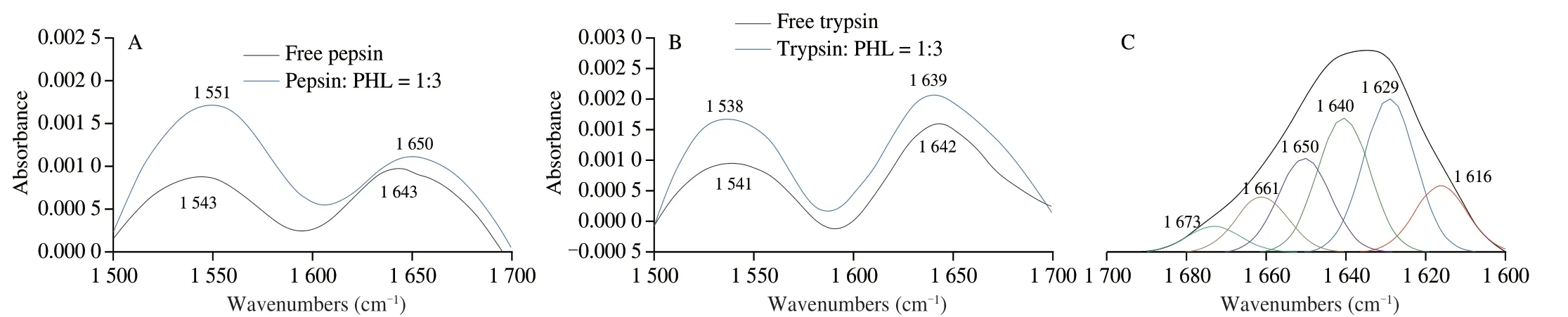
Fig.6 FT-IR spectra of pepsin (5.0 × 10-5 mol/L)/trypsin (2.0 × 10-5 mol/L) in the presence and absence of PHL at the region of 1 500-1 700 cm-1 (A,B).Secondary structural changes of free pepsin/trypsin (C,D) and pepsin/trypsin complex system with PHL (E,F).

Fig.6 (Continued)
Overall,the addition of PHL had a slight effect on the structure of the protease,but the main structure of the protease was not disrupted,which may affect some of the physiological functions of the protease.The comparative analysis provides theoretical guidance on PHL binding to proteases during digestion and absorption to understand the interaction of PHL with proteases during digestion and absorption.
3.6 Antioxidant studies
The antioxidant capacity of PHL was detected using the ABTS and FRAP methods.The antioxidant capacity of flavonoids is related to the hydroxyl group,and deprotonation of the hydroxyl group affects the free radical scavenging of the flavonoid[50].Fig.7 showed that PHL has a better ABTS cation radical scavenging rate and FRAP reducing ability.The antioxidant capacity showed a significant quantitative-effect relationship with its concentration.As we all know that the scavenging activity of flavonoid compounds is also related to pH,and higher pH will increase the scavenging[51,52].The ABTS cation radical scavenging rate of the PHL and trypsin system is higher than PHL alone (P<0.05),which was attributed to the deprotonation of PHL.The bound of PHL with trypsin may increase its antioxidant capacity.The ABTS cation radical scavenging rate was significantly reduced after the binding of PHL and pepsin(P<0.05).The binding of PHL with pepsin may interfere with the hydroxyl ionization of PHL,leading to a decrease in its antioxidant capacity.The results of the FRAP methods to determine the antioxidant capacity were similar to the ABTS assay.Ren[53]also reported similar results that the antioxidant activity of aloe-emodin was reduced after aloeemodin was combined with pepsin.
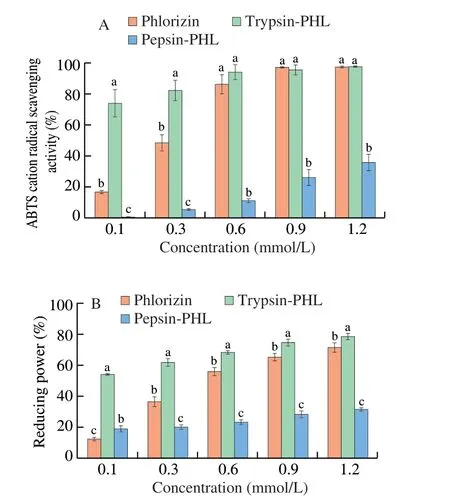
Fig.7 ABTS scavenging rate (A) and reduction capacity (B) of PHL and PHL with pepsin/trypsin complex system.Different letters (a-c) in the figure indicate significant differences (P < 0.05).
4. Conclusions
This work comparatively analyzed the binding modes and antioxidant changes of PHL to pepsin/trypsin.Molecular docking experiments demonstrated the binding of PHL to amino acids Asp-32 and Asp-215 of pepsin.PHL interacts with amino acids Ser-195,the catalytically active amino acid of trypsin.The binding of PHL to pepsin/trypsin was spontaneous.Synchronous fluorescence spectra revealed that the polarity of Trp declined after PHL interacted with pepsin,and the polarity of Trp was enhanced after PHL was bound to trypsin.CD and FT-IR spectra results proved a conformation change of the protease after PHL binding to pepsin/trypsin.In addition,it provides an explanation for the changes in the antioxidant activity of PHL after PHL binding to pepsin/trypsin.Ingested fruit flavonoids are generally absorbed in the small intestine.PHL binds more readily and firmly to trypsin than pepsin,which may facilitate the digestion and absorption of PHL.Moreover,the digestion and absorption of fruit flavonoids are also related to the regulation of the microbiota and other factors,which are affected by various factors.All experiments in this study were performedinvitro,and the results provide a reference forinvivostudies of PHL.Invivostudies will provide more information on the digestion and absorption of PHL in future work.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (21808020) and the Applied Basic Research Program of Science &Technology Department of Sichuan Province(2018JY0151).
- 食品科学与人类健康(英文)的其它文章
- Modifications in aroma characteristics of ‘Merlot’ dry red wines aged in American,French and Slovakian oak barrels with different toasting degrees
- Effect of different drying methods on the amino acids,α-dicarbonyls and volatile compounds of rape bee pollen
- Dynamic changes in physicochemical property,biogenic amines content and microbial diversity during the fermentation of Sanchuan ham
- A comparison study on structure-function relationship of polysaccharides obtained from sea buckthorn berries using different methods:antioxidant and bile acid-binding capacity
- Yolk free egg substitute improves the serum phospholipid profile of mice with metabolic syndrome based on lipidomic analysis
- Underlying anti-hypertensive mechanism of the Mizuhopecten yessoensis derived peptide NCW in spontaneously hypertensive rats via widely targeted kidney metabolomics

