Codominance Functional Marker of Bacterial Blight Resist-ance Gene Xa7 in Rice
Jian PENG Jun LIU Jianghui YU Youlun XIAO Lin JIA Xiaomei TANG Xiaoping ZHOU Cheng YU Jia LIU


Abstract[Objectives] A codominance functional marker of the broad-spectrum bacterial blight resistance gene, Xa7, of rice was identified for accurate detection, generation tracking, and differentiation between homozygous and hemizygous genotypes of the gene. [Methods] A potential functional marker containing four primers was designed using Premier 5 software and based on the differences on the sequences of Xa7, xa7, and allele-free genomes. The molecular distinctness of the marker in different materials was verified by PCR. Three crossbreed lines of Xa7 and their parents were inoculated with seven bacterial blight strains at the booting stage to examine the affected agronomic traits at maturation. [Results] The homozygous R084 of Xa7 could be amplified into a 91 bp band and the Nip free of allele with a 153 bp band, while the heterozygote Nip/R084, 91 bp and 153 bp bands. The candidate codominance marker, Xa7fun, amplified fragments that matched the predicted target bands. No 91 bp fragment was amplified from 18 germplasms of varied types, indicating a lack of Xa7 in them. Whereas Ry1, Ry2 and Ry3 had a 91 bp band, suggesting the inclusion of homozygous Xa7. Under an elevated temperature, Huazhan responded to the seven bacterial blight pathogens as highly susceptible (HS), intermediate susceptible (MS), or susceptible (S); R084 to six of the seven pathogens (HNA1-4, FuJ, GDA2, GD1358, PX086, and YN24) as highly resistant (HR), intermediate resistant (MR) or resistant (R); Ry-1 to five pathogens (GDA2, HNA1-4, FuJ, GD1358, and YN24) as HR or MR; Ry-2 to five pathogens (GDA2, GD1358, HNA1-4, PXO86, and YN24) as HR or R; and Ry-3 to 6 pathogens (HNA1-4, FuJ, GDA2, GD1358, PXO86, and YN24) as HR or MR. Therefore, the infiltration of Xa7 in the improved crossbred lines RY-1, RY-2, and RY-3 significantly accentuated the blight resistance of Huazhan. [Conclusions] Homozygous or hemizygous Xa7 could be accurately differentiated by the currently identified codominance functional marker Xa7fun. The Xa7 introgression did not significantly alter the critical agronomic traits in the hybridization from generation to generation and could be safely applied in breeding rice varieties with bacterial blight resistance.
Key wordsRice; Bacterial blight; Xa7 gene; Molecular maker; Resistance
DOI:10.19759/j.cnki.2164-4993.2023.05.001
Rice (Oryza sativa L.) is the main food crop for nearly half of the worlds population, as well as a main grain ration second only to corn in China, and it is widely distributed in the six major rice regions[1]. Rice cultivation and production have been affected by various diseases. Bacterial blight caused by the Gram-negative bacterium Xanthomonas oryzae pv. oryzae (Xoo) is a worldwide bacterial disease, particularly severe in Asia such as China, Japan, and India. The rice area south of the Yangtze River in China is a high incidence area of rice bacterial leaf blight, with a wide range of occurrence, fast epidemic speed, great harm, and high mutation, and the affected rice fields generally cause 20%-40% of yield loss, and in severe cases, the loss is even higher or total crop failure occurs[2-5]. Long-term practice has shown that although chemical bactericides can effectively control the occurrence of bacterial blight. However, it is a bacterial vascular disease, bactericides cannot directly contact the lesion, resulting in poor control effects, increased planting costs, environmental pollution, and disruption of ecological balance[6-7]. Therefore, using resistance (R) genes to select rice varieties with genetic resistance is the most economical, effective, and environmentally friendly method for controlling bacterial blight[8-9]. Up to now, at least 47 resistance genes for bacterial blight have been studied and reported[10-11], and sixteen genes, including Xa1, Xa2, Xa14, Xa31 (t), Xa45 (t), Xa3/Xa26, Xa4, xa5, Xa7, Xa10, xa13, Xa21, Xa23, xa25, Xa27, and xa41 (t), have been cloned, providing a wide range of genetic resources for cultivating disease-resistant varieties[6,11]. Among them, Xa7 is a dominant R gene located on chromosome 6 of rice, exhibiting resistance during the adult-plant stage, with broad spectrum and persistence. Moreover, under high temperature environments, gene Xa7 is more prominent in inducing defense responses to prevent bacterial invasion[11-12]. Since the identification of gene Xa7 from the Bangladesh indica rice variety DV85 in the 1970s, it has been highly concerned, applied, or studied[13-19]. It was not until 2021 that Chen et al.[11] successfully cloned this gene from rice variety R084 (Zhenhui 084). Research has shown that TALE AvrXa7, encoded by the avirulent gene AvrXa7, triggers resistance to gene Xa7 in rice. Gene AvrXa7 has been found in various bacterial blight strains, endowing gene Xa7 with the characteristic of broad-spectrum resistance. It exhibits resistance to 52 different bacterial blight strains from China and Japan, and is induced faster and higher at high temperatures[11,14]. Before the cloning of gene Xa7, many linkage markers of gene Xa7 were developed for breeding rice with bacterial blight resistance, such as linkage markers G1091 (with a genetic distance of 6.0 cM from Xa7), AFLP-31-10 (3 cM), STSP3 (0.9 cM), M3 (0.5 cM), M4 (1.8 cM), M5 (0.5 cM), GDSSR02-RM20593 (0.21 cM), RM20582 (0.14 cM), and fluorescent molecular marker PM-Xa7[15-19]. Research has shown that there may be recombinational crossover between linkage markers and target genes, resulting in the loss of target genes due to false positives, while functional markers of target genes themselves are fully coupled with them, avoiding genetic redundancy and recombination problems existing in linkage markers and improving the efficiency and accuracy of MAS selection[1,20]. After the cloning of gene Xa7 in 2021, Liu et al.[21] designed a codominant marker M6 closely linked to gene Xa7 and a functional dominant marker MX7 designed based on the sequences of the promoter and CDS regions of gene Xa7. M6 is used for detection and tracking of genes Xa7 and xa7 in lower generations, effectively differentiating between homozygous or heterozygous genotypes of genes Xa7 or xa7, while MX7 is used for differentiating Xa7 and xa7 genotypes in higher generations. Because MX7 is a dominant marker, it is impossible to confirm the homozygous or heterozygous genotypes of xa7 or xa7 when detecting materials of lower generations. Although M6 can directly identify homozygous or heterozygous genotypes, it cannot differentiate Xa7 or xa7 genotypes, and it is a linkage marker. Further research is needed on using one set of functional marker to directly detect heterozygous or homozygous genotypes of genes Xa7. In this study, a functional marker of a pair of primers was designed in the promoter region based on the differences between DNA sequences of genes Xa7 and xa7, to differentiate Xa7 or xa7 genotypes, and a linkage marker of a pair of primers was designed at a distance of approximately 202 kb downstream of gene Xa7, to differentiate Xa7/xa7 genotypes from allele-free genomes. Consequently, homozygous and heterozygous genotypes of gene Xa7 or xa7 or allele-free genomes could be effectively differentiated through a single PCR carried out in one PCR tube using the four primers at the same time, and the electrophoretic bands used to identify genes Xa7 and xa7 were functional bands. This study aimed to develop functional markers that could provide technical support for MAS breeding of breeding materials with bacterial blight resistance.
Materials and Methods
Experimental materials
Functional marker verification materials: Xa7 gene donor R084 (Zhenhui 084), Nip, Nip (female parent)/R084 (male parent) F1 material; verification materials for different rice germplasm resources: 18 rice germplasm resources (six japonica rice, four indica wild abortion maintainer lines, eight indica rice); two F3 lines Y629 and Y192, selected through hybridization and multiple crossing and other methods using R084 as the donor; high-generation hybrid breeding and resistance identification materials: including three lines of Ry1, Ry2, and Ry3 genetically modified with gene Xa7, sourced from BC1F8 generation of Huazhan (female parent)∥Huazhan/R084 (male parent) BC1F8 generation, with Huazhan as the recipient material and R084 as the donor material.
The above materials were all preserved or selected by the Rice Research Institute of Changde Agricultural and Forestry Research Institute. The experiment was carried out at the Rice Breeding Base of Changde Agricultural and Forestry Research Institute in 2021.
Design and synthesis of Xa7fun functional markers
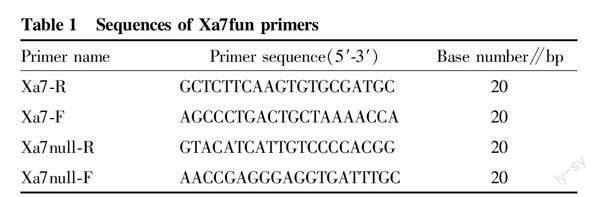
The hybrid and backcross offspring of Huazhan/Huazhan/R084 were tested for the Xa7 gene generation by generation using Xa7 gene linkage marker M5. The forward and reverse primers of M5 were (5′-3′): CGATTACTGGGCTCTGCAACTCTGT and GCATGTCTGTCGATTCGCCGTACGA[15]. Based on the differences in the sequences of gene Xa7 in rice variety R084 and in laozaogu[11,21], two primers were designed using Premier 5 software, namely Xa7-F and Xa7-R. Based on the comparison of the position at a distance of approximately 202 464 bp from gene Xa7 in R084 with the sequence of Nip, the deletion of a DNA sequence was found, and two primers were designed using Premier 5 software, namely Xa7null-R and Xa7null-F. The new functional marker designed with four primers was named Xa7fun (Table 1). The primers were synthesized by Beijing Tsingke Biotech Co., Ltd.
Verification of functional marker Xa7fun
During the tillering stage of rice, DNA was extracted from individual leaves of the test materials using the CTAB method. A PCR amplification system (10 μl) was adopted to amplify target DNA fragments of the testing materials: 2 × RapidTaq Master Mix 5 μl, 100 μmol/L primer 0.1 μl (4 primers of Xa7fun mixed equally, 2 primers of M5 mixed equally), DNA template 1 μl, and ddH2O 3.9 μl. The reaction program of Xa7fun started with pre-denaturation at 94 ℃ for 5 min, followed by cycle of denaturation at 94 ℃ for 30 s, annealing at 56-63 ℃ for 15 s to explore the optimal annealing temperature which was determined to be in the range of 59.0-61.0 ℃, and extension at 72 ℃ for 15 s, and completed with extension at 72 ℃ for 1 min. The PCR products were detected by 3.0% agarose gel electrophoresis. Meanwhile, the designed functional marker Xa7fun was used to verify germplasm resources such as six japonica rice, four indica wild abortion maintainer lines, eight indica rice, and improved lines Y629 and Y192 from different sources. The specificity of electrophoretic bands was verified on the high-generation crossbred lines Ry-1, Ry-2, and Ry-3 of gene Xa7 using linkage marker M5 and functional marker Xa7fun of gene Xa7, respectively.
Identification of resistance to bacterial blight in high-generation crossbred lines of Xa7
gene
Lines Ry1, Ry2, Ry3, Huazhan and R084 for inoculation were seeded on May 15, and the seedlings were manually transplanted in a manner of single parent planting on June 13 with a row spacing of 16.5 cm × 26.4 cm, 100 plants for each line. Conventional cultivation methods were adopted for field management.
The Rice Blast Identification Center of Hunan Institute of Plant Protection provided seven strains of bacterial blight for inoculation identification, namely FuJ, YN24, HNA1-4, GDA2, PXO86, PXO99, and GD1358, of which PXO86 and PXO99 are Philippine races No. 2 and No. 6, respectively, while others are representative strains in Chinas southern rice region[22]. The experiment used the method of leaf cutting inoculation for identification[5,22], with three replicates. During the booting stage, the seven strains of bacterial blight were inoculated to Jingang 30 (susceptible control), R084, Huazhan, Ry1, Ry2, and Ry3. Each strain was inoculated to three plants per material, five leaves per plant. After 21 d of inoculation, when the conditions of the inoculated materials were stabilized, the lengths of disease spots were measured to investigate the incidence and identify the disease resistance of tested plants. Meanwhile, a ZDR-20 continuous temperature and humidity recorder was used to record the natural temperature in the field during the onset of the disease.
Investigation on Xa7
introgression into agronomic traits of high-generation crossbred lines
The initial heading dates of R084, Huazhan, Ry1, Ry2 and Ry3 were recorded to calculate the duration from sowing to heading. After maturation, five plants were continuously measured for plant height in the middle row of R084, Huazhan, Ry1, Ry2, and Ry3, and three plants were continuously mixed in the middle row of each line for the determination of agronomic traits including panicle length per plant, effective panicles per plant, total grains per plant, full grains per plant, 1 000-grain weight, and yield per plant, and calculation of seed setting rate, in three replicates.
Data processing
The lesion length data of different strains on different individual plants of the test materials were analyzed, and based on the incidence and disease level of the susceptible control material Jingang 30, the pathogencity of seven bacterial blight strains to different rice strains were analyzed. The resistance and susceptibility were graded according to following resistance grading standards[5]: high resistance (HR) with average lesion length less than 1 cm, resistance (R) 1.1-3.0 cm, moderate resistance (MR) 3.1-5.0 cm, moderate susceptibility (MS) 5.1-12.0 cm, susceptibility (S) 12.1-20.0 cm, and high susceptibility (HS) greater than 20.1 cm. Microsoft Excel 2020 was used for data statistics, and DPS 15.10 was used to analyze the significance of differences in agronomic traits among different materials.
Jian PENG et al. Codominance Functional Marker of Bacterial Blight Resistance Gene Xa7
in Rice
Results and Analysis
Design and development of codominant functional marker of gene Xa7
Based on the cloned genomic sequences of Xa7 (R084) and xa7 (Laozaogu)[11,21], a mutation in gene Xa7 in EBEAvrXa7, consisting of 11 bp base insertion and SNP base substitution (G → T), resulted in the loss of Xa7 resistance, namely the xa7 gene, while Nip free of allele lacked the CDS and promoter region of the Xa7 or xa7 genes. Therefore, a forward primer Xa7-F was designed upstream of the Xa7 gene promoter, and a reverse primer Xa7-R was designed downstream. According to the primer design strategy in Fig. 1-A, R084 containing the Xa7 gene could be amplified into a 91 bp band, Laozaogu containing the xa7 gene could be amplified into present a 102 bp band, while Nip free of allele could not show any band through amplification. GenBank was used for sequence alignment of R084 and Nip, a 133 bp base deletion was found in R084 at a distance of approximately 202 444 bp from the promoter region of gene Xa7. A pair of primers was designed to differentiate R084 from Nip, including a forward primer of Xa7nullf and a reverse primer of Xa7nullr. According to the primer design strategy in Fig. 1-B, Nip without genes Xa7 or xa7 could be amplified into a 153 bp band, while R084 with Xa7 gene could not show any band after amplification. Hence, it could be seen that based on the design strategies of the four primers, R084 containing gene Xa7 could only show a 91 bp band after amplification, and Laozaogu containing gene xa7 could only present a 102 bp band, and Nip (Xa7null) free of alleles Xa7 or xa7 could give a 153 bp band, while the heterozygote of the Xa7/Xa7null genotype could show two bands of 91 and 153 bp after amplification, and heterozygotes of the xa7/Xa7null genotype could present two bands of 102 bp and 153 bp. Therefore, this marker could identify homozygous and heterozygous Xa7 and allele-free genomes through one time of PCR, thereby improving the efficiency of Xa7 gene pyramiding breeding or variety improvement.
Verification of functional marker Xa7fun
Verification of band sizes obtained with functional marker Xa7fun
The results showed (Fig. 2) that at annealing temperatures ranging from 56.0 to 63.0 ℃, a 153 bp band could be amplified from Nip and Nip/R084 using primers Xa7null-R/Xa7null-F, and a 91 bp band could be amplified from Nip/R084 using primers Xa7-F/Xa7-R. However, R084 did not produce any bands using primers Xa7null-R/Xa7null-F, while primers Xa7-F/Xa7-R could amplify a 91 bp band at 56.0-61.0 ℃. At 62.0 and 63 ℃, the bands was lighter and the amplification efficiency was lower. Therefore, the amplification efficiency of functional marker Xa7fun is optimal at annealing temperatures in the range of 59.0-61.0 ℃. Because there were no susceptible materials containing gene xa7 in this study, the unique 102 bp band sequence of gene xa7 could not be verified, but it did not affect the identification of resistance gene Xa7. According to the results of PCR using the four primers of Xa7fun and agarose electrophoresis, homozygous R084 containing gene Xa7 was amplified into a 91 bp band, Nip without gene Xa7 was amplified into a 153 bp band, and both 91 and 153 bp bands were amplified from heterozygous Nip/R084. It could be seen that the results were consistent with the sizes of target fragments predicted during the design of codominant marker and primers.
Verification of different rice germplasm resources using marker Xa7funIn order to verify the distribution of gene Xa7 in different rice germplasm resources using functional marker Xa7fun, PCR amplification and electrophoresis detection were performed on six japonica rice, four indica wild abortion maintainer lines and eight indica rice germplasm resources from different sources using this marker. From Fig. 3-A, it can be seen that none of the 18 different types of germplasm resources could be amplified into the 91 bp functional band, indicating that these materials do not contain gene Xa7. Previous studies analyzed the genetic diversity of 3 010 rice varieties using the Xa7 sequence. Although 493 varieties from the indica rice and aus subgroup were found to contain the same CDS as Xa7/xa7, only 27 of them were the same in the Xa7 gene promoter region with R084, belonging to the Xa7 genotype. Moreover, they were all resistant to bacterial blight pathogen PXO86, indicating that there are not many materials containing resistance gene Xa7[11,23]. The non-functional bands amplified from the 18 germplasm resources using primers Xa7null-R/Xa7null-F showed different polymorphisms, indicating differences in the DNA sequences at a distance of approximately 202 kb from the Xa7 gene promoter region among different rice materials, but this linkage marker does not affect the identification of functional bands of gene Xa7. Meanwhile, in order to further verify the genotyping of functional marker Xa7fun in low-generation improved crossbred lines, 20 individual plants were randomly selected from two F3 lines selected through hybridization and multiple cross with R084 as the donor for PCR amplification and electrophoresis detection. The results showed that two individual plants numbered 22 and 37 of Y629 were homozygous for gene Xa7, while the remaining 18 individual plants were all heterozygous for gene Xa7 (Fig. 3-B); and the two individual plants numbered 39 and 44 in Y192 were heterozygous for gene Xa7, while the remaining 18 individual plants were of the allele-free genotype (Fig. 3-C). In summary, functional marker Xa7fun could effectively differentiate between homozygous and heterozygous genotyping of gene Xa7 in different rice germplasm resources and improved crossbred lines.
Detection of high-generation crossbred lines of gene Xa7 using functional marker Xa7fun
The hybrid and backcross offspring of Huazhan/Huazhan/R084 were tested generation by generation using linkage marker M5 of gene Xa7. Starting from the BC1F3 generation, individual plants homozygous for gene Xa7 with good agronomic traits were selected for seed collection and generation addition, until three plant lines were retained in the BC1F4 generation, named Ry-1, Ry-2, and Ry-3, respectively (Fig. 4-A). In 2021, six random individual plants of Ry-1, Ry-2, and Ry-3 were detected using M5, and all of them contained gene Xa7 and were homozygous (Fig. 4-B). Meanwhile, the six individual plants corresponding to Ry-1, Ry-2 and Ry-3 were detected using the designed codominant functional marker Xa7fun, and all of them only showed the 91 bp functional band (Fig. 4-C), indicating that they were homozygous for gene Xa7, which was consistent with the results detected using M5.
Identification of resistance to bacterial blight in high-generation crossbred lines of gene Xa7
Seven bacterial strains of bacterial blight were inoculated to Ry-1, Ry-2, Ry-3, R084, and Huazhan, all of which had entered the booting stage (July 23). After 21 d of inoculation, the lengths of diseased spots on the test materials were measured. ZDR-20 continuous temperature and humidity recorders showed that the daily maximum temperatures during the period were in the range of 31-39 ℃, with an average of 34.5 ℃, and twelve days showed daily maximum temperatures exceeding 35 ℃. The daily mean temperatures were in the range of 29.5-35.7 ℃, and there were 19 d with daily mean temperatures exceeding 30 ℃ (Fig. 5). Therefore, the inoculation and induction period of bacterial blight disease was prolonged under high temperature stress. The identification results of resistance to bacterial blight disease showed that Ry-1 was highly resistant (HR) to strains GDA2 and HNA1-4, moderately resistant (MR) to strains FuJ, GD1358, and YN24, and moderately susceptible (MS) and susceptible (S) to strains PXO86 and PXO99, respectively. Ry-2 was highly resistant to strains GDA2, GD1358, and HNA1-4, moderately resistant to strains PX086 and YN24, and moderately susceptible and susceptible to strains FuJ and PXO99, respectively. Ry-3 was highly resistant to strain HNA1-4, moderately resistant to strains FuJ, GDA2, GD1358, PXO86, and YN24, and only susceptible to PXO99. However, the recipient parent Huazhan showed high susceptibility, medium susceptibility or susceptibility to the seven strains, while the donor parent R084 of gene Xa7 showed high resistance, medium resistance, or resistance to six of the seven strains except for strain PXO99, to which R084 was susceptible (Table 2). The above results indicated that the infiltration of gene Xa7 significantly improved the resistance to bacterial blight of improved lines Ry-1, Ry-2, and Ry-3, with Ry-3 having the best resistance to bacterial blight. However, Ry-1, Ry-2, Ry-3 and R084 containing gene Xa7 were all susceptible to strain PX099, indicating that gene Xa7 was not resistant to strain PXO99, which is consistent with previous studies[11,21,24].
Effects of Xa7 introgression on agronomic traits of high-generation crossbred lines
In order to further investigate the impact of Xa7 introgression on the agronomic traits of rice, we analyzed the agronomic traits of the three improved lines, Huazhan, and R084. The results (Table 3) indicated that Ry-1, Ry-2 and Huazhan had the same growth period, while Ry-3 had a growth period longer than Huazhan but shorter than R084. The plant heights of the three improved lines were extremely significantly higher than that of Huazhan (P<0.01, the same below), while Ry-1 and Ry-2 were significantly lower than R084 (P<0.05, the same below), and no significant difference was found between Ry-3 and R084. Ry-1 and Ry-3 had significantly more effective panicles per plant than Huazhan and R084, while Ry-2 had no significant differences from Huazhan and R084. The panicle lengths of the three improved lines were significantly smaller than that of Huazhan, but significantly (Ry-1) or extremely significantly (Ry-2, Ry-3) larger than R084. The values of total grains per plant were between R084 and Huazhan. Specifically, Ry-3 was significantly lower than Huazhan and had no significant difference from R084, and Ry-1 and Ry-2 had no significant differences from Huazhan, but were significantly or extremely significantly higher than R084, respectively. Previous studies have shown that 25-30 ℃ is the optimal temperature for the heading and flowering stage of rice. When the daily mean temperature reaches 32 ℃ or above or the daily maximum temperature reaches 35 ℃ or above, it will have extremely adverse effects on flowering and fertilization, leading to abnormal flowering and pollination, resulting in a large number of empty grains. However, there are significant differences among varieties, and if rice can still maintain a high seed setting rate under high temperature conditions, it indicates its strong adaptability to high temperature[25-26]. The tested lines in this study completed heading and flowering from July 27 to August 8, during which the temperature records showed that the daily maximum temperatures were in the range of 35.0-39.0 ℃, and the daily mean temperatures were in the range of 32.0-35.7 ℃ (Fig. 5), indicating that the tested lines were all subjected to natural high-temperature stress during the heading and flowering stage. The seed setting rates of the three improved lines containing gene Xa7 were significantly higher than that of Huazhan, but had no significant differences from that of R084. In addition to being more resistant to bacterial blight under high temperature, gene Xa7 might also enhance the heat resistance of rice materials during heading and improve seed setting rate, but further verification is needed. The three improved lines were inconsistent in the changes of 1 000-grain weight, with Ry-3 being significantly higher than Ry-1, Ry-2 and Huazhan, but Ry-1 and Ry-2 being significantly lower than R084, respectively. There was no significant difference between Ry-3 and R084. The three improved lines had no significant differences in yield per plant from Huazhan and R084. In summary, the growth periods, plant heights, panicle lengths, total grains per plant, seed setting rates and 1 000-grain weights of the three improved lines were between R084 and Huazhan, and the yields per plant were not significant different from Huazhan or R084, indicating that the infiltration of gene Xa7 did not cause significant changes in important agronomic traits of rice materials, suggesting good application prospects.
Discussion and Conclusions
Previous studies have used codominant marker M6, which is closely linked to gene Xa7, to differentiate homozygous and heterozygous materials of genes Xa7 and xa7 and allele-free genomes in low-generation improved lines, or to select homozygotes of Xa7 and xa7 genotypes. However, marker M6 cannot differentiate Xa7 and xa7 genotypes, which are differentiated by dominant functional marker MX7. However, MX7 cannot differentiate homozygous or heterozygous materials of Xa7 and allele-free genomes. Therefore, homozygous improved lines of gene Xa7 can be accurately selected by performing PCR using two sets of markers, M6 and MX7, respectively[21]. In this study, a functional marker Xa7fun, which was designed based on the comprehensive development strategy of linkage marker M6 and functional marker MX7 of gene Xa7, can be used to identify functional primers for Xa7 genotypes or linkage primers for allele-free genomes. The band obtained by electrophoresis using functional primers was a 91 bp functional band of gene Xa7, which could be detected accurately with a high detection efficiency, avoiding false positives due to the loss of the target gene caused by recombinational crossover between linkage markers and the target gene. The band type of the linkage primers was a linkage marker located approximately 202 kb away from gene Xa7. Nip, free of allele, could be amplified into a 153 bp band, while heterozygous F1 of Nip/R084 was amplified into 91 and 153 bp bands. Therefore, it is mainly used to identify allele-free genomes and heterozygotes of gene Xa7. Xa7fun combines both functional and linkage markers in one set of marker, effectively differentiating homozygous or heterozygous genotypes of gene Xa7 in each hybrid generation through a single PCR, making breeding work more efficient and practical.
In this study, due to the lack of donor materials for gene xa7, the Xa7fun marker was not verified for xa7 genotypes during PCR. However, materials containing xa7 are generally used for cloning or functional research of the Xa7 and xa7 genes, as well as the design and verification of Xa7 and xa7 functional markers[11,21]. When using MAS to aggregate target genes, the background selection of donors and receptors is more important, and improved hybrid offspring with complementary traits is often further selected through the hybridization and aggregation of two materials with excellent traits. Therefore, the susceptible recessive gene Xa7 has no aggregation significance for variety improvement. R084 harbors the broad-spectrum resistance gene, Xa7, to bacterial blight disease, and has good agronomic traits. In this study, we used high-quality and widely-promoted restorer line Huazhan as the receptor and resistant material R084 as the donor and selected Ry-1, Ry-2 and Ry-3 through hybridization, backcross, and MAS breeding. Their resistance to bacterial blight disease was significantly improved compared with the recipient parent Huazhan, and important agronomic traits were between or higher than Huazhan and R084, so they combined the advantages of Huazhan and R084.
Research has shown that R084 and Zhe-kang 1, which contain gene Xa7, have no resistance to the physiological race PX099 of Philippine bacterial blight[11,21,24]. The results of this study indicated that the three hybrid improved lines Ry-1, Ry-2, Ry-3 and R084 containing gene Xa7 had no resistance to strain PXO99, which is consistent with previous studies. In addition, the expression or regulation of certain genes from the recipient parent Huazhan might have an impact on the resistance of gene Xa7, resulting in Ry-1 having no resistance to strain PXO86 and Ry-2 having no resistance to strain FuJ. Therefore, the resistance of all offspring of an improved hybrid line is not the same as that of its donor material, and it is closely related to the material background. Further screening is needed in multiple lines of hybrid offspring. For instance, gene Xa23 has characteristics such as wide resistance spectrum, strong resistance, complete dominance, and resistance throughout the entire growth period, its improved crossbred lines are not all resistant, and further resistance screening needs to be carried out by inoculating bacterial blight strains[27-30]. Although the widely-used Xa21 gene has strong resistance, it has no resistance to 96% of bacterial blight strains in Zhejiang, Yunnan, and South Korea, and has little resistance to bacterial blight induced by strain FuJ[5,14,31,32]. Therefore, in the context of global warming, the aggregation of three dominant genes, gene Xa7, which is resistant during the adult stage and have resistance which can be enhanced by high temperature, and genes Xa23 and Xa21 with resistance throughout the entire growth period, is of great significance for resistance breeding against bacterial blight in southern rice regions of China. We believes that the breeding of conventional rice can aggregate two (Xa7+Xa23 or Xa7+Xa21) or three (Xa7+Xa23+Xa21) genes, but the aggregation of too many genes increases the difficulty of breeding or the time for material stabilization. However, in the breeding of hybrid rice, different resistance genes can infiltrate into the parents based on the characteristics of hybrid F1, and then new combinations with complementary resistance genes to bacterial blight disease in the parents of hybrid rice can be bred. A large amount of research has been conducted on the aggregation breeding of gene Xa7 and other bacterial blight resistance genes, and the resistance effects after aggregation are good. Huang et al.[32] aggregated four genes, including Xa7, Xa21, Xa22 and Xa23, into the hybrid rice restorer line Huahui 1035 by the MAS method, and the offspring materials showed varying degrees of resistance to 11 representative strains of bacterial blight in China. Liu et al.[14] showed that Baixiangzhan, which carries genes Xa7 and Xa23, had high resistance to type IV bacteria, while IRBB7, which only carries Xa7, was resistant to type IV bacteria, and Xinhuangzhan, Juhezhan and H120-2-9, which carry Xa23, were resistant to type IV bacteria, and other 28 resources such as Pengdao 2, which carries Xa23, were all moderately resistant to type IV bacteria. Therefore, the aggregation of genes Xa7 and Xa23 into the same rice material increased the resistance level of the material to type IV bacteria. Lan et al.[24] developed two lines containing gene Xa21, resistant to four strains including PXO61, PXO99, ZHE173 and GD1358, but not to strains FuJ and YN24, two lines containing gene Xa7, resistant to five strains including PXO61, ZHE173, GD1358, FuJ and YN24, but not to strain PXO99, and three lines containing genes Xa21 and Xa7, resistant to all six strains. Therefore, the interaction and resistance complementation between different resistance genes can broaden the resistance spectrum, enhance resistance levels, and further increase resistance persistence by broadening the resistance spectrum.
References
[1] YU JH, LI YY, QIN GN, et al. Development and validation of functional markers of low-cadmium accumulation mutation sites in rice OsNRAMP5 gene[J]. Jiangsu Journal of Agricultural Sciences, 2022, 38(2): 289-295. (in Chinese).
[2] CHEN FD, YAN BX, HE ZH. Mechanisms of disease resistance to bacterial blight and perspectives of molecular breeding in rice[J]. China Industrial Economics, 2020, 56(12): 2533-2542. (in Chinese).
[3] LIU WD, LIU JL, TRIPLETT L, et al. Novel insights into rice innate immunity against bacterial and fungal pathogens[J]. Annual Review of Phytopathology, 2014(52): 213-241.
[4] YAN CY, LIU Y, MOU TTM. Improvement of rice bacterial blight resistance of hybrid rice Jinyou 207 by molecular marker-assisted selection[J]. Chinese Journal of Rice Science, 2013, 27(4): 365-372. (in Chinese).
[5] YU JH, LIU TC, WENG LS, et al. Analysis of Xa21 and Xa23 of indica rice varieties in different genetic background to broad spectrum bacterial blight pathogens[J]. Chinese Journal of Tropical Crops, 2021, 42(12): 3433-3442. (in Chinese).
[6] CHEN XF, MEI L, JI ZD, et al. Exploration of new bacterial-blight resistance genes from rice Landrace resources in China[J]. Journal of Zhejiang Normal University (Natural Sciences), 2020, 43(1): 8-12. (in Chinese).
[7] XIANG X, CHEN LL, ZHANG DD, et al. Physical mapping and functional markers of bacterial blight resistance genes in rice[J]. Molecular Plant Breeding, 2019, 17(2): 509-516. (in Chinese)
[8] RAO KK, LAKSHMINARASU M, JENA KK. DNA markers and marker-assisted breeding for durable resistance to bacterial blight disease in rice[J]. Biotechnology Advances, 2002, 20(1): 33-47.
[9] JIANG N, YAN J, LIANG Y, et al. Resistance genes and their interactions with bacterial blight/leaf streak pathogens (Xanthomonas oryzae) in rice (Oryza sativa L.): An updated review[J]. Rice (New York, N Y), 2020, 13(1): 3.
[10] CHEN S, WANG CY, YANG JY, et al. Identification of the novel bacterial blight resistance gene Xa46(t) by mapping and expression analysis of the rice mutant H120[J]. Scientific Reports, 2020, 10(1): 1-11.
[11] CHEN XF, LIU PC, MEI L, et al. Xa7, a new executor R gene that confers durable and broad-spectrum resistance to bacterial blight disease in rice[J]. Plant Communications, 2021, 5(10): 1-14.
[12] ZHOU JH, PENG Z, LONG JY, et al. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice[J]. The Plant Journal:for Cell and Molecular Biology, 2015, 82(4): 632-643.
[13] SIDHU GS, KHUSH GS, MEW TW. Genetic analysis of bacterial blight resistance in seventy-four cultivars of rice, Oryza sativa L.[J]. Theoretical and Applied Genetics, 1978, 53(3): 105-111.
[14] LIU F, LIANG Q, CHEN WX, et al. Analysis of molecular marker on rice germplasms of bacterial blight resistant gene Xa7 and Xa23[J]. Molecular Plant Breeding, 2016, 14(4): 935-940.(in Chinese).
[15] PORTER BW, CHITTOOR J, M YANO M, et al. Development and mapping of markers linked to the rice bacterial blight resistance gene Xa7[J]. Crop Science, 2003, 43(4): 1484-1492.
[16] HE YQ, LI X, ZHANG JF, et al. Gene pyramiding to improve hybrid rice by molecular marker techniques[A]. Brisbans, Queensland, Augstralia: 4th International Crop Science Congress, 2004.
[17] CHEN S, HUANG ZH, ZENG LX, et al. High-resolution mapping and gene prediction of Xanthomonas Oryzae pv. oryzae resistance gene Xa7[J]. Molecular Breeding, 2008, 22(3): 433-441.
[18] LI D Q, ZHONG Q F, ZENG M, et al. Progress in mapping, cloning and application of resistance genes to bacterial blight disease in rice[J]. China Rice, 2017, 23(5): 19-27. (in Chinese).
[19] WU H, DENG GF, GAO LJ, et al. Development and breeding application of the fluorescence molecular marker of rice bacterial blight resistance gene Xa7[J]. Molecular Plant Breeding, 2021, 19(12): 4024-4031. (in Chinese).
[20] SUN PY, ZHANG WH, ZHANG L, et al. Development and application of functional marker for high nitrogen use efficiency and chilling tolerance gene OsGRF4 in rice[J]. Acta Agronomica Sinica, 2021, 47(4): 684-690. (in Chinese).
[21] LIU PC, MEI L, HE LM, et al. Development of markers for identification and maker assisted breeding of Xa7 gene in rice (Oryza sativa L.) [J]. Euphytica, 2021(217): 134.
[22] XIAO YL, LI JJ, YU JH, et al. Improvement of bacterial blight and brown planthopper resistance in an elite restorer line Huazhan of Oryza[J]. Field Crops Research, 2016(186): 47-57.
[23] WANG WS, MAULEON R, HU ZQ, et al. Genomic variation in 3, 010 diverse accessions of Asian cultivated rice[J]. Nature, 2018, 557(7703): 43-49.
[24] LAN YR, WANG JY, WANG Y, et al. Improvement of rice bacterial blight resistance of Hua 201S, an elite photo-thermo sensitive genicmale sterile line, by molecular marker-assisted selection[J]. Chinese Journal of Rice Science, 2011, 25(2): 169-174. (in Chinese).
[25] KUI LM, TAN LB, TU J, et al. Identification of QTLs associated with heat tolerance of Yuanjiang common wild rice (Oryza rufipogon Griff.) at flowering stage[J]. Journal of Agricultural Biotechnology, 2008, 16(3): 461-464.(in Chinese).
[26] YU JH, ZHAO S, ZHOU H, et al. Effect of interval sowing on agronomic traits and thermo-tolerance of japonica rice from Northeast China[J]. Chinese Journal of Eco-Agriculture, 2012, 20(8): 1037-1042. (in Chinese).
[27] YANG DW, YE N, YE XF, et al. Study on enhancing bacterial blight resistance of hybrid rice restorer lines through marker assisted selection of the Xa23 gene[J]. Fujian Journal of Agricultural Sciences, 2015, 30(4): 351-356. (in Chinese)
[28] YU J, WANG YW, MA WQ, et al. The MAS research in the population of bacterial blight resistance gene Xa23 in rice[J]. Journal of South China Agricultural University, 2010, 31(4): 1-5. (in Chinese).
[29] FAN HH, WANG LY, ZHANG LX, et al. Breeding of rice lines with bacterial blight resistance gene Xa23 by using marker-assisted selection[J]. Chinese Journal of Rice Science, 2011, 25(3): 331-334. (in Chinese)
[30] ZHENG KL, ZHUANG JY, WANG HR. Performan
- 农业生物技术(英文版)的其它文章
- Effects of Seed Dressing with Microbial Inoculum on Nutrient Composition and Biological Yield of Silage Corn
- Study on Factors Affecting Aerobial Plate Count in Raw Milk
- Discussion on the Main Reasons for a Large Number of Père David’s Deer (Elaphurus davidianus) with Abnormal Antler Shedding
- Role and Effects of Eucommia ulmoides Leaves in the Growth Process of Livestock and Poultry
- A Study on the Interest Linkage Mechanism of the Taxus Industry Chain in Pengshui Miao and Tujia Autonomous County
- Therapeutic Effect and Mechanism of Jipei Dilong Ointment on Acute Soft Tissue Injury in Rats

