Characterization of blueberry exosome-like nanoparticles and miRNAs with potential cross-kingdom human gene targets
Yngfn Leng,Liubin Yng,Siyi Pn,b,Leilei Zhn,Fng Yun,b,
a College of Food Science and Technology, Huazhong Agricultural University, Wuhan 430070, China
b Hubei Key Laboratory of Fruit &Vegetable Processing &Quality Control, Huazhong Agricultural University, Wuhan 430070, China
c Wuhan GeneCreate Biological Engineering Co., Ltd., Wuhan 430070, China
Keywords: Edible plant derived exosomes-like nanoparticles Size exclusion chromatography miRNA Target gene prediction Blueberry
ABSTRACT Edible plant derived exosome-like nanoparticles (ELNs) have been shown to have multiple nutraceutical functions.However,the diversity of plant materials makes the plant derived ELN study challenging.More efforts are still needed to explore the feasible isolation methods of edible plant derived ELNs and the possible roles of food-derived ELNs in improving human health.In this study,a size exclusion chromatography based method was compared with the traditional ultracentrifugation method to isolate blueberry derived ELNs(B-ELNs),and the miRNA prof ile of B-ELNs was analyzed by high-throughput sequencing.A total of 36 miRNAs were found to be enriched in B-ELNs compared with berry tissue,and their potential cross-kingdom human gene targets were further predicted.Results showed that size exclusion chromatography was effective for B-ELN isolation.The most abundant miRNAs in B-ELNs mainly belonged to the miR166 family and miR396 family.Target gene prediction indicated that B-ELNs could potentially regulate pathways related to the human digestive system,immune system and infectious diseases.
1.Introduction
The vesicles released by eukaryotic cells into the extracellular space are called extracellular vesicles (EVs),which are usually divided into different types according to their size[1].With a diameter from 30 nm to 150 nm,exosomes are nanoscale vesicles containing lipids,membrane and cytosolic proteins,as well as nucleic acids (RNA/miRNA),and play important roles in intercellular communication[2].In 2013,Ju et al.[3]identif ied edible plant derived exosomes-like nanoparticles (ELNs) from grapes.After that,ELNs were successively isolated from many other edible plants,such as grapefruit[4],apple[5],ginger[6],carrot[7],lemon[8],and coconut[9].Plant derived ELNs were found to be nano-sized vesicles in the saucer or cup shape with the phospholipid bilayer structure,which had a diameter ranging from 50 nm to 300 nm[10],containing up to hundreds of different lipids,miRNAs,and proteins[11],and were also found to have potential positive functions for human health.For example,ginseng-derived ELNs could promote the tumor-supportive M2-like phenotype transform into a tumoricidal M1-like state in the tumor-associated macrophages to inhibit melanoma growth[12].Another study reported that ginger derived ELNs could improve intestinal barrier function and alleviate colitis[13].Ju et al.[3]found that grape ELNs could accelerate intestinal epithelial proliferation to promote recovery from colitis.Broccoli-derived ELNs played a crucial role in inhibiting mouse colitis[14].ELNs from sweet oranges could modulate the permeability of intestinal epithelial cell and the expression of anti-inflammatory genes and tight junction genes[15].Likewise,ELNs from lemon could be an edible chemotherapeutics delivery vehicle and be applied for gastric cancer therapy[16].Abraham et al.[17]demonstrated that the ELNs in cucumbers had a skin-improving effect.These studies provided evidence that some edible plant derived ELNs had potential human health-promoting functions.Because of the favorable biosafety and targeting capability,edible plant derived ELNs are promising new bioactive components to be applied in food industries.
However,since plant derived ELNs are different from animal exosomes in many biological aspects,several basic problems are still unsolved.Firstly,the reported extraction and isolation methods for plant derived ELNs were mainly derived from the exosome extraction procedures used for animal biofluids,including ultracentrifugation(UC),ultrafiltration,size exclusion chromatography (SEC)[18],and microfluidic technique[19].Among them,UC was one of the most widely used methods to extract plant derived ELNs[20-22].Recently,a PEG 6000-based precipitation method has been reported to selective enrich ginger derived ELNs,offering a new cost-effective way to isolate edible plant derived ELNs[23].Because of the diverse species of plants,the compositions of plant tissue or juice are more complicated than mammalian cells.The isolation and purification of plant derived ELNs remain challenging.Moreover,there is still a need to evaluate these techniques for edible plant derived ELNs extraction,which is crucial to the downstream analysis and application.
The second problem is that little transcriptomic and proteomic information about plant derived ELNs is available,which makes it hard to understand plant derived ELNs’ biological functions[24].Among the cargos of plant derived ELNs,microRNAs (miRNAs),a class of small (19-24 nt) noncoding RNAs,have gained the most interest because they can act as efficient gene expression regulators[25]by directing mRNA cleavage[26].For example,rice-derived miR168a could specifically target and regulate LDLRAP1 expression in mouse livers[27].Plant miR159 could inhibit breast cancer cell growth by targetingTCF7gene[28].It is highly possible that the functions of edible plant derived ELNs dependon the miRNA they carried.Moreover,miRNA profiles in plant derived ELNs are different between plant species or even cultivars,indicating that ELNs from different edible plant sources may have different regulating functions.
Blueberry (Vacciniumspp.) is a popular small fruit,which is rich in vitamins,dietary fiber,and biologically active substances[29].Blueberries have been reported to have multiple health-benefit functions such as oxidative stress regulation,anti-inflammation,and cholesterol regulation[30].Blueberry derived ELNs (B-ELNs),which naturally exist in blueberry,could be a promising bioactive component of blueberry for mammals’ health,and as a potential nano delivery system for nutraceutical compounds in the future.This study aimed to identify and characterize ELNs from blueberries.Two extraction methods,UC and SEC,were compared and small RNA sequencing was used to identify miRNAs with potential cross-kingdom human gene targets.
2.Materials and methods
2.1 Pretreatment of blueberry juice
Blueberry samples were picked from a local blueberry orchard in Wuhan,Hubei,China (N 31°60’,E 114°05’).Fruits from northern highbush blueberry (Vaccinium corymbosumL.) ‘Legacy’ were hand harvested in June of 2021.The fresh berries (250 g) were washed with distilled water and blended into a puree.The puree was diluted with 1 000 mL phosphate buffered saline solution (PBS) and well mixed.The mixture was sequentially centrifuged at 3 000 ×gfor 10 min,8 000 ×gfor 30 min,and 10 000 ×gfor 60 min at 4 °C to remove large particles,dead cells,and cell debris respectively (Fig.1).The supernatant was passed through a 0.22 µm filter and concentrated with the 10 000 MWCO membrane,and the clear juice obtained was used for further B-ELN isolation.
2.2 B-ELN isolation by ultracentrifugation (UC)
B-ELNs in the clear juice were isolated by ultracentrifugation at 4 °C at 150 000 ×gfor 90 min.The precipitant was washed with PBS,and then resuspended in 1 mL PBS and stored at -80 °C for further analysis.Isolation was performed in triplicates.
2.3 B-ELN isolation by size exclusion chromatography (SEC)
A commercial qEV column (Izon Science,Canterbury,New Zealand) was used for B-ELNs isolation.The column was firstly conditioned with 69.3 mL of PBS.Then,10 mL of clear blueberry juice was loaded,and 10 mL of the eluate was collected as fraction 1.Then,21.4 mL PBS was added to the column and the 21.4 mL eluate was collected as fraction 2.Finally,50 mL PBS was added to the column and 20 mL eluate was collected as fraction 3.Since B-ELNs were most abundant in fraction 2,so fraction 2 was further concentrated by 10 000 MWCO membrane and stored at -80 °C for further analysis.Isolation was performed in triplicates.
2.4 Dynamic light scattering (DLS) analysis
The size distribution profile of B-ELN isolates by UC and SEC was determined by DLS using a Zetasizer Nano-ZS 90 size analyzer(Malvern Instruments Ltd.,Malvern,UK) at 25 °C.Backscatter detection was used.The scattered light is collected at 173 °C.Decay times are used to determine the distribution of the diffusion coefficientsD0of the particles,which,in turn,can be converted into a distribution of apparent hydrodynamic diameter,Dh,using the Stokes-Einstein relationshipDh=KBT/3πηD0,where KBis the Boltzmann constant,Tis the absolute temperature andηis the solvent viscosity.The water was assumed that of the solvent viscosity[15].Intensity particle size distribution was converted into Volume particle size distribution (volume distribution) through Mie theory,and this volume distribution was further converted into Number particle size distribution (number distribution) using the Zetasizer software.
2.5 Transmission electron microscopy (TEM)
TEM was employed to characterize the morphology of B-ELNs.TEM analysis was performed according to the published method with minor modifications[31].Briefly,the B-ELNs were added to copper grids at room temperature.Then negative staining was applied to B-ELNs for 30 s,using 1% aqueous phosphotungstic acid (PTA)solution (pH 6.4-7.0),positioned at room temperature until the PTA solution was dried completely.Measurements were carried out by using an FEI TECNAI 12 G2 Twin,operating at 80 kV and equipped with an electron energy loss filter and a slow-scan charge-coupled device camera (Hitachi,Tokyo,Japan).
2.6 Total protein analysis
The total protein concentration of B-ELN isolates was determined using the BCA protein assay kit (Nanjing Jiancheng Bioengineering Institute,Jiangsu,China) according to the manufacturer’s instructions.Briefly,BCA reagent A was mixed with reagent B (A:B=50:1) to prepare a BCA working solution.Then the B-ELNs suspension was mixed with 250 μL BCA working solution and incubated for 30 min at 37 °C.Absorption was measured by a Multskan GO microplate reader (Thermo,MA,USA) at the wavelength of 562 nm.Each sample was assayed in triplicate.
2.7 Total RNA preparation
Total RNAs in B-ELNs and blueberry tissue were prepared for small RNA sequencing.RNA of both B-ELNs and tissue was extracted using the TRIzol method.Briefly,TRIzol reagent(Invitrogen,Carlsbad,CA,USA) was added to the B-ELN isolates,and chloroform was added to the mixture.Then,the sample was centrifuged at 12 000 ×gfor 15 min and the aqueous phase was carefully collected.Isopropanol and ethanol were added to let RNAs settle.After RNA extraction,the degradation and contamination of RNA were verified using 1% agarose gels.The purity was determined using a NanoPhotometer spectrophotometer (IMPLEN,CA,USA).The integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies,CA,USA).
2.8 Small RNA sequencing for B-ELNs
The small RNA libraries were constructed using NEBNext®Multiplex Small RNA Library Prep Set for Illumina®(NEB,USA).The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TRuSeq SR Cluster Kit v3-cBot-HS (Illumina Inc.CA,USA) and sequenced on a Novaseq 6000 platform.To ensure the quality of information analysis and obtain clean reads,raw reads of FASTQ format were filtered by custom Perl and Python scripts to remove reads containing ploy N,with 5’ adapter contaminants,without 3’ adapter or the insert tag,containing ploy A or T or G or C,reads smaller than 18 nt,and low-quality reads.Clean reads with a certain length range were chosen for downstream analyses.
2.9 Known miRNA alignment and novel miRNA prediction
Filtered reads (18-30 nt) were mapped to their reference genome and analyzed using the bowtie package.The mapped reads were aligned with the miRBase22.0 database to identify known miRNA.MirDeep2’s quantifier.pl was used to obtain the miRNA counts.Custom scripts were used to obtain base bias on the first position of identified miRNA with a certain length and each position of all identified miRNA respectively.
Before novel miRNA prediction,reads originated from protein-coding genes,repeat sequences,rRNA,tRNA,snRNA,and snoRNA were removed.Small RNA reads were identified as ncRNA,including rRNA,tRNA,snRNA,and snoRNA,based on genome annotation or Rfam database,and repeat sequences were identified and removed using RepeatMasker.Some small RNA were actual mRNA fragments,as so reads mapped to the intron or exon of genes were removed.The hairpin structure of miRNA precursors can be used to predict novel miRNA.The software miREvo[32]and miRDeep2[33]were integrated to predict novel miRNA through exploring the secondary structure,the Dicer cleavage site and the minimum free energy of the small RNA reads unannotated in the former steps.At the same time,MirDeep2’s quantifier.pl was used to obtain the miRNA counts,and custom scripts were used to obtain base bias on the first position of identified miRNA with a certain length and each position of all identified miRNA respectively.
2.10 Differential expression of miRNA
The miRNA expression levels were estimated by TPM (transcript per million) through the normalization formula as described previously[34].Differential expression analysis of B-ELNsvs.Tissue was performed using the DEGseq (1.38.0) R package.P-value was adjusted usingq-value[35].The tissue sample was set as a reference.CorrectedP-value < 0.05 and log2(fold change) > l were set as the threshold for screening differentially expressed miRNAs.
2.11 miRNA target gene prediction and A67nnotation
The human genome was used as a reference genome for target gene prediction.Miranda3.3 software was employed to predict the target human genes.MiRanda algorithm is based on a comparisone of miRNAs complementarity to 3’UTR regions.The binding energy of the duplex structure,evolutionary conservation of the whole target site and its position within 3’’UTR are calculated and account for a final result which is a weighted sum of match and mismatch scores for base pairs and gap penalties.Only differentially expressed miRNAs(B-ELNsvs.Tissue) were selected for prediction with the following software parameters: -sc168-en-10-scale4-strict-out.OMICSHARE cloud platform (http://www.omicshare.com/tools/Home/Soft/pathwaygsea) was used to perform Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis for all miRNA target genes.
2.12 Quantitative real-time PCR
Six miRNAs were selected to verify the sequencing and computational results,and quantitative real-time PCR (qRT-PCR)was performed.TheGAPDHgene was used as control and specific primers were designed based on cDNA sequences (Table S1).The qRT-PCR was performed on a Bio-Rad S1000 with Bestar SYBR Green RT-PCR Master Mix (TOYOBO).Relative gene expression was calculated using the Livak and Schmittgen 2-ΔΔCtmethod[36],normalized with the reference gene GAPDH.PCR amplifications were performed in triplicate for each sample.
2.13 Statistical analysis
Statistical analysis was performed using SPSS 22.0 (SPSS,Chicago,IL,USA).Heatmaps and correlation coefficient dendrograms were visualized by TBtools (v1.09873)[37].All data were representative of three independent experiments and statistical differences were considered significant at*P< 0.05 or**P< 0.01.Image processing was performed using Origin Pro 2019b (Origin Lab Corporation,Northampton,MA,USA).
3.Results
3.1 B-ELNs extracted by UC and SEC methods
In this study,B-ELNs were isolated from blueberry juice through UC and SEC methods respectively (Fig.1).B-ELNs isolated by both methods were compared in the aspects of size distribution,morphology,and protein content.Since there were no published methods of SEC based isolation of plant derived ELNs,we preliminarily collected three fractions from SEC to determine the best fraction.The results showed that SEC could effectively separate the particles from blueberry juice according to the particle size.The average diameter of fraction 1 was 289 nm,fraction 2 was 119 nm,and fraction 3 was 18.6 nm (Fig.2).The TEM imaging showed that the vesicular structures mainly existed in fraction 2 (Fig.3F).So fraction 2 was collected for further experiments.
Characterizations of B-ELNs obtained via UC and SEC methods were compared (Fig.3).Both intensity and number distributions of B-ELNs were compared because they can provide different information.Specifically,since we were focusing on a smaller size range,and number distribution could provide a better estimation of B-ELNs’ diameter.While considering the possible aggregation in the isolates,intensity distribution could provide additional information.From the number distribution,the average diameter of B-ELNs extracted through UC and SEC was (169 ± 12) and (119 ± 52) nm(n=3),respectively,which was more comparable with the observation by TEM imaging (Figs.3C and F).When considering the intensity distribution of the isolates obtained by UC and SEC,both methods resulted in bimodal intensity distribution.The results indicated that number distribution was a better way for B-ELN size estimation.But for both methods,it was inevitable to include some larger particles,which were speculated to be the aggregates of proteins.The number of aggregates was obviously more in the UC extract than in SEC.Protein analysis also showed that the protein content in UC samples was approximately ten times higher than that of SEC,possibly because the SEC method was more effective to remove protein contaminants (Fig.3G).
3.2 Small RNA sequencing result
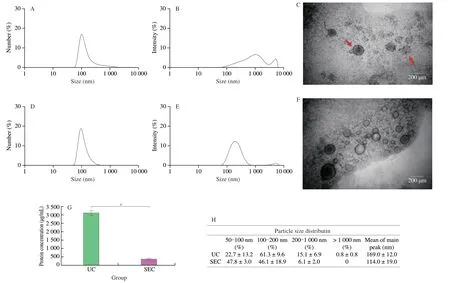
Fig.3 Characterization of B-ELNs extracted by UC and SEC.(A) Size distribution by number of B-ELNs isolated by UC;(B) Size distribution by intensity of B-ELNs isolated by UC;(C) TEM image of the B-ELNs isolated via UC (the red arrows showed aggregated B-ELNs);(D) Size distribution by number of B-ELNs isolated by SEC;(E) Size distribution by intensity of B-ELNs isolated by SEC;(F) TEM image of the B-ELNs isolated via SEC;(G) Protein concentration of B-ELNs as measured via BCA Protein Assay Kit;(H) Particle size distribution of B-ELNs after UC and SEC isolation.Data are represented as the mean ± standard error,statistical differences were considered significant at *P < 0.05.
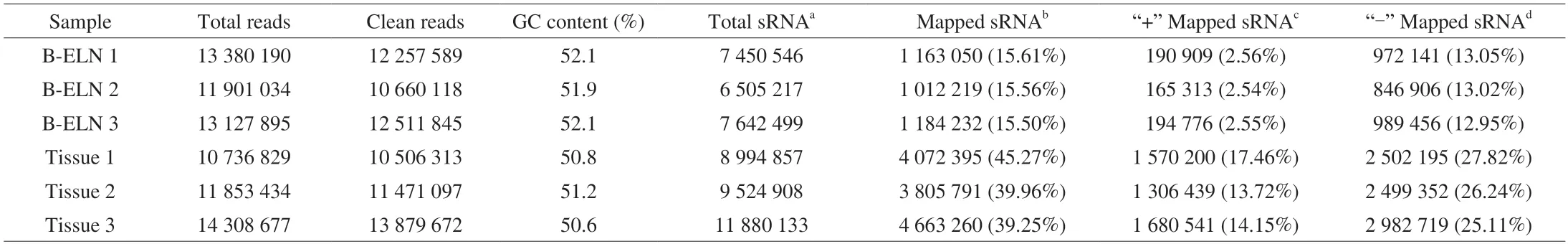
Table 1 Summary of small RNA sequencing data.
The extracted B-ELNs were subjected to small RNA sequencing.To better understand the unique miRNAs in B-ELNs,we use the berry tissue RNA as a reference.Six small RNA libraries (two samples and three biological replicates) were prepared (Table 1).Filtered reads of length at 18-30 nt were mapped.The mapped small RNA in B-ELNs ranged from 1 012 219 to 1 184 232,while mapped small RNA in tissue ranged from 3 805 791 to 4 663 260.The small RNAs in blueberry tissue were dominantly distributed between 24 and 21 nt,which was in accordance with the length distribution of general plant small RNAs[38-39].However,in B-ELNs,the most abundant small RNAs were distributed in 29-30 nt range (Fig.4A).Small RNA annotation revealed several types of small RNAs in the samples,which were distributed differently in B-ELNs and tissue (Figs.4B and C).Among the small RNAs in B-ELNs,0.25%-0.31% were annotated as known miRNAs and 0.08%-0.09% were annotated as novel miRNAs.In blueberry tissue,0.13%-0.17% of small RNAs were annotated as known miRNAs and while 0.15%-0.16% were novel.
3.3 Differential expression analysis of miRNA between tissue and B-ELNs
A total of 318 miRNAs were identified in B-ELN and tissue samples.Among them,126 miRNAs were commonly found in both samples.While 14 miRNAs were only present in B-ELNs,178 were only found in tissue (Fig.5A and Table S2).There were 89 differentially expressed miRNAs between B-ELNs and tissue,including 36 upregulated and 53 downregulated miRNAs (Fig.5 and Table S3).36 upregulated miRNAs were significantly higher in B-ELNs compared with tissue samples.
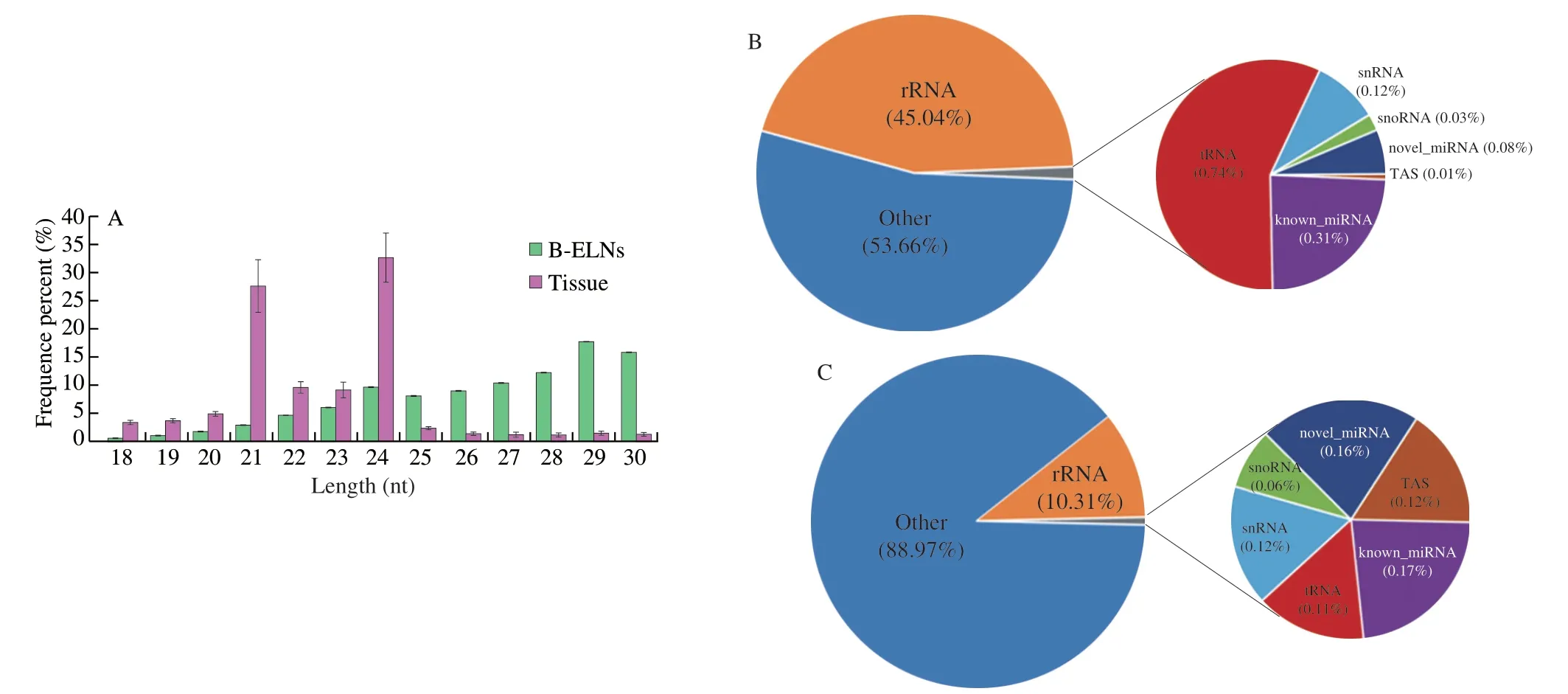
Fig.4 Data was measured through high-throughput sequencing.(A) Length distribution of clean reads of B-ELNs and blueberry tissue small RNA libraries;(B) Statistics of sRNA classification of B-ELNs;(C) Statistics of sRNA classification of Tissue.
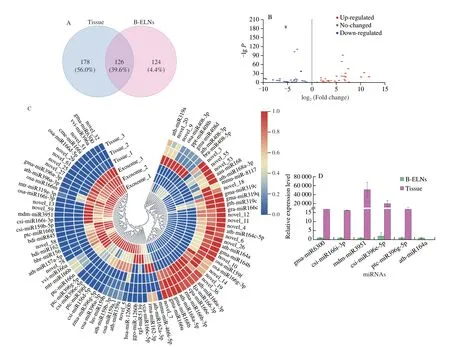
Fig.5 Differential expression of miRNA between tissue and B-ELNs.(A) Venn diagram of known miRNAs and novel miRNAs in B-ELNs and Tissue;(B) Volcano plot of the different expression patterns between B-ELNs and Tissue;(C) Heatmap of differential miRNAs between B-ELNs and Tissue.The heatmap is based on the TPM;(D) Expression profiles of six miRNAs in B-ELNs and Tissue.
3.4 MiRNA expression validation by qRT-PCR
Five upregulated miRNAs (gma-miR6300,csi-miR166i-3p,csi-miR396e-5p,mdm-miR3951 ptc-miR396g-5p) and 1 downregulated miRNA (ath-miR164a) were selected from the differentially expressed miRNAs and were analyzed by qRT-PCR to verify the RNA-seq results (Fig.5D).The results showed that the qRT-PCR expression results were highly correlated with RNA-seq results,and the RNA-seq data were reliable and suitable for further analysis.
3.5 Target genes of upregulated miRNAs
In this study,we mainly focused on the 36 miRNAs that were enriched in B-ELNs.Among the 36 upregulated miRNAs,miR166 family and miR396 family accounted for the largest proportion (Fig.6).All 36 upregulated miRNAs were posited onto miRanda software to predict their target genes in humans.A total of 3 662 target genes were obtained and proceeded with the GO and KEGG annotation.Pathway enrichment analysis revealed that the predicted human target genes were related to 57 GO biological processes (Table S4) and 336 KEGG pathways (Table S5).GO annotation of the target genes indicated that miRNA of B-ELNs could possibly regulate the human genes with various functions.The GO terms were divided into 3 categories,including biological process (BP),cellular component(CC),and molecular function (MF) (Fig.7).In the BP,the predicted targets were mainly associated with the cellular processes,biological regulation,regulation of biological processes,and metabolic processes.The most abundant terms in the CC were cell,cell part,organelle,organelle part,membrane,and membrane part.Further,the most abundant GO terms in MF were binding and catalytic activity.The KEGG results showed that the most enriched pathways were related to signal transduction,such as the cAMP signaling pathway,Ras signaling pathway,and TNF signaling pathway.There were also a lot of human diseases related terms,among which the infectious disease and cancer related terms were most abundant (Fig.8).16 KEGG pathways were closely associated with human digestion systems,includingIL-10,WNTtarget genes,indicating the possible roles of B-ELNs in human gastrointestinal health (Table 2).

Fig.6 Upregulated miRNAs between B-ELNs and blueberry tissue.
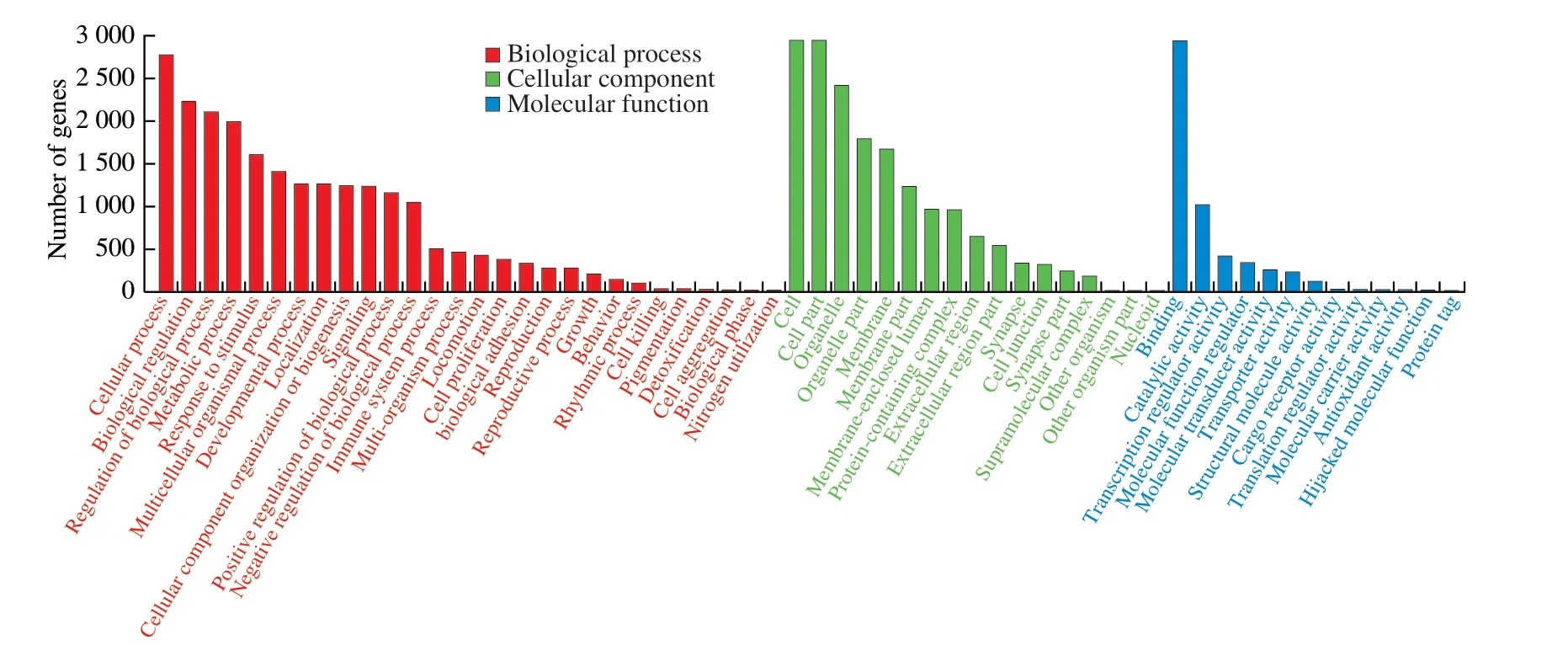
Fig.7 GO classification of 36 upregulated miRNAs in the B-ELNs.
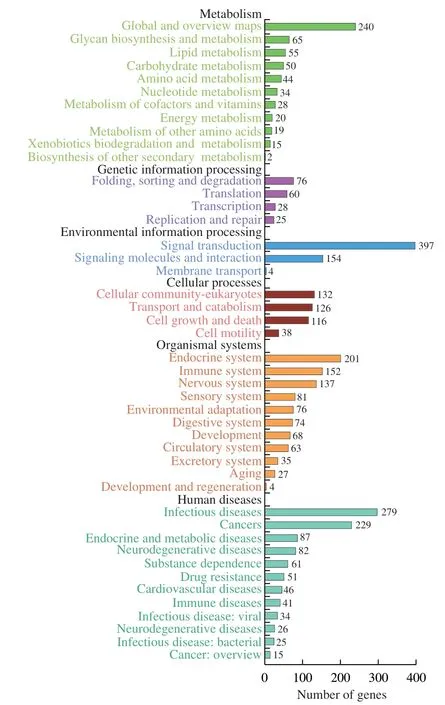
Fig.8 KEGG enrichment analysis based on the 36 upregulated miRNAs in the B-ELNs.
4.Discussion
In the past few years,more and more research indicated the presence of ELNs in vegetables and fruits,which resembled exosomes in structure and function,had potential beneficial functions in human health[14-15,40].The plentiful origins and apparent biosafety of these edible plant derived ELNs made them attractive not only to researchers but also to food industries.To date,the published edible plant derived ELNs extraction methods included UC[41],electrophoretic technique with 300 kDa cut-off dialysis bag (ELD)[16]and PEG 6000-based precipitation method[23].The ELD method is time-saving and requires no special equipment,but is difficult to operate because the electrophoretic direction has to be reversed,and the electrophoretic buffer needs to be replaced manually every 30 min[16].The PEG 6000-based precipitation method also eliminates the need for expensive ultracentrifugation,but the PEG cannot be fully removed from the ELN isolates[23].Nevertheless,extracting edible plant derived ELNs with differential UC was still the major technique,so we chose this method to compare with the SEC.The UC separation is based on differences in density and size of the particles,but the resulting ELN isolates often contain contaminants[42].This problem became particularly obvious as we found that the B-ELN isolates extracted by the UC method had an obvious pink color and gel-like texture.Additional purification steps were needed to obtain a pure B-ELN isolate.The reproducibility was also poor possibly because of the high pectin content in blueberry.It was reported that the centrifugal force of ultracentrifugation may damage the isolated vesicles,reducing their quality[43],which was also observed in our study that more gathered or polygon shaped vesicles were observed by TEM.Therefore,we employed a size exclusion chromatography(SEC) based technique to isolate B-ELNs,which had been successfully applied in exosome extraction in animal biofluids[44].In our study,the feasibility of this method on plant derived ELNs was confirmed via the characterization of B-ELNs by the nanoparticle size distribution and morphology observation.
Compared with UC,SEC required less preparation time because ELN isolation and purification could be achieved at the same time.B-ELN suspension obtained by SEC was colorless,indicating there were fewer contaminations and plant pigments.B-ELNs extracted by SEC also contained fewer proteins and the vesicles were well separated,round shaped with a clear membrane structure.Our results suggested that SEC was able to achieve higher B-ELN purity and a more complete vesicle structure compared with UC.Moreover,SEC did not rely on expensive instruments and had good repeatability,which was inconsistent with previous reports[45].For SEC,the lifetime of the separation column was a determinant of cost.From our experience,the separation was still effective when the column was reused 15 times.The SEC method described here can provide a lowcost and time-effective alternative to extracting and purifying edible plant derived ELNs.If performed on a large volume column,it can likely be adopted for preparative scale purification of therapeutically valuable ENPs.
Plant miRNAs are universally 2’-O-methylated at the 3’-terminal nucleotide,which endows them with resistance against degradation[46].Once encapsulated in ELNs,many of the miRNAs could go through harsh conditions within the gastrointestinal tract and in the circulation of the host[47].Some recent studies have also shown that plant miRNAs,especially dietary plant miRNAs,may reach host circulation and target cells to regulate gene expression[7].But due to the diversity of plants,the function of these miRNAs is still largely unknown.In our study,304 miRNAs were identified in blueberry tissue RNA and miR166 family was the most abundant family.Similarly,Yue et al.[48]identified 412 conserved miRNAs in blueberry with a distribution ranging in 29 families.Another study showed that 84 known miRNAs and 16 novel miRNAs were identified in blueberry fruit[49].No obvious consistency was found between these results,indicating the plant miRNAs are versatile and maybe depend on the cultivar.In B-ELNs,we totally identified 140 miRNAs.To investigate the miRNAs that were specifically incorporated into B-ELNs,we compared the miRNAs in B-ELNs and the berry tissue and found that 36 miRNAs were significantly enriched in B-ELNs,which were presumed to be related to the potential function of B-ELNs.Among the 36 miRNAs,miRNAs from miR396 family and miR166 family accounted for the highest expression level,which was similar with another report about B-ELNs that a total of 104 miRNAs were identified and miR166 family accounted for the highest expression level[50].
The prediction of target genes for miRNA is significant to study miRNA’s function and mechanism.As expected,GO annotation,and KEGG pathway analysis indicated that miRNAs in B-ELNs were possibly involved in a variety of metabolic-related biological pathways and had potential roles in regulating inflammatory,tumor,and cancer disease,which was in consistent with the anti-inflammatory effect of B-ELNs reported in human cells (EA.hy 926)[22].
What has to be mentioned is that the concept of miRNAs through dietary intake was still subjected to intense debate.There was little direct evidence that the dietary miRNAs could be general uptake[51].From the existing research results,we hypothesize that B-ELNs are more likely to act through direct contact with the digestive tract in dietary intake.Hence,we further selected the prediction results that related to the digestive tract.We found that some interesting target genes such asIL-10,andWNT,which were often reported to be regulated by edible plant derived ELNs in other studies.For example,Zhang et al.[52]reported that ginger derived ELN anti-inflammatory effects against DSS-induced colitis could be conferred by enhancing the production of pro-healing anti-inflammatory cytokines IL-10.Mu et al.[7]reported that a role for edible plant derived ELNs mediated interspecies communication by inducing the expression of genes for activation of WNT signaling pathway,which is crucial for maintaining intestinal homeostasis.It has to be mentioned that target gene predictions here were based on the software.But identifying the genes regulated miRNAs is a key step in developing a comprehensive understanding of gene regulation by edible plant derived ELNs.Morein vivoandin vitrostudies are still needed to explore the functions of miRNAs and B-ELNs towards human wellness.
5.Conclusion
In summary,the size exclusion chromatography could be used for edible plant derived ELNs extraction with desirable purity and structure,which was promising for applications in the future research about edible plant derived ELNs.The highly expressed miRNAs in the B-ELNs possessed potential cross-kingdom human gene targets.These results provide a theoretical basis for further exploration of the cross-kingdom regulation function of plant-derived genes.
Conflicts of interest
The authors declared that there is no conflict of interest.
Acknowledgement
This work was supported by the National Natural Science Foundation of China (31701561).
Appendix A.Supplementary data
Supplementary data associated with this article can be found,in the online version,at http://doi.org/10.26599/FSHW.2022.9250074.
- 食品科学与人类健康(英文)的其它文章
- Protective effects of oleic acid and polyphenols in extra virgin olive oil on cardiovascular diseases
- Diet and physical activity inf luence the composition of gut microbiota,benef it on Alzheimer’s disease
- Inf luence of nitrogen status on fermentation performances ofnon-Saccharomyces yeasts: a review
- Ganoderma lucidum: a comprehensive review of phytochemistry,eff icacy,safety and clinical study
- Resveratrol combats chronic diseases through enhancing mitochondrial quality
- Demonstration of safety characteristics and effects on gut microbiota of Lactobacillus gasseri HMV18

