Analysis of the potential biological value of pyruvate dehydrogenase E1 subunit β in human cancer
Yao Rong,Song-Hua Liu,Ming-Zheng Tang,Zhi-Hang Wu,Guo-Rong Ma,Xiao-Feng Li,Hui Cai
Abstract BACKGROUND The pyruvate dehydrogenase E1 subunit β (PDHB) gene which regulates energy metabolism is located in mitochondria.However,few studies have elucidated the role and mechanism of PDHB in different cancers.AIM To comprehensive pan-cancer analysis of PDHB was performed based on bioinformatics approaches to explore its tumor diagnostic and prognostic value and tumor immune relevance in cancer.In vitro experiments were performed to examine the biological regulation of PDHB in liver cancer.METHODS Pan-cancer data related to PDHB were obtained from the Cancer Genome Atlas (TCGA) database.Analysis of the gene expression profiles of PDHB was based on TCGA and Genotype Tissue Expression Dataset databases.Cox regression analysis and Kaplan-Meier methods were used to assess the correlation between PDHB expression and survival prognosis in cancer patients.The correlation between PDHB and receiver operating characteristic diagnostic curve,clinicopathological staging,somatic mutation,tumor mutation burden (TMB),microsatellite instability (MSI),DNA methylation,and drug susceptibility in pan-cancer was also analyzed.Various algorithms were used to analyze the correlation between PDHB and immune cell infiltration and tumor chemotaxis environment,as well as the co-expression analysis of PDHB and immune checkpoint (ICP) genes.The expression and functional phenotype of PDHB in single tumor cells were studied by single-cell sequencing,and the functional enrichment analysis of PDHB-related genes was performed.The study also validated the level of mRNA or protein expression of PDHB in several cancers.Finally,in vitro experiments verified the regulatory effect of PDHB on the proliferation,migration,and invasion of liver cancer.RESULTS PDHB was significantly and differently expressed in most cancers.PDHB was significantly associated with prognosis in patients with a wide range of cancers,including kidney renal clear cell carcinoma,kidney renal papillary cell carcinoma,breast invasive carcinoma,and brain lower grade glioma.In some cancers,PDHB expression was clearly associated with gene mutations,clinicopathological stages,and expression of TMB,MSI,and ICP genes.The expression of PDHB was closely related to the infiltration of multiple immune cells in the immune microenvironment and the regulation of tumor chemotaxis environment.In addition,single-cell sequencing results showed that PDHB correlated with different biological phenotypes of multiple cancer single cells.This study further demonstrated that down-regulation of PDHB expression inhibited the proliferation,migration,and invasion functions of hepatoma cells.CONCLUSION As a member of pan-cancer,PDHB may be a novel cancer marker with potential value in diagnosing cancer,predicting prognosis,and in targeted therapy.
Key Words: Cuprotosis;Pyruvate dehydrogenase E1 subunit β;Pan-cancer;Prognosis;Liver cancer
INTRODUCTION
Cancer currently remains a problem that threatens the public health security of the world's population[1].Although the medical community has made many advances in the treatment of cancer,the improvement in prognosis has not been as good as expected.In addition,the lack of timely diagnosis of cancer patients,delays in treatment,and an aging population have also contributed to the continuous increase in cancer incidence and mortality[2].Therefore,it is particularly important to find effective cancer diagnostic markers and potential therapeutic targets.
The pyruvate dehydrogenase (PDH) complex is a nucleus-encoded mitochondrial complex consisting of PDH (E1),dihydrolipoamide acetyltransferase (E2) and lipoamide dehydrogenase (E3) copies,while the E1 enzyme subunit contains a heterotetramer of PDH E1 subunit alpha 1 (PDHA1) and PDH E1 subunit β (PDHB)[3,4].Unlike PDHA1,which is highly mutant,PDHB has a mutation rate of only 10% of PDHA1 in the general population[5].However,the risk posed by the absence or mutation of PDHB is not negligible.PDHB is found in mitochondria and has the ability to catalyze the conversion of glucose-derived pyruvate to acetyl coenzyme A,thereby regulating oxidative phosphorylation[6].Some studies have described PDHB as a glycolysis regulatory gene that can play a role in the development of cancer[7].In addition,based on previous descriptions and identifications in the literature,PDHB has been identified as a new member of the cuprotosis-related genes in cancers and other diseases[8-11].Cuprotosis is an emerging form of copper-dependent mediated apoptosis in tumor cells,and the effect of cuprotosis on cell death is largely dependent on abnormalities in the tricarboxylic acid (TCA) cycle in tumor cells and,interestingly,the Warburg effect is inextricably linked to it[12].In fact,the Warburg effect is a metabolic modality that is prevalent in tumor cells,which are normally able to dramatically increase glucose uptake and ferment to lactate while reducing the occurrence of the mitochondrial TCA cycle,even under hyperoxia.This upregulation of glycolysis provides a more advantageous environment for cancer cells to survive[13,14].
It has been shown that in some mitochondrial differentially expressed proteins,tumor cells show a significantly enhanced dependence on the Krebs cycle and oxidative phosphorylation.Long non-coding RNA (lncRNA) SNHG3 can affect the prognosis of ovarian cancer and affect energy metabolism by regulating miRNAs.Analysis of the interaction regulatory network of microRNA (miRNA)-lncRNA SNHG3 and miRNA-PDHB showed that SNHG3 was able to act as a potent sponge for hsa-miR186-5p and hsa-miR-590-3p to target PDHB,especially hsa-miR-186-5p.Interestingly,in this mechanism,PDHB namely serves as a key target during the Krebs cycle and glycolytic pathway[15].Another report has also shown that PDHB can play an important role in copper-dependent cell death through crosstalk between the Krebs cycle and mitochondrial enzymes[8].PDHB is a regulatory molecule of energy metabolism and may have potential mechanisms of action involved in and regulating the cuprotosis pathway in cancer.Therefore,there is a need for a comprehensive understanding of the landscape of PDHB's role in cancer development,and this could also provide a basis for further exploration of the relevance of PDHB in the cancer cuprotosis pathway.
The study of PDHB as a cancer marker is a new translational research direction.Through the combination of bioinformatics mining and basic research,we explore the unique potential of PDHB as a cancer biomarker.At the same time,based on previous research theories,we attempt to determine the potential regulatory role and associated biological processes of PDHB in cancer.The focus of this work is to provide evidence for PDHB as a potential biomarker for cancers of which liver cancer is the predominant type.
MATERIALS AND METHODS
Analysis of different expression differences
The sample type,size,clinical information (including race,sex,age,etc.),and miRNA data of all types of cancer patients in this study were obtained from The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/) database.Based on sample analysis from TCGA and Genotypic Tissue Expression Dataset (GTEX) databases,PDHB expression differences between 33 cancers and the corresponding paracancerous normal tissues were evaluated.The abbreviations for these 33 cancers are shown in Table 1.The TISIDB (http://cis.hku.hk) database evaluates the molecular subtypes of PDHB in different cancers,selects and obtains statistically significant analysis results (P>0.05) in the "Subtype" module.The UALCAN (http://ualcan.path.uab.edu/index.html) database was used to evaluate methylation levels and differences in protein expression of PDHB in pan-cancer samples based on the TCGA database to further screen for statistically significant cancer expression types (P>0.05).The HPA (https://www.proteinatlas.org) database was used to demonstrate immunohistochemical staining to reflect PDHB expression levels in normal and corresponding cancer tissues (screening criteria for cancer groups were moderate or high staining intensity and cell number ≥ 25%-75%).

Table 1 Abbreviations for the 33 cancers included in this study
Survival analysis, pathological correlation, receiver operating characteristic (ROC) curves and genetic alterations
Survival information data were retrieved and downloaded for each sample from the TCGA database,and the main aspects analyzed included overall survival (OS),disease-specific survival (DSS),disease-free survival (DFS) and progression-free survival (PFS).Univariate Cox regression analysis was used to assess and analyze the expression levels and prognostic relevance of PDHB in pan-cancer and forest plots were plotted using the R packages "survivor" and "forestplot" andP-values,hazard ratio and 95% confidence interval (CI) are displayed.Survival rates of the two groups were then analyzed and compared using the Kaplan-Meier method,and Kaplan-Meier curves were plotted using the R packages "survminer" and "survivor".The R packages "pROC" and "ggplot2" were used to evaluate the RNAseq data of TCGA and GTEX and to visualize the results.The area under the ROC curve (AUC) was calculated to confirm the diagnosis and prognosis of cancer.The cBioPortal (https://www.cbioportal.org/) database enables visual analysis of cancer genomics including somatic mutations.We were able to assess genetic alterations in PDHB genes in different cancers,including missense mutations,deletions,splicing proteins,gene mutations,and deletions.
Correlation analysis of tumor mutational load (TMB), microsatellite instability (MSI) and methylation levels
Correlations between TMB and MSI and PDHB gene expression were calculated separately by Spearman's statistical method and the results were visualized by creating radar plots using the R package "fmsb" (aP<0.05,bP<0.01,cP<0.001).In addition the variability of the methylation levels of PDHB in multiple cancer types was analyzed by the UALCAN database (aP<0.05,bP<0.01,cP<0.001).
Tumor microenvironment (TME) analysis and immune infiltration assessment
We calculated the correlation between PDHB expression and stromal scores and immune scores in the TME of different cancer types by running the R packages "ESTIMATE" and "LIMMA".To reliably assess the correlation between PDHB gene expression and immune regulation in pan-cancer,we used immunedeconv,an R package that integrates six state-ofthe-art algorithms such as TIMER,xCell,MCP-counter,CIBERSORT,EPIC and quanTIseq[16],allowing us to quantify the association of PDHB with the abundance of immune cell infiltration.The Pearson method was used to perform correlation analysis between PDHB expression and chemokines and chemokine receptors,and immune checkpoint (ICP) genes in TCGA cancers.
Single cell sequencing data analysis
The correlation between PDHB and different biological functions of multiple cancers was analyzed at the single-cell levelusing the CancerSEA (http://biocc.hrbmu.edu.cn/CancerSEA/) database,a specialist single-cell sequencing platform.Data on the correlation between PDHB expression and different tumor functional states were downloaded from the CancerSEA dataset and heatmaps were created using R software.t-Distributed Stochastic Neighbor Embedding (t-SNE) plots were used to identify PDHB expression in individual cancers and were derived directly from the CancerSEA database.
Enrichment analysis of PDHB-related genes
The PDHB protein co-expression network was analyzed using the "Network" section of the BioGRID database (https://thebiogrid.org/).The GEPIA2.0 database was used to obtain the top 100 PDHB-related genes in the pan-cancer.The "Gene_Corr" analysis module in the TIMER2.0 database was used to obtain a heat map of the association between PDHB and its related genes in pan-cancer.In addition,the Kyoto Encyclopedia of Genes and Genomes enrichment analysis of PDHB and related genes was performed by R software.
Drug sensitivity of PDHB in pan-cancer
The NCI-60 compound activity data and RNA-seq expression files with CallMinerTM were downloaded to analyze and visualize PDHB chemotherapeutic drug sensitivity in pan-cancer (https://discover.nci.nih.gov/cellminer/home.do).We mainly selected Food and Drug Administration or clinically approved drugs for analysis.
Cell culture
The human normal gastric epithelial cell line GES-1 and human gastric cancer cell lines HGC-27,MKN-45,MGC-803;human normal hepatocyte line L-O2 and human liver cancer cell lines SMMC-7721,HEPG2,H-97,Huh7;human normal colon epithelial cell line NCM460 and human colon cancer cell lines SW620 and HCT116;human normal breast cell line MCF-10A and human breast cancer cell lines MDA-MB-231 and MCF-7;human normal prostate cell line RWPE-2 and human prostate cancer cell lines PC-3,22Rv1 and DU145,were purchased from the Chinese Academy of Sciences Cell Resource Centre and stored in the central laboratory of Gansu Provincial People's Hospital.Cells were cultured in RPMI-1640 containing 10% fetal bovine serum (FBS),1% double antibodies (streptomycin and penicillin) in a humidified 5% CO2incubator at 37 ℃.
RNA isolation and quantitative reverse transcription PCR (qRT-PCR)
RNA was extracted according to the instructions of the M5 Universal RNA Mini Kit.To determine the RNA concentration and purity in accordance with the experimental requirements,absorbance values were measured at 260 nm and 280 nm.RNA was reverse transcribed to cDNA according to the instructions of the M5 Sprint qPCR RT Kit with gDNA Remover Reverse Transcription kit.2 × M5 HiPer SYBR Premix EsTaq (with Tli RNaseH) was used as a fluorescent dye for the qRT-PCR assay.The primers for PDHB and the internal reference GAPDH were designed and synthesized by Bioengineering (Shanghai) Co.The sequences used for PDHB were forward:5′-GACACTCCCATATCAGAGATGG-3′ and reverse:5′-CTTGGCAGCTGAGTTTATAACC-3′;The sequences used for GAPDH were forward:5′-GGAAGCTTGTCATCAATGGAAATC-3′ and reverse:5′-TGATGACCCTTTTGGCTCCC-3′.mRNA expression levels of PDHB were calculated and analyzed by the 2-ΔΔCtformula.
Western blot analysis
Liver cancer cell lines and gastric cancer cell lines with a culture density of approximately 80% were washed twice with phosphate-buffered saline (PBS) buffer.Cells were completely lysed with phenylmethylsulfonyl fluoride-spiked highperformance lysis solution (RIPA Lysis Buffer) (Boster,Wuhan),centrifuged (12000 × rpm,4 ℃,10 min) and the supernatant collected.Protein concentration was measured using a BCA protein assay kit (Solarbio,Beijing).Each protein sample of 20 μg was separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore,United States).The membranes were subsequently closed with protein-free fast closure solution (Boster,Wuhan) and rinsed three times for 5 min each time with TBST rinse solution.The membranes were incubated with primary antibodies of PDHB (Proteintech Group,Wuhan) and β-Tubulin (Boster,Wuhan) overnight on a 4 ℃ shaker,followed by incubation with secondary antibodies for 1 h,three rinses with TBST rinse solution for 10 min each time and then with an enhanced chemiluminescence kit (Boster,Wuhan) to visualize the blots.The total grey values of the protein bands were analyzed using ImageJ software to quantify the protein expression levels of the genes.
Transfection
When the confluency of Huh7 hepatoma cells reached approximately 70%,PDHB siRNA and its control NC siRNA (Gemma Gene,China) were transfected according to Lipo 2000 instructions,and the relevant experiments were performed 48 h after transfection.Supplementary Table 1 shows the sequence information for the four siRNAs used in this study.
Cell proliferation and colony formation assays
The siRNA-transfected Huh7 hepatoma cells were completely digested and seeded into 96-well plates with approximately 2500 cells per well.According to the instructions of the CCK-8 (APExBIO,United States) reagent,the proliferation level of Huh7 hepatoma cells at different time points (0 h,24 h,48 h,72 h) was detected.Finally,GraphPad Prism 6.0 software was used to generate the proliferation curve of hepatoma cells.Huh7 cells successfully transfected siRNA PDHB/NC were placed on fresh six-well plates and cultured for one week,during which time the medium was changed every 2-3 d.The colonies formed were fixed with 4% paraformaldehyde and stained with crystal violet (Sigma-Aldrich) after reaching a predetermined time period,then photographed and counted.
Migration, invasion, and wound healing experiments
Cell migration and invasion assays were performed using the Transwell chamber method,as described below.The successfully transfected Huh7 cell suspensions were counted,and serum-free Huh7 (siRNA-PDHB) and Huh7 (siRNANC) cell suspensions at concentrations of 5 × 105/100 μL were seeded on a Transwell chamber with or without Matrigel,respectively,for invasion and migration experiments,and medium containing 20% FBS was loaded into the lower chamber.Cells were incubated in an incubator for 24 h-48 h and then removed and fixed with 4% paraformaldehyde,stained with 0.1% crystal violet dye for 30 min,placed under a microscope for imaging and counted.Transfected Huh7 cells were seeded in 6-well plates at a culture density close to 100%.Cells were then scraped with a sterile 10 μL pipette tip along the midline of each well,counted to 0 h,and labeled.Cells were incubated for 72 h,cell migration was observed at 0 h,24 h,48 h,and 72 h,respectively,and the cells were washed with PBS buffer before each observation to calculate mobility.
Statistical analysis
All gene expression data were normalized by log2.Gene expression data from TCGA and GTEX were examined by the Wilcoxon test.Univariate Cox regression analysis was performed to detect differences between the two data sets by the rank sum test.Correlation analysis of PDHB gene expression with immune cell infiltration was assessed by the Spearman method.Correlation analysis of PDHB with ICP genes,chemokines and chemokine receptors was performed by the Pearson method.All analyses of pan-cancer-related data were based on the application of R software (version 4.2.0,www.R-project.org) or automated measurements through an online database.Pvalues <0.05 were considered statistically significant.
RESULTS
Analysis of PDHB expression in pan-cancer
The technical process of this study is shown in Figure 1.We obtained the expression profile of PDHB in pan-cancer through the TCGA database.The ranking of PDHB expression levels in different types of cancer showed that it had the highest expression levels in TCGT and the lowest expression levels in esophageal carcinoma (ESCA) (Figure 2A).We found that PDHB expression was downregulated in most cancers,including colon adenocarcinoma (COAD),ESCA,head and neck squamous cell carcinoma (HNSC),kidney renal clear cell carcinoma (KIRC),kidney renal papillary cell carcinoma (KIRP),LUSC,rectum adenocarcinoma (READ),Sarcoma (SARC),while PDHB expression increased in breast invasive carcinoma (BRCA),liver hepatocellular carcinoma (LIHC),lung adenocarcinoma (LUAD),and uterine corpus endometrial carcinoma (UCEC) (Figure 2B).It is worth noting that some types of cancer in the TCGA database lack control group samples [such as adrenocortical carcinoma (ACC),lymphoid neoplasm diffuse large B-cell lymphoma (DLBC),brain lower grade glioma (LGG),testicular germ cell tumors (TGCT),etc.],in order to ensure more accurate and comprehensive analysis results,the sample size was expanded in conjunction with the TCGA database and GTEX database to further explore the differential expression of PDHB in cancer and normal tissues.The results showed that PDHB showed up-or down-regulated expression in some emerging cancer types.Specifically,PDHB was up-regulated in glioblastoma multiforme (GBM),DLBC,THCA,LGG,ovarian serous cystadenocarcinoma (OV),pancreatic adenocarcinoma (PAAD),prostate adenocarcinoma (PRAD),skin cutaneous melanoma (SKCM),TGCT,THYM and uterine carcinosarcoma,and down-regulated in ACC,BLCA,and stomach adenocarcinoma (STAD) (Figure 2C).The UALCAN platform was also used to analyze the expression differences of PDHB at the protein level in different cancer tissues.The results showed that protein expression levels of PDHB were downregulated in breast cancer,colon cancer,GBM,clear cell carcinoma,HNSC,and PAAD,and up-regulated only in hepatocellular carcinoma (HCC) (Figure 2D).As there is a great deal of heterogeneity between different types of cancer,the presence of molecular subtypes can deepen the understanding and thinking of cancer.We examined the effect of PDHB on the molecular subtypes of different cancers based on the TISIDB database,and as shown in Supplementary Figure 1,PDHB expression was significantly associated with different molecular subtypes of multiple cancers.In BRCA,PDHB expression was significantly correlated with Basal,Her2,LumA,and LumB subtypes.In LIHC,PDHB expression was significantly correlated with iCluster 1/2/3 subtype and significantly upregulated in iCluster 2.The cancer with the highest number of tumor subtypes significantly associated with PDHB was LGG (n=6),whose associated tumor subtypes were classic-like,code,G-CIMP-high,G-CIMPlow,mesenchymal-like,PA-like.In addition,PDHB was also expressed to varying degrees in other tumor subtypes,including atypical,basal,immunoactive,Wnt-altered,etc.
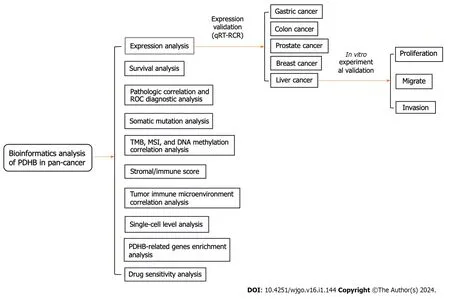
Figure 1 The predominant methodical workflow in the current study. PDHB: Pyruvate dehydrogenase E1 subunit β;ROC: Receiver operating characteristic;TMB: Tumor mutation burden;MSI: Microsatellite instability;qRT-RCR: Real-time quantitative PCR.
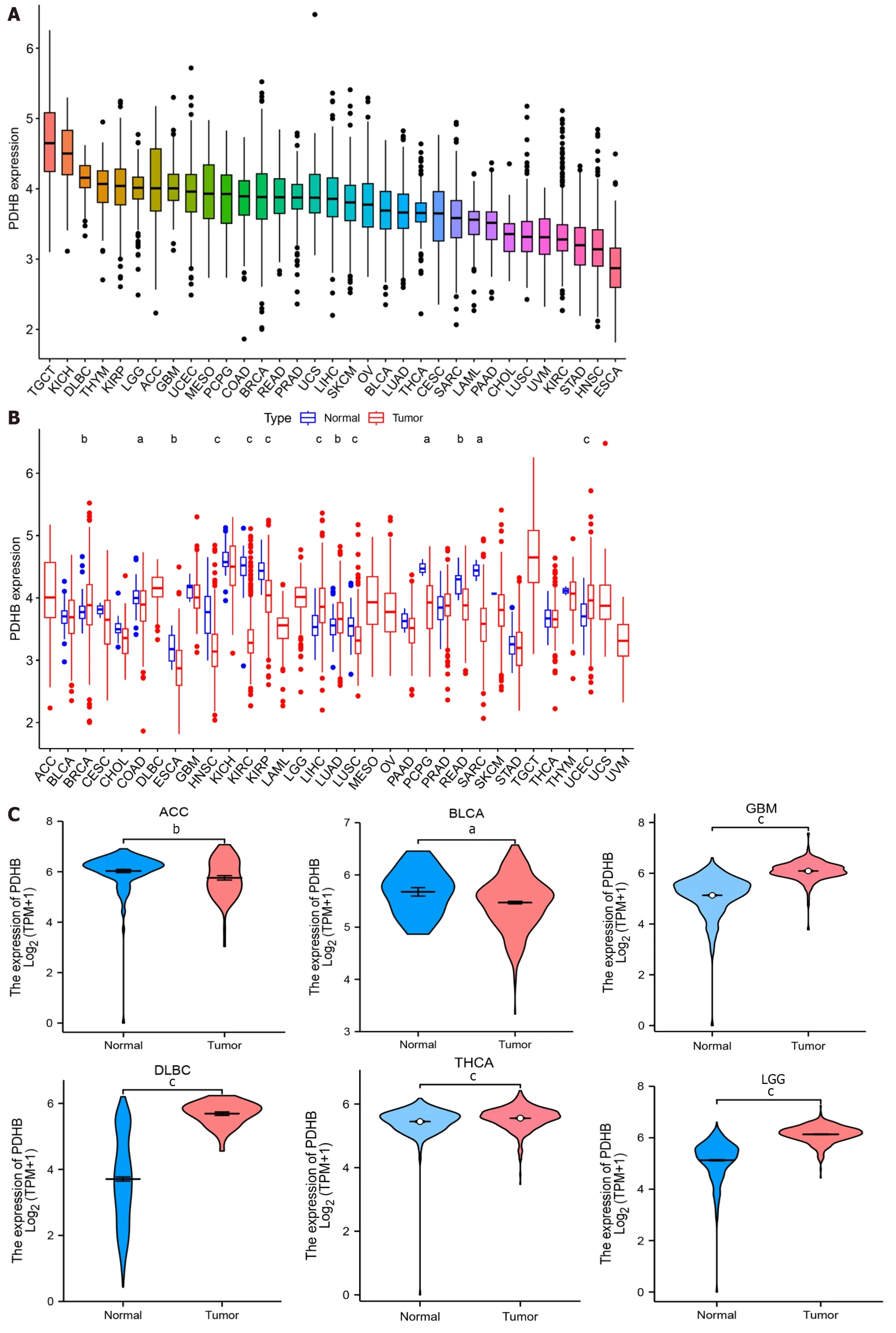
Figure 2 Expression levels of pyruvate dehydrogenase E1 subunit β in pan-cancer. A: Comparison of pyruvate dehydrogenase E1 subunit β (PDHB)expression levels in different cancers based on The Cancer Genome Atlas (TCGA) database;B: Expression of PDHB in cancer vs normal tissues in the TCGA database;C: Differential expression of PDHB in cancer and normal tissues in the TCGA joint Genotype Tissue Expression Dataset database;D: Protein levels of PDHB in tumors and normal tissues from the UALCAN database.aP <0.05,bP <0.01,cP <0.001.PDHB: Pyruvate dehydrogenase E1 subunit β;ACC:Adrenocortical carcinoma;BLCA: Bladder urothelial carcinoma;BRCA: Breast invasive carcinoma;CESC: Cervical squamous cell carcinoma and Endocervical adenocarcinoma;CHOL: Cholangiocarcinoma;COAD: Colon adenocarcinoma;DLBC: Lymphoid neoplasm diffuse large B-cell lymphoma;ESCA: Esophageal carcinoma;GBM: Glioblastoma multiforme;HNSC: Head and Neck squamous cell carcinoma;KICH: Kidney chromophobe;KIRC: Kidney renal clear cell carcinoma;KIRP: Kidney renal papillary cell carcinoma;LAML: Acute myeloid leukemia;LGG: Brain lower grade glioma;LIHC: Liver hepatocellular carcinoma;LUAD: Lung adenocarcinoma;LUSC: Lung squamous cell carcinoma;MESO: Mesothelioma;OV: Ovarian serous cystadenocarcinoma;PAAD: Pancreatic adenocarcinoma;PCPG: Pheochromocytoma and Paraganglioma;PRAD: Prostate adenocarcinoma;READ: Rectum adenocarcinoma;SKCM: Skin cutaneous melanoma;STAD:Stomach adenocarcinoma;TGCT: Testicular germ cell tumors;THCA: Thyroid carcinoma;THYM: Thymoma;UCEC: Uterine corpus endometrial carcinoma;UCS:Uterine carcinosarcoma;UVM: Uveal melanoma.
Cancer types with differences in PDHB expression were then screened out using the UALCAN database and their protein expression was visually analyzed through the HPA database.PDHB expression was moderately or highly stained with ≥ 25%-75% stained cells in tumor tissues of BRCA,COAD,KIRC,LIHC,LUAD,THCA,and UCEC,whereas it was moderately stained or unstained in their corresponding normal tissues (Figure 3).
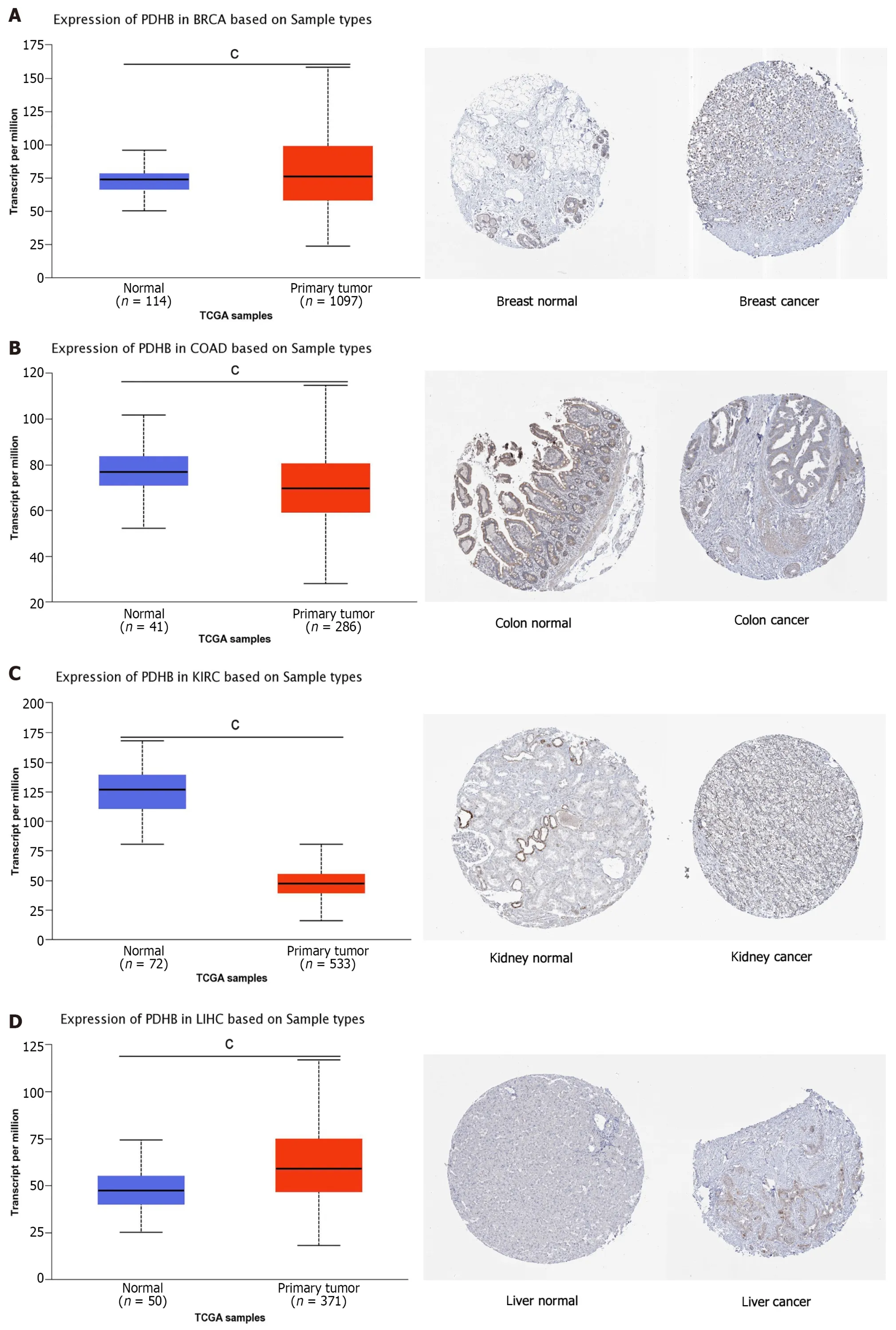
Figure 3 Pyruvate dehydrogenase E1 subunit β expression in the UACLAN and HPA databases. A: Breast invasive carcinoma;B: Colon adenocarcinoma;C: Kidney renal clear cell carcinoma;D: Liver hepatocellular carcinoma;E: Lung adenocarcinoma;F: Thyroid carcinoma;G: Uterine corpus endometrial carcinoma.BRCA: Breast invasive carcinoma;COAD: Colon adenocarcinoma;KIRC: Kidney renal clear cell carcinoma;LIHC: Liver hepatocellular carcinoma;LUAD: Lung adenocarcinoma;THCA: Thyroid carcinoma;UCEC: Uterine corpus endometrial carcinoma.
Survival analysis of PDHB in pan-cancer
Univariate COX regression analysis was used to understand the correlation between PDHB expression and survival in pan-carcinoma patients.From the forest plot describing the correlation between PDHB expression and OS,DSS,DFS,and PFS,we found that PDHB expression was significantly associated with good survival outcomes in patients with KIRC (Figure 4A-C),KIRP (Figure 4),MESO (Figure 4A-C),LGG (Figure 4A-C),and BRCA (Figure 4B and D).Conversely,PDHB expression levels correlated with adverse prognosis in patients with PRAD (Figure 4C) and HNSC (Figure 4D).In addition,the Kaplan-Meier survival curve associated with PDHB (Figure 4) was analyzed using the R software package,and the patient population was divided into two groups,the PDHB low expression group and the PDHB high expression group.We found that highly expressed PDHB was negatively correlated with DFS in patients with HNSC.In patients with KIRP,MESO,and LGG,overexpressed PDHB was positively correlated with OS,PFS,and DFS.Similarly,highly expressed PDHB was positively correlated with PFS in STAD patients,OS in THCA patients,and DSS in BRCA patients.
Clinicopathological analysis and ROC diagnostic analysis related to PDHB in pan-cancer
From the results of clinicopathological analysis associated with PDHB,PDHB expression was significantly correlated with tumor stage in patients with several types of cancer,including ESCA,KIRC,KIRP,LUAD,SKCM,TGCT,and THCA (Figure 5A).Specifically,in ESCA and LUAD,the expression of PDHB was lower in stage I-II and higher in stage III-IV,whereas the opposite results were observed in KIRC,KIRP and THCA.In addition,in order to further explore the potential diagnostic value of PDHB in pan-cancer,the ROC curves of PDHB in different cancers were drawn.The results showed that PDHB had better diagnostic accuracy for oral squamous cell carcinoma,HNSC,ESCA,KIRC,TGCT,OV,and LIHC (AUC between 0.7 and 0.9) in diagnosing tumors (Figure 5B) and was more accurate (AUC >0.9) for PAAD,DLBC,LUSC,LGG,and GBM (Figure 5B).This suggests that the PDHB gene has a strong ability to diagnose tumors.

Figure 5 Correlation of pyruvate dehydrogenase E1 subunit β expression with tumor stage in multiple cancers and receiver operating characteristic diagnostic analysis. A: Correlation between pyruvate dehydrogenase E1 subunit β (PDHB) gene expression and tumor staging;B: Receiver operating characteristic curve analysis of PDHB in various cancers (AreaUnderROC >0.7).ACC: Adrenocortical carcinoma;BLCA: Bladder urothelial carcinoma;BRCA: Breast invasive carcinoma;CHOL: Cholangiocarcinoma;COAD: Colon adenocarcinoma;ESCA: Esophageal carcinoma;HNSC: Head and Neck squamous cell carcinoma;KICH: Kidney chromophobe;KIRC: Kidney renal clear cell carcinoma;KIRP: Kidney renal papillary cell carcinoma;LIHC: Liver hepatocellular carcinoma;LUAD: Lung adenocarcinoma;LUSC: Lung squamous cell carcinoma;MESO: Mesothelioma;PAAD: Pancreatic adenocarcinoma;PRAD: Prostate adenocarcinoma;SKCM: Skin cutaneous melanoma;STAD: Stomach adenocarcinoma;TGCT: Testicular germ cell tumors;THCA: Thyroid carcinoma;UVM: Uveal melanoma.
Analysis of genetic mutations in PDHB in pan-cancer
The somatic mutation frequency of the PDHB gene in pan-cancer was subsequently mapped using the cBioPortal platform.Figure 6A shows the mutation frequency of the PDHB gene in different cancers,with PDHB having the highest mutation frequency in uterine sarcoma at more than 3%.In addition,we investigated the types of mutations and mutation sites triggered by PDHB.The major mutation type was missense mutation (n=37),which accounted for the largest proportion of mutations,while other types included truncating mutation (n=3),splice mutation (n=2) and sv/fusion mutation (n=1) (Figure 6B).

Figure 6 Mutation frequency of pyruvate dehydrogenase E1 subunit β in pan-cancer. A: Somatic mutation analysis of pyruvate dehydrogenase E1 subunit β (PDHB) in different cancers;B: CBioPortal shows the mutation type and mutation frequency of PDHB sequences.
Correlation of PDHB expression with TMB, MSI and methylation levels
TMB and MSI are gradually becoming important potential predictive tumor markers;thus,the correlation analysis of PDHB with TMB and MSI co-expression was performed in pan-cancer.Among them,PDHB expression was significantly positively correlated with TMB in 2 cancers,namely UCEC and STAD,while it was negatively correlated with TMB in THYM,LGG,KIRC,cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) and BRCA (Figure 7A).It was also shown that PDHB expression was significantly correlated with MSI in 12 cancers,and this correlation was positive in UCEC,STAD,READ and KIRC and negative in ACC,PRAD,OV,LUSC,LUAD,kidney chromophobe (KICH),GBM and BRCA (Figure 7B).DNA methylation is an important epigenetic modification,and alterations in its steady state are an important factor in promoting tumorigenesis.Therefore,we analyzed the methylation levels of PDHB in pan-cancer from the UALCAN database.The analysis showed that PDHB exhibited a hypomethylation state in BLCA,KIRP,and LIHC,in contrast,PDHB was highly methylated in ESCA,HNSC,KIRC,LUSC,and PRAD (Figure 7C).In general,DNA methylation is often inversely proportional to the transcription level of a gene,and the level of promoter methylation can reflect the expression trend of a gene in cancer,thereby helping to determine its expression level in different cancers.Thus,combined with the results shown in Figure 1B,that PDHB has lower transcriptional expression levels in ESCA,HNSC,KIRC,and LUSC,this suggests that PDHB may be a methylation driver gene.
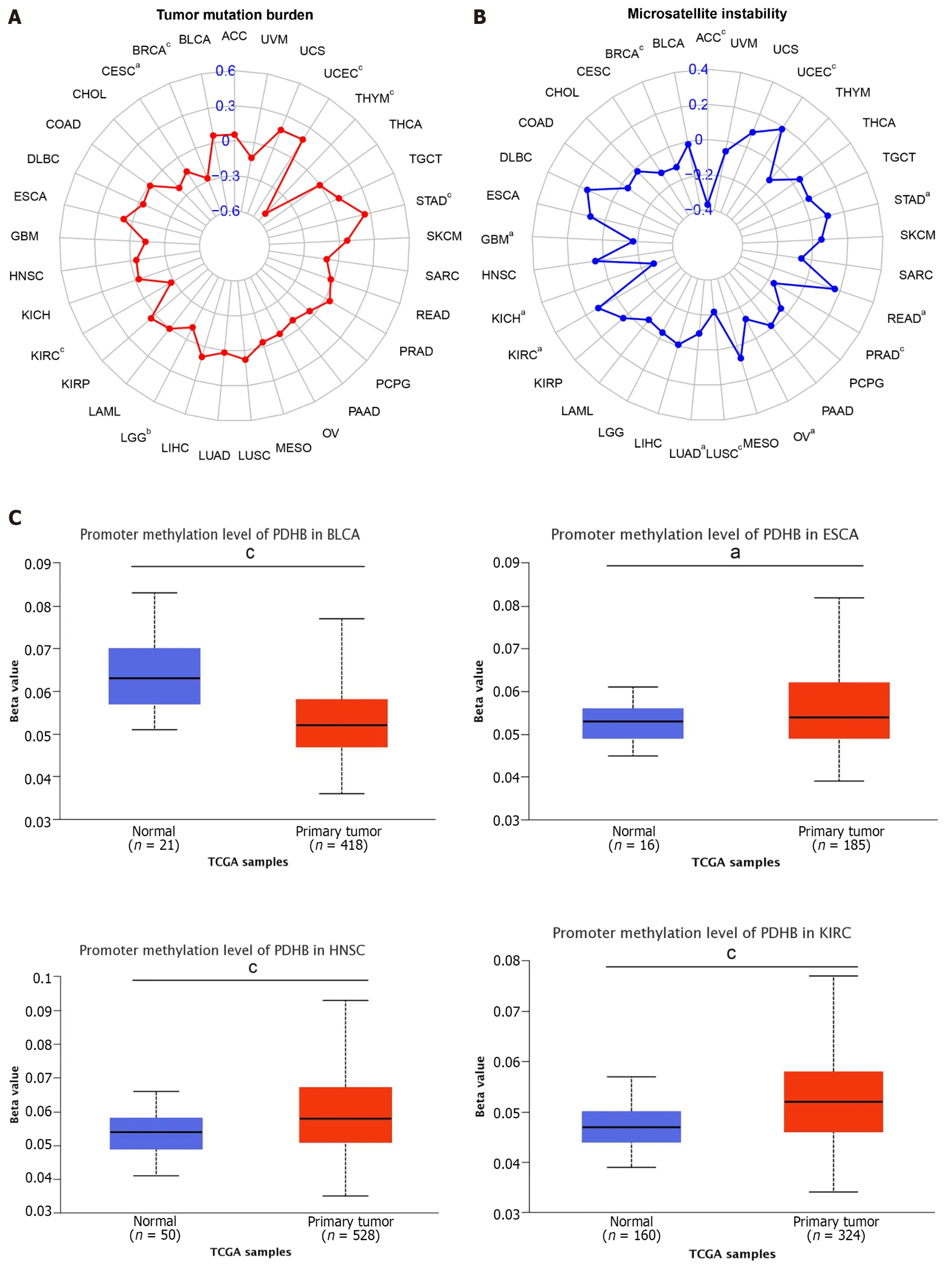
Figure 7 Correlation analysis of pyruvate dehydrogenase E1 subunit β gene expression with DNA methylation levels in tumor mutation burden,microsatellite instability and pan-cancer. A: Radar plot demonstrating the relationship between tumor mutation burden and pyruvate dehydrogenase E1 subunit β (PDHB) gene expression in various malignancies.Correlation coefficients are indicated by red curves and ranges are indicated by blue values;B: Radar plot showing the relationship between microsatellite instability and PDHB gene expression and various malignancies.Correlation coefficients are shown by blue curves and ranges are shown by green values;C: PDHB methylation levels in the UALCAN database on different cancers.aP <0.05,bP <0.01,cP <0.001.ACC: Adrenocortical carcinoma;BLCA: Bladder urothelial carcinoma;BRCA: Breast invasive carcinoma;CESC: Cervical squamous cell carcinoma and Endocervical adenocarcinoma;CHOL: Cholangiocarcinoma;COAD: Colon adenocarcinoma;DLBC: Lymphoid neoplasm diffuse large B-cell lymphoma;ESCA:Esophageal carcinoma;GBM: Glioblastoma multiforme;HNSC: Head and Neck squamous cell carcinoma;KICH: Kidney chromophobe;KIRC: Kidney renal clear cell carcinoma;KIRP: Kidney renal papillary cell carcinoma;LAML: Acute myeloid leukemia;LGG: Brain lower grade glioma;LIHC: Liver hepatocellular carcinoma;LUAD:Lung adenocarcinoma;LUSC: Lung squamous cell carcinoma;MESO: Mesothelioma;OV: Ovarian serous cystadenocarcinoma;PAAD: Pancreatic adenocarcinoma;PCPG: Pheochromocytoma and Paraganglioma;PRAD: Prostate adenocarcinoma;READ: Rectum adenocarcinoma;SKCM: Skin cutaneous melanoma;STAD:Stomach adenocarcinoma;TGCT: Testicular germ cell tumors;THCA: Thyroid carcinoma;THYM: Thymoma;UCEC: Uterine corpus endometrial carcinoma;UCS:Uterine carcinosarcoma;UVM: Uveal melanoma.
Analysis of the correlation between PDHB in pan-cancer and immune infiltration
The multicellular environment during tumor development is known as the TME,which consists of immune cells,stromal cells,special signaling molecules,and blood vessels around the tumor[17].The TME plays a unique role in tumor occurrence and drug resistance;therefore targeted TME is a more advantageous cancer treatment.In order to explore the correlation between PDHB expression and pan-cancer TME,the immune score and stromal score of PDHB in different cancers were calculated,respectively.The results showed that PDHB expression was negatively correlated with the immune scores of seven cancers,including BLCA,BRCA,LGG,LUAD,PRAD,THCA and UCEC,and only positively correlated with LUSC immune scores (Figure 8A).Furthermore,PDHB expression was negatively correlated with stromal scores for BLCA,BRCA,LGG,LIHC,LUAD,MESO,OV,PRAD,SARC,TGCT,THCA,THYM and UCEC,and only positively correlated with stromal scores for PAAD (Figure 8B).
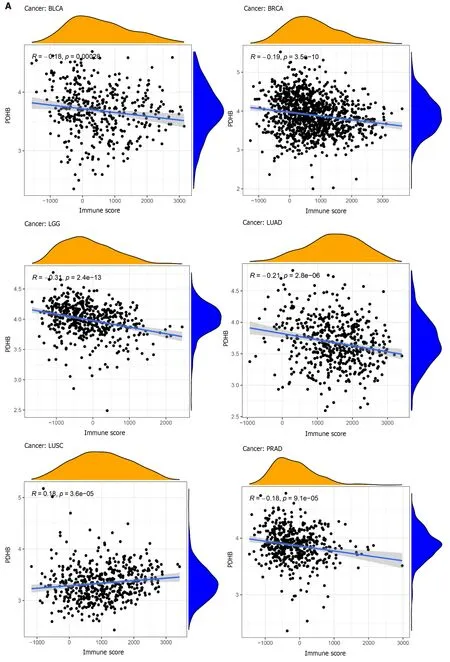
Figure 8 Correlation of pyruvate dehydrogenase E1 subunit β gene expression with stromal and immune scores in different cancers. A:Correlation analysis of pyruvate dehydrogenase E1 subunit β (PDHB) gene expression with immune scores of bladder urothelial carcinoma (BLCA),breast invasive carcinoma (BRCA),brain lower grade glioma (LGG),lung adenocarcinoma (LUAD),lung squamous cell carcinoma,pancreatic adenocarcinoma (PAAD),thyroid carcinoma (THCA),uterine corpus endometrial carcinoma (UCEC);B: Correlation analysis of PDHB gene expression with stromal scores of BLCA,BRCA,LGG,liver hepatocellular carcinoma,LUAD,mesothelioma,ovarian serous cystadenocarcinoma,PAAD,prostate adenocarcinoma,sarcoma,testicular germ cell tumors,THCA,thymoma,UCEC.aP <0.05,bP <0.01,cP <0.001.BRCA: Breast invasive carcinoma;BLCA: Bladder urothelial carcinoma;LGG: Brain lower grade glioma;LUAD:Lung adenocarcinoma;LUSC: Lung squamous cell carcinoma;PAAD: Pancreatic adenocarcinoma;THCA: Thyroid carcinoma;UCEC: Uterine corpus endometrial carcinoma;BRCA: Breast invasive carcinoma;LIHC: Liver hepatocellular carcinoma;MESO: Mesothelioma;OV: Ovarian serous cystadenocarcinoma;PRAD:Prostate adenocarcinoma;SARC: Sarcoma;TGCT: Testicular germ cell tumors;THCA: Thyroid carcinoma;THYM: Thymoma;UCEC: Uterine corpus endometrial carcinoma.
To further examine the role of PDHB in tumor immunity,we selected several algorithms to study the correlation between PDHB gene expression and immune cell infiltration in pan-cancer.The results showed that in all cancer types,PDHB expression was significantly associated with the infiltration of at least one immune cell (Figure 9A and B).The TIMER analysis showed that PDHB expression in 11,14,13,13,13,and 12 cancers was significantly associated with CD4+T cells,CD8+T cells,neutrophils,macrophages,dendritic cells (DCs),and B cells,respectively (Figure 9A).CIBERSORT analysis showed that in 22 immune cells and subtypes,PDHB expression was significantly associated with CD8+T cells,regulatory T cells (Tregs),M2 macrophages,activated natural killer (NK) cells,memory B cells,naïve B cells,and activated mast cells in most cancers (Figure 9B).Co-expression analysis of PDHB and immune cells showed that the expression of this gene was associated with the infiltration and penetration of a variety of immune cells.

Figure 9 Correlation analysis of pyruvate dehydrogenase E1 subunit β expression and tumor immunity.A: TIMER method to analyze the correlation between pyruvate dehydrogenase E1 subunit β (PDHB) and immune cell infiltration;B: CIBERSORT method to analyze PDHB correlation with immune cell infiltration;C: Co-expression analysis of PDHB with tumor chemokines;D: Co-expression analysis of PDHB with tumor chemokine receptors;E: Co-expression analysis of PDHB with immune checkpoint gene.aP <0.05,bP <0.01,cP <0.001.PDHB: Pyruvate dehydrogenase E1 subunit β.ACC: Adrenocortical carcinoma;BLCA: Bladder urothelial carcinoma;BRCA: Breast invasive carcinoma;CESC: Cervical squamous cell carcinoma and Endocervical adenocarcinoma;CHOL:Cholangiocarcinoma;COAD: Colon adenocarcinoma;DLBC: Lymphoid neoplasm diffuse large B-cell lymphoma;ESCA: Esophageal carcinoma;GBM: Glioblastoma multiforme;HNSC: Head and Neck squamous cell carcinoma;KICH: Kidney chromophobe;KIRC: Kidney renal clear cell carcinoma;KIRP: Kidney renal papillary cell carcinoma;LAML: Acute myeloid leukemia;LGG: Brain lower grade glioma;LIHC: Liver hepatocellular carcinoma;LUAD: Lung adenocarcinoma;LUSC: Lung squamous cell carcinoma;MESO: Mesothelioma;OV: Ovarian serous cystadenocarcinoma;PAAD: Pancreatic adenocarcinoma;PCPG: Pheochromocytoma and Paraganglioma;PRAD: Prostate adenocarcinoma;READ: Rectum adenocarcinoma;SKCM: Skin cutaneous melanoma;STAD: Stomach adenocarcinoma;TGCT:Testicular germ cell tumors;THCA: Thyroid carcinoma;THYM: Thymoma;UCEC: Uterine corpus endometrial carcinoma;UCS: Uterine carcinosarcoma;UVM: Uveal melanoma.
ICP genes play an important role in influencing immune cell infiltration and immunotherapy[18].Therefore,this study focused on analyzing the correlation between PDHB expression and ICP genes (Figure 9C).The analysis showed that PDHB expression in cancer showed good correlation with 46 ICP genes.PDHB was significantly associated with the expression of multiple ICP genes in most cancers,such as PDCD1/PD-L1,CTLA4,IDO2,CD274,CD96,TGFB1,LAG3,CD276,BTNL2,CD48 and CD80,etc.Thus,PDHB may be involved in the activation of ICP genes and play an important role in the corresponding signaling pathways.Chemokines and chemokine receptors have been shown to mediate immune escape and the recruitment of immunosuppressive cells in tumors,and thus to pro-tumorigenesis[19].Therefore,it is necessary to explore the relevance of PDHB in the tumor chemotactic environment.We found that PDHB expression in BRCA,KIRC,KIRP,LGG,LUAD,and PRAD was significantly and negatively correlated with the chemokine CCL22 and its receptors,CCR4,CCR5,and CCR10,in Treg cells (Figure 9D and E).Furthermore,in most cancers,PDHB expression was predominantly negatively correlated with the chemokine CCL2 and its receptor CCR2 of tumorassociated macrophages (TAM) (Figure 9D and E).Taken together,PDHB mediates different immune regulatory mechanisms in a variety of cancers to achieve divergent outcomes.
Single-cell level analysis of PDHB in pan-cancer
Single-cell sequencing enables the differential analysis of genetic information such as the genome and transcriptome of cancer cells at the single-cell level,allowing us to better understand the heterogeneity of different tumor cells exhibiting phenotype differences such as immune characteristics,growth invasion and drug resistance[20].The correlation of PDHB expression with biological functions in different cancer single cells was analyzed by the CancerSEA database.The results showed that PDHB expression was positively correlated with DNA damage and negatively correlated with angiogenesis in most cancers.Specifically,in uveal melanoma (UVM),PDHB expression was negatively correlated with multiple biological effects such as apoptosis,cell cycle,differentiation,DNA damage,DNA repair,etc.In OV,PDHB expression was negatively correlated with invasion and stemness.PDHB gene expression was positively correlated with functions such as angiogenesis,inflammation,and metastasis in retinoblastoma (RB) (Figure 10A).We separately analyzed the correlation of PDHB expression with metastasis in OV,angiogenesis in RB,and DNA repair biological functions in UVM (Figure 10B).In addition,we demonstrated the distribution of PDHB expression at the single cell level in OV,RB,and UVM as t-SNE plots (Figure 10C).
Enrichment analysis of PDHB-related genes in pan-cancer
We used the BioGRID platform to collate and analyze the functional interaction network of PDHB-associated genes to explore potential markers linked to PDHB (Figure 11A).In addition,data on the top 100 genes associated with PDHB were obtained through the GEPIA2.0 database,as detailed in Supplementary Table 2.The results of the analysis showed that PDHB was positively correlated with the expression levels of ACTR8 (r=0.57),KCTD6 (r=0.55),MLH1 (r=0.52),PSMD6 (r=0.6),RPP14 (r=0.59),and USP19 (r=0.52) in pan-cancer (Figure 11B).Cluster analysis then showed that PDHB gene expression was significantly and positively correlated with the levels of six genes,ACTR8,KCTD6,MLH1,PSMD6,RPP14,and USP19,in the vast majority of cancer types (Figure 11C).In addition,Enrichment analysis showed that PDHB-related genes were mainly enriched in functions related to cellular energy metabolism,such as mitochondrial inner membrane,mitochondrial matrix,respiratory chain,mitochondrial intermembrane space,etc.(Figure 11D).

Figure 11 Enrichment analysis of pyruvate dehydrogenase E1 subunit β-associated genes in pan-cancer. A: Interaction network of pyruvate dehydrogenase E1 subunit β (PDHB)-associated biomarkers derived from the BioGRID database;B: GEPIA2.0 showed that PDHB expression was positively correlated with actin related protein 8 (ACTR8),potassium channel tetramerization domain containing 6 (KCTD6),mutl homolog 1 (MLH1),proteasome 26s subunit,non-ATPase 6 (PSMD6),ribonuclease P/MRP subunit p14 (RPP14),and ubiquitin specific peptidase 19 (USP19) genes;C: Heat map showing PDHB expression positively correlated with 6 genes (ACTR8,KCTD6,MLH1,PSMD6,RPP14,USP19);D: Enrichment analysis of PDHB-related genes.PDHB: Pyruvate dehydrogenase E1 subunit β.ACC: Adrenocortical carcinoma;BLCA: Bladder urothelial carcinoma;BRCA: Breast invasive carcinoma;CESC: Cervical squamous cell carcinoma and Endocervical adenocarcinoma;CHOL: Cholangiocarcinoma;COAD: Colon adenocarcinoma;DLBC: Lymphoid neoplasm diffuse large B-cell lymphoma;ESCA: Esophageal carcinoma;GBM: Glioblastoma multiforme;HNSC: Head and Neck squamous cell carcinoma;KICH: Kidney chromophobe;KIRC:Kidney renal clear cell carcinoma;KIRP: Kidney renal papillary cell carcinoma;LAML: Acute myeloid leukemia;LGG: Brain lower grade glioma;LIHC: Liver hepatocellular carcinoma;LUAD: Lung adenocarcinoma;LUSC: Lung squamous cell carcinoma;MESO: Mesothelioma;OV: Ovarian serous cystadenocarcinoma;PAAD: Pancreatic adenocarcinoma;PCPG: Pheochromocytoma and Paraganglioma;PRAD: Prostate adenocarcinoma;READ: Rectum adenocarcinoma;SKCM:Skin cutaneous melanoma;STAD: Stomach adenocarcinoma;TGCT: Testicular germ cell tumors;THCA: Thyroid carcinoma;THYM: Thymoma;UCEC: Uterine corpus endometrial carcinoma;UCS: Uterine carcinosarcoma;UVM: Uveal melanoma.
Drug sensitivity study
Analysis of PDHB expression and drug susceptibility showed that the IC50s of Chelerythrine,Nelarabine,Fludarabine,Fenretinide,Lapachone and Voranistat were positively correlated with the expression of PDHB.The higher the IC50value,the weaker the effect of drugs that induce cancer cell death,indicating that PDHB can enhance cancer cell resistance to certain drugs.Conversely,PDHB expression was negatively correlated with IC50s of Dasatinib and Dolastatin 10 (Figure 12).In summary,PDHB expression is closely related to the susceptibility of certain drugs.

Figure 12 Drug sensitivity analysis.Correlation of IC50 with pyruvate dehydrogenase E1 subunit β expression for different drugs. A:Chelerythrine;B: Nelarabine;C: Fludarabine;D: Fenretinide;E: Lapachone;F: Vorinostat;G: Dasatinib;H: Dolastatin 10.PDHB: Pyruvate dehydrogenase E1 subunit β.
Validation of PDHB expression
As shown in Figure 13A-E,PDHB was differentially expressed at the mRNA level in gastric cancer cell lines (HGC-27,MGC-803,MKN-45,AGS),colon cancer cell lines (SW480,SW620,HCT116),hepatoma cell lines (SMMC-7721,HepG2,H-97,Huh7),breast cancer cell lines (MDA-MB-231,MCF-7),prostate cancer cell lines (PC-3,22Rv1,DU145) and the corresponding normal cell lines.The results showed that the mRNA expression levels of PDHB in gastric and colon cancer cells were significantly lower than those in normal cells (Figure 13B and C) and upregulated in liver and prostate cancer cells (Figure 13A-E),which was consistent with the results of bioinformatics analysis (P<0.05).However,mRNA expression of PDHB in breast cancer cell lines was lower than in normal breast cells,contradicting the bioinformatics analysis results (Figure 13D).In addition,in order to further understand the difference in protein expression of PDHB in liver and gastric cancer cells,the western blot method was used for analysis and data quantification.As shown in Figures 5E and F,PDHB showed a high expression trend in SMMC-7721,HepG2,H-97,and Huh7 hepatoma cell lines compared to normal hepatocytes L-O2,which is consistent with the results of bioinformatics analysis and qRT-PCR (Figure 13F and G).
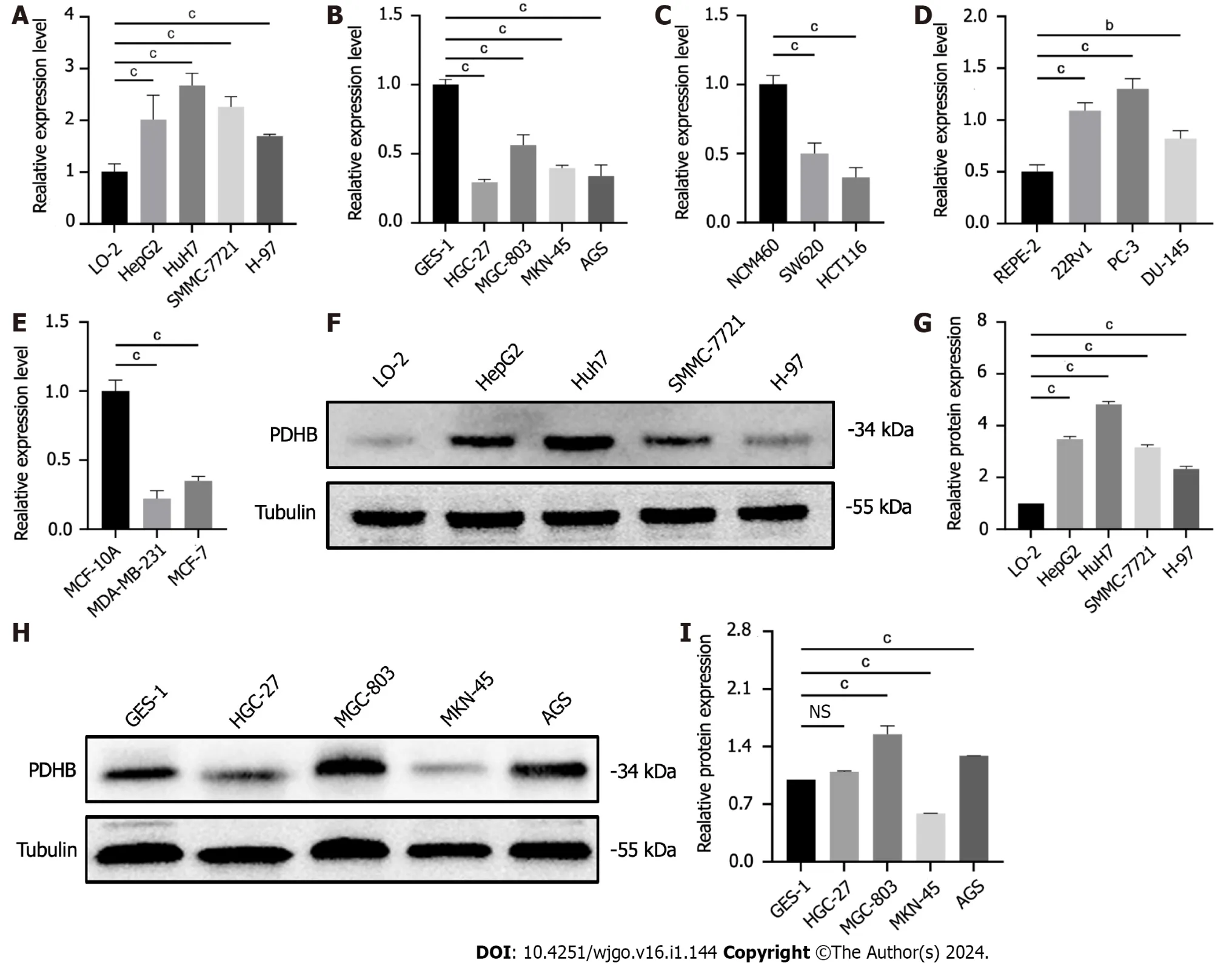
Figure 13 Results of pyruvate dehydrogenase E1 subunit β expression validation. A: Pyruvate dehydrogenase E1 subunit β (PDHB) expression in the human normal hepatocyte cell line (L-O2) and human hepatoma cell lines (SMMC-7721,HepG2,Huh7,H-97);B: PDHB expression in the human normal gastric mucosal cell line (GES-1) and human gastric cancer cell lines (HGC-27,MGC-803,MKN-45);C: PDHB expression in the human normal colonic epithelial cell line(NCM460) and human colon cancer cell lines (SW620,HCT116);D: PDHB expression in the human normal breast cell line (MCF-10A) and breast cancer cell lines(MDA-MB-231,MCF-7);E: PDHB expression in the human normal prostate cell line (RWPE-2) and prostate cancer cell lines (PC-3,22Rv1 and DU145);F: Validation of PDHB protein expression in the L-O2 and hepatoma cell lines (HepG2,SMMC-7721,Huh7,H-97);G: Quantitative plots;H: Validation of PDHB protein expression in the GES-1 and gastric cancer cell lines (MKN-45,AGS,HGC-27,MGC-803);I: Quantitative plots.aP <0.05,bP <0.01,cP <0.001.PDHB: Pyruvate dehydrogenase E1 subunit β;L-O2: Human normal hepatocyte cell line;GES-1: Gastric mucosal cells;NS: Not significant.
Compared with normal gastric mucosal cells,PDHB had higher protein expression levels in AGS and MGC-803 gastric cancer cells,but its protein expression was significantly downregulated in HGC-27 cell lines.Differences in transcription and translation levels between cells of the same type of cancer require further investigation (Figure 13H and I).
PDHB as a biomarker for liver cancer
The above analysis showed that PDHB had significantly increased mRNA and protein expression levels in liver cancer patient samples and hepatoma cells,and had good diagnostic accuracy for liver cancer (AUC=0.801).In addition,in order to facilitate the clinical application of predicting PDHB-related prognosis,we performed multivariate Cox regression analysis of clinical information and genetic characteristics of TCGA liver cancer patients,and included age,sex,tumor node metastasis stage,pathological stage,and PDHB gene to form a linear spectrum (Supplementary Figure 2A).Differentiated and calibrated methods were used for survival outcomes,and calibration curves showed good agreement between predicted and observed survival for years 1,3,and 5 (Supplementary Figure 2B).This cancer-promoting correlation of PDHB was also reflected in liver cancer immunity.As shown in Supplementary Figure 2C,overexpressed PDHB was negatively correlated with most effector immune cells in liver cancer.
The regulatory effect of PDHB on proliferation, migration and invasion of hepatoma cells
In view of the unique diagnostic potential of PDHB for liver cancer,further functional studies were carried out to examine the potential regulatory role of PDHB in liver cancer.Due to the high expression levels of PDHB in Huh7 hepatoma cells,we selected these cells for subsequent experimental interventions.Four PDHB siRNA knockout vectors were transferred to Huh7 hepatoma cells to detect transfection efficiency by qRT-PCR and western blotting.As shown in Figure 14A-C,PDHB knockout was successful after transfection,and the results showed transfection efficiency greater than 70%.The effect of PDHB on the proliferative capacity of hepatoma cells was detected by the CCK-8 method and plate cloning assay (Figure 14D-F).The results showed that when PDHB expression was down-regulated,the proliferative capacity of Huh7 hepatoma cells decreased significantly compared to the control group.By measuring the degree of wound healing to reflect the effect of PDHB on tumor cell migration,the results clearly showed that PDHB significantly inhibited the wound healing effect of tumor cells when its expression in Huh7 cells was downregulated (Figure 14G and H).We subsequently investigated the effect of PDHB on the migration and invasion capacity of hepatoma cells with Transwell migration/invasion experiments,and the results again showed that when PDHB expression was inhibited,the migration ability of Huh7 cells was significantly inhibited (Figure 14I and J).Furthermore,PDHB knockout was observed in the invasion assay resulting in a statistically significant reduction in the number of Huh7 cells passing through the Matrigel (Figure 14I,K).The overall results of this study are shown in Supplementary Table 3.

Figure 14 Effects of siRNA-pyruvate dehydrogenase E1 subunit β on proliferation,migration and invasion of Huh7 hepatoma cells. A:Transfection efficiency of siRNA-pyruvate dehydrogenase E1 subunit β (PDHB) in Huh7 cell lines;B: The relative expression of mRNA reflects the efficiency of siRNA-PDHB transfection;C: siRNA-PDHB transfection efficiency by protein expression level;D: The CCK-8 method detected the proliferation capacity of Huh7 cell lines;E: Colony formation assay to determine the proliferation capacity of Huh7 cell lines;F: The histogram shows the number of colonies formed;G: Cell scratch assay to detect the migration capacity of Huh7 cell lines;H: Histogram of quantification of cell scratch assay results;I: Transwell method was used to detect the migration and invasion capacity of Huh7 cell lines;J: The histogram shows the number of migrating cells;K: The histogram shows the number of invading cells.aP <0.05,bP <0.01,cP <0.001.PDHB: Pyruvate dehydrogenase E1 subunit β.
DISCUSSION
PDHB is highly likely to be an emerging cancer diagnostic marker and potential therapeutic target,but its potential association with cancer has not been clarified;therefore,we conducted comprehensive pan-cancer data mining.This study provides new and strong evidence that improves the current understanding of the characteristics and potential role of PDHB in various cancers.
According to gene expression profiling analysis,PDHB expression varies in different types of cancer,suggesting that PDHB may have a two-way regulatory mechanism and function of promoting or suppressing cancer.For example,Zhuet al[21] found that in colorectal cancer cells,PDHB can act as a sponge of miR-146b-5p,which plays a role in promoting cancer cell growth,invasion and glycolysis,but when PDHB is overexpressed,this cancer-promoting effect will be weakened.In addition,ourin vitroresults show that at the mRNA level,PDHB expression is downregulated in gastric,colon,and breast cancer cells,while up-regulated in hepatoma cells and prostate cancer cells.Among them,the mRNA expression results and protein expression results of PDHB in liver cancer were significantly consistent,and the results of bioinformatics analysis were consistent,and the expression was up-regulated.It is worth noting that the regulation of PDHB expression in tumor cells may be affected by cellular heterogeneity.Cellular heterogeneity is a ubiquitous biological phenomenon,and tumor tissue also has strong cellular heterogeneity[22].Recent studies have shown that even the same cells may have significant heterogeneity.This heterogeneity exists at all levels of DNA,RNA,proteins,etc[23].The transcriptome expression levels of gastric cancer cells showed that PDHB expression was significantly downregulated in AGS,MKN-45,HGC-27,and MGC-803 cells.However,at the proteome expression level,PDHB was overexpressed in AGS and MGC-803 cells.Therefore,the heterogeneity of gastric cancer cells may lead to inconsistent expression of PDHB at the mRNA and protein levels in conspecific cell lines,and whether it can lead to differences in protein function still needs further investigation.
Survival analysis showed that the expression of PDHB was down-regulated in KIRC,KIRP,and STAD,which played a role as a tumor suppressor gene to protect patient prognosis.Notably,combined with the cancer expression profile analysis of PDHB,the overexpression of PDHB also played a protective role in the prognosis of patients with LGG and BRCA,while low expression of PDHB in ACC,ESCA,and HNSC led to a poor prognostic outcome.This may be due to the different expression levels of PDHB in various cancers.When PDHB is mutated,deleted,or affected by genetic environmental factors,the pro-or anti-tumor effects of PDHB will be changed,resulting in different prognostic outcomes.Therefore,more in-depth studies on PDHB are needed in the future to understand the specific mechanisms of this phenomenon and to develop more precise treatment strategies for specific types of tumors.Previous studies have also shown that high expression of PDHB is closely related to a good prognosis in KIRC patients,and is a potential prognostic marker for KIRC[10].PDHB may reflect the prognosis of certain cancers.In addition,ROC curve analysis showed that PDHB had good tumor diagnostic value in a variety of cancers.
Among the 33 malignancies,the number of nonsynonymous variants within the somatic genomic region is known as pan-cancer TMB[24].Changes in tumor microsatellite sequence length caused by insertion or deletion mutations during DNA replication are called MSI and are mainly caused by defects in the deficient MisMatch Repair (dMMR)[25].MSI-H/dMMR and TMB-H reflect the frequency of genomic mutations and may indicate the efficacy of ICP inhibitor therapy[26].In addition,tumor samples of MSI-H tend to have a TMB-H phenotype[27].Our findings show that PDHB was significantly associated with TMB expression levels in UCEC,THYM,STAD,LGG,KIRC,CESC,and BRCA.Whereas,the expression levels of PDHB in UCEC,STAD,READ,KIRC,ACC,PRAD,OV,LUSC,LUAD,KICH,GBM,and BRCA were significantly correlated with MSI expression.Among them,the expression of PDHB was positively correlated with the expression of TMB and MSI in UCEC and STAD,but negatively correlated with the expression of both in BRCA.This suggests that PDHB expression is potentially linked to TMB and MSI in a variety of cancers.Altered DNA methylation status is an important driver of cancer development[28].DNA methylation occurs at different regions of the gene,resulting in significant differences in function.Hypermethylation levels at gene promoters and transcriptional start sites inhibit gene expression,while hypermethylation levels at gene bodies act to promote gene expression,which will confer new genetic traits on cancers[29].Gene expression is controlled by a complex set of dynamic network systems,and the role of DNA methylation in regulating gene expression is equally complex and reversible[30].Combined with the results of PDHB gene expression,it may be a potent methylation driver gene.The correlation analysis of DNA methylation reflected its potential regulatory effect on the expression of PDHB in different cancers.
In recent years,studies using cancer-associated cells in the TME as therapeutic targets have become increasingly popular[31].StromalScore and ImmuneScore are valuable for the diagnosis and prognosis of tumors.We used the StromalScore and ImmuneScore to predict the correlation of PDHB genes with stromal and immune cell content in different tumors.PDHB expression was negatively correlated with stromal cell or immune cell abundance in a variety of cancers,suggesting a potential association between PDHB and tumor purity in multiple cancers.The composition of the tumor immune microenvironment is very complex,ranging from effector immune cells such as CD4+T cells and CD8+T cells to immunosuppressive cells such as Tregs,CAFs,and TAMs,which together constitute the immune mechanism regulatory network of tumors[32-34].The tumor's immune machinery ensures a balance between tumor immune activation and immune suppression,which is essential.When a factor upsets this balance,the inactivation of anti-tumor effects and the occurrence of immune escape will greatly benefit the shaping of the tumor growth environment[35].At the same time,understanding the composition of immune cells in the TME can lead to better investigation of effective immunotherapy strategies.We found that in most cancers,PDHB expression was significantly correlated with the infiltration of multiple immune cells.However,it is worth noting that in some cancers,the expression of PDHB had the same correlation trend as effector immune cells and suppressor immune cells,that is,it is simultaneously positively or negatively correlated.This suggests that PDHB may have a dominant selectivity for the expression of immune cells,thereby playing the role of dominant immune cells.On the other hand,immune cells in the TME may have a dual role,and different immune cells may determine the final immune process through tandem action.Studies have suggested that when the TME is induced by tolerant immune cells such as Tregs and TAMs to become an immunosuppressive environment,the function of effector immune cells such as NK cells will be hindered,and the immunosuppressive effect of the TME will cause NK cells to exert tumor-promoting functions[36].Furthermore,PDHB expression is associated with chemokines and chemokine receptors,which in the TME can have a recruiting effect on immune cells and further regulate their migration and localization[37].We found that the chemokine receptors CCR4,CCR5,and CCR10 of Tregs and the response chemokine CCL22 were negatively correlated with the expression of PDHB in cancers with a better prognosis.Tregs have been shown to play an important adjuvant role in the development of many types of cancer[38],suggesting that the inhibitory effect of PDHB on chemokines and their receptors may contribute to a good prognosis in cancer patients.In addition,PDHB expression was significantly negatively correlated with the expression of ICP genes such as PD-L1,CTLA4 and TIGIT.High expression of PD-L1 or CTLA-4 is an important cause of uncontrolled T lymphocyte-mediated specific immune responses or the occurrence of immune escape[39].In conclusion,our study shows a potential association between PDHB and tumor immunomodulation,but the mechanism by which it affects cancer immune function is complex and unclear,which needs to be further explored by additional studies.
Single-cell sequencing has profound implications for cancer treatment and makes it easier to explore the biological function of single-cell targets in cancer[40].Single-cell transcription sequencing of pan-carcinoma showed a clear association between PDHB and stemness in OV,angiogenesis in RB,and DNA repair in UVM.It has been reported that downregulation of the DNA repair gene was able to hinder the growth and metastasis of UVM cells in zebrafish xenograft models and nude mice[41].Another study showed that exosomes secreted from RB cells can promote tumor angiogenesis through microRNAs[42].This suggests that PDHB may play an important role in the development of cancer by operating different phenotypic functions.In addition,we performed co-expression analysis and functional enrichment analysis of PDHB and its related genes,and found that they were mainly related to cellular energy metabolism components and methylation-related functions,such as mitochondrial inner membrane,mitochondrial matrix,mitochondrial intermembrane space,respiratory chain,methyltransferase activity,tRNA methyltransferase activity,etc.Mitochondria play a key role in cell signaling,metabolism and energy metabolism,and abnormalities in their metabolism can lead to a variety of diseases including cancer[43].The correct structure of the inner mitochondrial membrane ensures efficient cell respiration and regulates the important balance of apoptosis and survival[44].It has been shown that the main sites of production of reactive oxygen species (ROS) are the mitochondrial matrix and the intermembrane space of mitochondria and that abnormal accumulation of ROS can also induce ferroptosis in cancer cells[45,46].In addition,methyltransferases are widely found in plants,animals and microorganisms and use S-adenosylmethionine as a methyl donor to catalyze methylation reactions and may lead to the inactivation of some tumor suppressor genes,driving tumor progression[47].The above analysis results suggest that PDHB may be potentially associated with changes in mitochondrial energy metabolism and methyltransferase activity during tumorigenesis and progression.
HCC is the most common primary liver cancer and is a highly aggressive malignancy[48].In recent years,it has been reported that the bidirectional effect of copper on liver cancer has been elucidated.Overloaded copper may promote the malignant transformation of HCC by regulating proliferation-related signaling pathways,ROS levels,and metabolic balance,and may also trigger cuproptosis and inhibit the development of HCC;however,these mechanisms are not well understood[49,50].One of the characteristics of cancer is the impaired energy metabolism process,which allows cancer cells to use recombination such as mitochondrial metabolism to promote the onset and progression of malignant tumors[51].As a potential cuproptosis-related gene,PDHB is also a regulatory molecule of energy metabolism,which may have the corresponding biological regulatory ability of HCC.Studies have shown that the adaptation of PDHB to glutamine depletion in HCC cells is particularly important in pyruvate metabolism.In glutamine deficiency conditions,when PDHB expression is downregulated,mitochondrial function and TCA metabolism of HCC cells are inhibited,and the proliferative capacity of HCC cells is significantly weakened[52].Therefore,the potential biological role of PDHB in hepatoma cells is worth further exploration.
Bioinformatics analysis based on PDHB in liver cancer showed that it may be a potential molecular marker for the diagnosis of liver cancer and may further affect the prognosis of patients.Therefore,we chose liver cancer as an entry point for further exploration.Throughin vitroexperiments,it was found that knocking down the expression level of PDHB could significantly reduce the proliferation,migration,and invasion ability of hepatoma cells,and confirmed that overexpressed PDHB has a unique cancer-promoting effect in some high-expression malignancies such as liver cancer,and can promote the development of multiple malignant phenotypes in tumor cells.
CONCLUSION
In summary,we analyzed the potential regulatory role and related biological processes of PDHB in cancer from multiple perspectives and aspects using a comprehensive pan-cancer analysis.In addition,we used a "broad to specific" model to explore the great potential of PDHB as a biomarker and therapeutic target for liver cancer.However,there are some limitations to this study.First,our study is based on a large number of existing microarray and sequencing results,which has the potential to lead to incorrect analysis of cellular markers.Second,we are not yet able to clearly understand the exact mechanism by which PDHB regulates tumor immunity,and this requires more in-depth research.This study preliminarily validates that PDHB may be used as a potential biomarker for cancers of which liver cancer is the predominant type,and may have great prospects in immunotherapy and targeted therapy.
ARTICLE HIGHLIGHTS
Research background
Cancer remains the most serious public health problem worldwide,with a high mortality rate.The best prognosis of cancer patients benefits from early detection and timely treatment.Therefore,early diagnosis and treatment are particularly important,and the discovery of new diagnostic and prognostic markers in addition to potential molecular therapeutic targets are essential.
Research motivation
Pyruvate dehydrogenase E1 subunit β (PDHB) is closely associated with the regulation of cancer metabolism and is significantly differentially expressed in multiple cancers.However,there is a lack of reports exploring the role of PDHB in cancer from multiple perspectives,and we expect to provide evidence for the use of PDHB as a potential biomarker for cancers,predominantly liver cancer.
Research objectives
In this study,we used bioinformatics methods and cell function experiments to investigate the diagnostic and prognostic value and tumor immune relevance of PDHB in pan-cancer,as well as its biological regulation in liver cancer.
Research methods
PDHB-related pan-cancer data were obtained from The Cancer Genome Atlas (TCGA) database,and gene expression profiles of PDHB were explored based on TCGA and Genotypic Tissue Expression Dataset databases.The correlation between PDHB and survival was analyzed using Cox regression analysis and Kaplan-Meier methods.Receiver operating characteristic diagnosis,tumor staging,mutation assessment,tumor mutation burden (TMB),microsatellite instability (MSI),DNA methylation and drug sensitivity were also assessed for PDHB.Correlation of PDHB with immune cell infiltration and tumor chemotactic environment,and co-expression analysis of PDHB with immune checkpoint (ICP) genes were analyzed using different algorithms.The expression and functional phenotypes of PDHB in tumor single cells were investigated by single-cell sequencing,and enrichment analysis of the potential oncogenic functions of PDHB was performed.The expression of mRNA or protein levels of PDHB in several cancers was validated.Finally,the regulatory effect of PDHB on the proliferation,migration and invasion of liver cancer was verified.
Research results
PDHB was clearly differentially expressed in most cancers.PDHB was significantly associated with the prognosis of patients with a variety of cancers.In some cancers,PDHB expression was clearly correlated with gene mutation,pathological stage,TMB,MSI,and ICP gene expression.The expression of PDHB was closely related to the infiltration of various immune cells in the immune microenvironment and the regulation of tumor chemotactic environment.In addition,single-cell sequencing results showed that PDHB was associated with different biological phenotypes of single cells in a variety of cancers.The present study further demonstrated that downregulation of PDHB expression inhibited the proliferation,migration and invasion of hepatoma cells.
Research conclusions
PDHB may be a novel cancer marker with potential value in tumor diagnosis,prognosis prediction,immunomodulation,and liver cancer-targeted therapy.
Research perspectives
The role of PDHB in specific types of cancer should be further investigated and the mechanisms by which PDHB is associated with cuproptosis in cancer require further examination.
ACKNOWLEDGEMENTS
We appreciate the help of the RCA citation tool (https://www.referencecitationanalysis.com/) in improving our article.We would like to thank the Key Laboratory of Molecular Diagnostics and Precision Medicine for Surgical Oncology in Gansu Province,and the General Surgery Clinical Medical Center of Gansu Provincial Hospital for their contributions.
FOOTNOTES
Co-first authors:Yao Rong and Song-Hua Liu.
Author contributions:Rong Y and Cai H conceived and designed the study;Rong Y wrote and revised the manuscript,and Rong Y and Liu SH jointly completed the methodology section,Rong Y mainly performed the operation of related software and the production of charts related to bioinformatics analysis,and Liu SH mainly carried out bioinformatics analysis and statistical analysis of the experimental data;Rong Y,Liu SH and Tang MZ jointly completed quantitative reverse transcription PCR,western blot,and cell function experiments;Rong Y and Liu SH made outstanding contributions to the manuscript revision process,with Rong Y rewriting the manuscript content and Rong Y and Liu SH correcting all figures;Rong Y and Liu SH contributed equally to this article.Wu ZH,Ma GR,Li XF performed all the data collection and analysis,and generated the charts;all authors read and approved the final manuscript.
Supported byThe 2021 Central-Guided Local Science and Technology Development Fund;Lanzhou COVID-19 Prevention and Control Technology Research Project,No.2020-XG-1;Gansu Province Outstanding Graduate Student "Innovation Star" Project,No.2022CXZX-748,No.2022CXZX-746.
Conflict-of-interest statement:The authors declare that they have no conflicts of interest to report regarding the present study.
Data sharing statement:The original contributions presented in the study are included in the article/supplementary material.Further inquiries can be directed to the corresponding author.
Open-Access:This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers.It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license,which permits others to distribute,remix,adapt,build upon this work non-commercially,and license their derivative works on different terms,provided the original work is properly cited and the use is non-commercial.See: https://creativecommons.org/Licenses/by-nc/4.0/
Country/Territory of origin:China
ORCID number:Hui Cai 0000-0001-5857-1744.
S-Editor:Qu XL
L-Editor:Webster JR
P-Editor:Zhang XD
 World Journal of Gastrointestinal Oncology2024年1期
World Journal of Gastrointestinal Oncology2024年1期
- World Journal of Gastrointestinal Oncology的其它文章
- Early gastric cancer recurrence after endoscopic submucosal dissection: Not to be ignored!
- Present situation and prospect of immunotherapy for unresectable locally advanced esophageal cancer during peri-radiotherapy
- Comprehensive evaluation of rare case: From diagnosis to treatment of a sigmoid Schwannoma: A case report
- Emerging role of liquid biopsy in rat sarcoma virus mutated metastatic colorectal cancer: A case report
- Application of neoadjuvant chemoradiotherapy and neoadjuvant chemotherapy in curative surgery for esophageal cancer: A metaanalysis
- Comprehensive analysis of the role of ubiquitin-specific peptidases in colorectal cancer: A systematic review
