Copy number variation sequencing for diagnosis of cytomegalovirus infection based low‑depth whole‑genome sequencing technology in fetus: Three cases and literature review
CHAI Shi‑wei, CHEN Ze‑jun, LIU Chun‑tao, CHEN Su, HE Gui‑lin, CHEN Yue‑fen,WANG Rui‑xia, ZHU Xin, LING Yi, GU Shuo
Department of Fetal Medicine, First Affiliated Hospital of Hainan Medical College, Haikou 570102,China
Keywords:
ABSTRACT Objective: To summarize the application value of copy number variant sequencing (CNV‑seq)in the detection of fetal chromosome and cytomegalovirus load.Methods: The study analyzed the clinical basic data, relevant laboratory tests, treatment process, and outcomes of three patients with positive cytomegalovirus load detected by CNV‑seq for fetal chromosomes and cytomegalovirus load, and literature review was done simutaneoubly.Results: In all three cases, the amniotic fluid cytomegalovirus load was less than 105 Copies/ml, and there were no significant neurological abnormalities observed during pregnancy or postpartum follow‑up.There is no literature review on the application of CNV‑seq technology in the detection of cytomegalovirus infection, only literature reports on genome analysis of CMV‑DNA in confirmed patients were available.Conclusion: CNV‑seq can be used to detect cytomegalovirus load, which may have a certain degree of predictive value for fetal outcome.CNV‑seq can simultaneously detect fetal chromosomes and pathogenic microorganisms, which is of great significance for the prevention and control of birth defects.
1.Introduction
Birth defects refer to abnormalities in the body structure, function,or metabolism of the fetus, which not only seriously endanger the survival and quality of life of children, but also cause serious economic burden to families and society.Pregnancy related pathogenic microbial infections are an important cause of birth defects[1].Cytomegalovirus (CMV) is one of the most common pathogens of congenital infection.CMV infection during pregnancy can be transmitted vertical transmission through the placenta,resulting in fetal cytomegalovirus intrauterine infection.It is a common cause of birth defects such as congenital neural deafness,visual abnormalities, and mental retardation after birth, and can seriously cause maternal abortion, stillbirth, premature delivery,and neonatal death[2-3].In recent years, the early detection and treatment of cytomegalovirus infection has gradually attracted high attention from obstetricians and prenatal diagnostic doctors.Due to limitations such as reliance on laboratory conditions and lack of standardization of experimental methods, traditional detection techniques still have controversy over their sensitivity and specificity, and traditional techniques cannot distinguish between congenital and postpartum cytomegalovirus infections[4].The genome copy number variant sequencing (CNV seq) based on next‑generation sequencing (NGS) technology provides a new means of detection.The American Society of Obstetricians and Gynecologists (ACOG) has recommended CNV seq as a first‑line diagnostic method for fetal structural abnormalities[5].CNV‑seq utilizes low depth whole genome sequencing to analyze 23 pairs of chromosomal aneuploidies and copy number variations above 100 kb at once[6-7].This study retrospectively analyzed the clinical basic data, relevant laboratory tests, treatment processes,and outcomes of 3 out of 900 patients with ultrasound abnormalities or serological abnormalities in the Fetal Medicine Department of the First Affiliated Hospital of Hainan Medical College who tested positive for cytomegalovirus load with CNV‑seq.Prenatal consultation and follow‑up were provided based on copy numbers to provide new ideas for the diagnosis and treatment of these patients,Provide relevant basis for the application of CNV‑seq technology in predicting neural system defects caused by fetal cytomegalovirus infection, as follows.
2.Case Data
Example 1: 20 years old, G1P0.On May 15, 2020, a nuchal translucency (NT) test of 1.1mm was performed in an external hospital.On June 12, 2020, non‑invasive prenatal DNA testing(NIPT) was performed in our hospital, indicating low risk.On September 3, 2020, no significant abnormalities were found in the fetal system ultrasound examination conducted in our hospital.On September 28, 2020, at 31+weeks of pregnancy, ultrasound revealed“oligohydramnios”.Amniotic fluid samples were taken from our prenatal diagnosis center for CNV‑seq to detect fetal chromosomes and CMV.The results showed that CMV was detected and fluorescence quantitative PCR was used to verify abnormal CMV load.At the same time, the pregnant woman’s CMV serological test showed negative CMV‑IgM and positive CMV‑IgG (in Table 1).On October 5, 2020, at 33+weeks of pregnancy, a live female baby was delivered by cesarean section due to “fetal distress, fetal growth restriction, oligohydramnios”.On October 13, 2020, the newborn underwent peripheral blood CMV testing and showed positive CMV‑IgM.The urine CMV‑DNA quantification was 4.54×104Copies/ml (in Table 2), indicating congenital cytomegalovirus infection.On October 15, 2020, a newborn underwent cardiac ultrasound examination that revealed patent foramen ovale, while a cranial ultrasound examination revealed the brain of a premature infant.The MRI examination of the fetal head showed no abnormalities.
Example 2: 21 years old, G3P1.On January 9, 2022, NT was found to be 1.8mm in the external hospital.On February 17, 2022, the Down syndrome screening results showed low risk, and fetal system ultrasound examination showed no abnormalities.On April 1, 2022,serum CMV testing showed CMV IgM negative and CMV IgG positive (see Table 1).On April 3, 2022, at 26+weeks of pregnancy,ultrasound showed that “the left nasal bone of the fetus is not shown, and the left ventricular bright spot”.amniotic fluid samples were taken from our prenatal diagnosis center for quantitative fluorescence PCR (QF), CNV‑seq detection of fetal chromatin and CMV.CNV‑seq detected CMV and fluorescence quantitative PCR confirmed abnormal CMV load.On July 4, 2022, a live male infant was delivered via vaginal route at 40 weeks of pregnancy.
Example 3: 33 years old, G1P0.On September 20, 2020, NT examination was conducted in our hospital, which was 1.1mm.On November 18, 2020, NIPT indicated low risk.On December 20,2020, obstetric ultrasound was conducted at 25+weeks of pregnancy,which indicated that “fetal heart proportion was increased and cardiac structure was abnormal, interventricular septum defect,aortic straddle, pulmonary atresia”.Considering complex congenital heart disease, amniotic fluid samples were taken from the prenatal diagnosis center of our hospital for QF detection CNV‑seq detection of fetal chromosomes and CMV.CNV‑seq detected CMV and fluorescence quantitative PCR confirmed abnormal CMV load.On January 11, 2021, at 27+weeks of pregnancy, the pregnant woman underwent CMV serum antibody testing, which indicated CMV‑IgG and CMV‑IgM (in Table 1).The patient delivered a live male infant on April 7, 2021 at 36+3 weeks of pregnancy due to “premature rupture of membranes” through cesarean section.The newborn’s postnatal CMV examination showed negative CMV‑IgG, and urine CMV‑DNA quantification was 4.8×104Copies/mL (in Table 2).
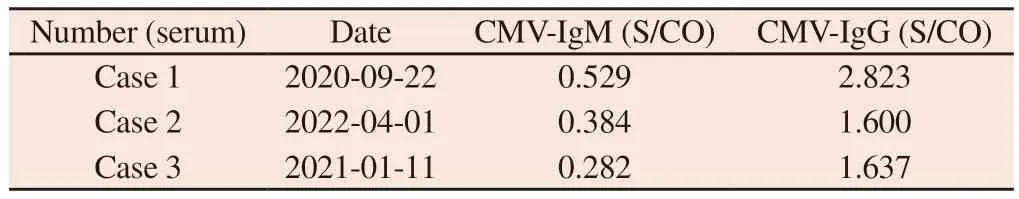
Tab 1 Results of CMV antibodies in pregnant women’s serum
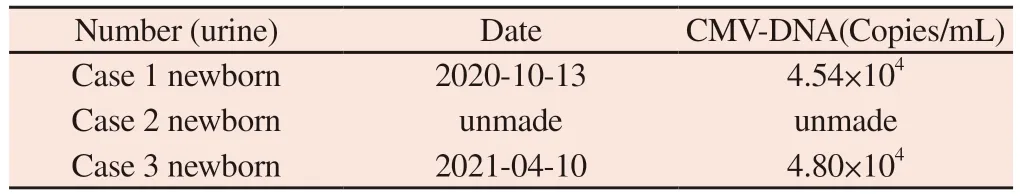
Tab 2 Newborn urine CMV‑DNA results
3.Results
3.1 Prenatal diagnosis and CMV detection of CNV‑seq
CNV‑seq results of amniotic fluid of 3 pregnant women all indicated that no pathogenic or suspected pathogenic variation of aneuploidy, conformity chromosome linked inheritance or autosomal dominant inheritance was detected in the samples (in Table 3).Case 3: Complete exon sequencing of amniotic fluid sample genome was performed for consideration of complex malformations of congenital heart disease in ultrasonic diagnosis, and the results suggested:DYNC2H1 gene c.3782G>T and c.1506T>C complex heterozygous mutations, which are of unknown clinical significance,are associated with short costothoracic dysplasia with or without multiple fingers syndrome type 3, which is inherited autosomal recessive or double‑gene recessive.CMV bioinformatics analysis was performed in the amniotic fluid of 3 pregnant women at the same time of CNV‑seq.The presence of CMV was determined by the ratio of detected sequence number to total sequence number.The ratio of case 1 was 179/2048166(0.000087), case 2 was 15619/1815815(0.008602) and case 3 was 13227/2736964(0.004833)(in Table 3).On this basis, PCR quantitative verification of CMV‑DNA in the amniotic fluid samples confirmed the presence of CMV was carried out.The results showed that CMV was detected in the amniotic fluid samples of all the 3 cases, and the viral load of case 1 was 1227.76Copies/ml, and that of case 2 was 11639.00Copies/ml.Case 3 had a viral load of 13754.60Copies/ml (in Table 3).
3.2 Pregnancy outcome and neonatal follow‑up
No obvious clinical symptoms were found in the three pregnant women during pregnancy, and no symptomatic antiviral treatment was given.Among them, the pregnant women of case 1 and case 3 gave birth to premature infants by cesarean section due to “fetal distress, fetal growth restriction, oligohydramnios” and “premature rupture of membranes”, and the pregnant women of case 2 had a vaginal vaginal vaginal delivery.All three newborns were born with full Apgar score and no obvious abnormalities in appearance.Combined with the results of 3 pregnant women or newborns with serologically specific CMV antibody, urine CMV‑DNA, CNV‑seq qualitative detection of CMV and PCR quantitative verification of CMV, the clinical diagnosis of pregnancy cytomegalovirus infection or congenital cytomegalovirus infection can be considered.Meanwhile, long‑term follow‑up has so far found no difference in growth and development between newborns and their peers.Meanwhile, long‑term follow‑up has so far found no difference in growth and development between newborns and their peers, and no obvious abnormalities were found in hearing detection and other nervous system detection (in Table 4).In Case 3, the patient received“stage‑Ⅰ and stage‑Ⅱ staging operations for whole‑pulmonary shunt and ventricular septal defect repair” in an external hospital due to “complex congenital heart disease malformation” in January 2022 and May 2022.The patient recovered well after surgery, and there was no difference in feeding, growth and development compared with his peers.
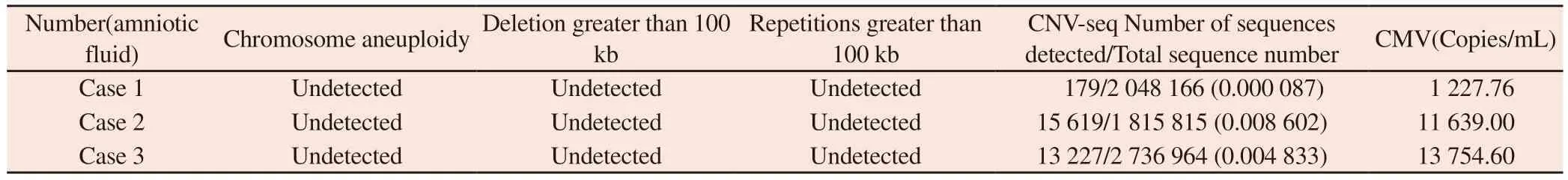
Tab 3 Prenatal diagnosis and CMV test results of CNV‑seq in amniotic fluid of pregnant women
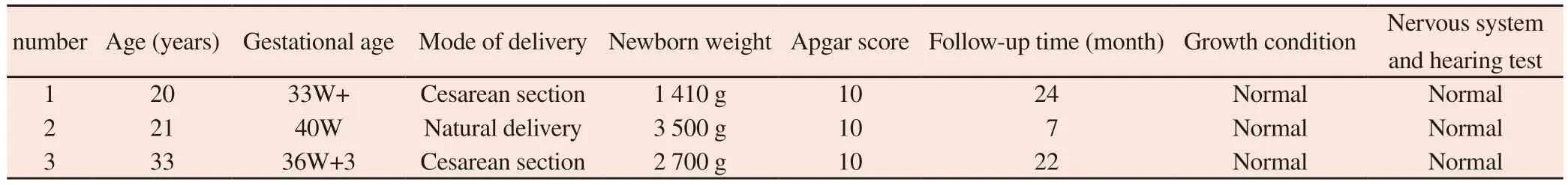
Tab 4 Pregnancy outcomes and neonatal follow‑up of patients
4.Discussion
In this study, the keywords “CNV‑seq and/or CMV”, “high‑throughput sequencing and/or cytomegalovirus” were searched in Pubmed, CNKI, Wanfang and other literature platforms, and a total of 65 CNV‑seq and CMV‑related literature were obtained.However,it mainly focuses on high‑throughput sequencing technology or CNV‑seq in the analysis of CMV genome map or the genetic diversity of CMV.Previous studies collected different samples of clinically diagnosed patients with CMV infection, isolated the CMV virus from different samples, and then sequenced the virus genome to obtain the virus genome map for bioinformatics analysis.Currently, no direct qualitative or quantitative detection of CMV by CNV‑seq or other high‑throughput sequencing techniques has been found.However, in this study, sequence alignment qualitative detection of CMV was performed at the same time of routine prenatal diagnosis of CNV‑seq.On this basis, quantitative PCR verification was conducted on CMV detected by CNV‑seq, and the PCR results were consistent with those of CNV‑seq.
CMV infection is spread from person to person and has obvious species‑specific characteristics to the host.The infection rate varies with regional culture and other factors.CMV is neurotropic and one of the most common viral infections, which can lead to adverse pregnancy outcomes and different degrees of birth defects[8-9].Since there is the possibility of CMV infection at all periods of pregnancy,and with the continuous promotion of the national two‑child and three‑child birth plan, the number of high‑risk pregnant women is gradually increasing, and the situation of CMV infection is not optimistic.However, most pregnant women infected with CMV during pregnancy or newborns with congenital CMV infection have no obvious clinical symptoms, which increases the difficulty in the diagnosis and treatment of CMV infection[10].
Previous studies have suggested that the risk of primary CMV infection in the first 3 months of pregnancy and related sequelae after birth of the fetus in women who are pregnant for the first time and women who have had reproductive history within 2 years is 19 times and 5 times higher than that of the general population,respectively[11].In newborns with clinical symptoms, about 25%will develop long‑term sequelae;65% to 80% of the survivors can be complicated with severe neurological sequelae[12-14].Currently,routine CMV detection methods mainly include detection of serum specific IgG/IgM antibody by ELISA, quantitative detection of viral DNA in urine and blood samples by Q‑PCR, detection of PP65 antigen, and imaging examination (such as ultrasound and MRI).Serological specific antibody is often used as the primary screening method for CMV, CMV‑DNA of urine and blood samples is the gold standard for the diagnosis of CMV, and imaging results are used as the auxiliary diagnostic basis.However, serological CMV‑ IgM positive rate is low, and the positive diagnosis value of CMV‑igg is not high.PCR detection of CMV‑DNA has a time limit for urine samples, and the above detection methods cannot avoid the missed diagnosis of CMV infection due to their respective limitations.In this study, the sera of pregnant women in case 1 and case 3 were positive for IgG, but negative for IgM.The CMV‑DNA loads of newborn urine were all higher than the lower limit of weak positive value range (2.50×104Copies/mL).Although the serological results of the patient were negative, it was not clear whether CMV infection existed or not, and there was still the possibility of missed diagnosis.In this study, cases 1 and 3 were further examined for CMV‑DNA in the urine of neonates after combining the results of hematologic specific antibody with abnormal ultrasound results, and the presence of congenital CMV infection was found in neonates.However, the results of serum specific antibodies of pregnant women in case 2 in this study were consistent with those of Case 1 and Case 3, and the ultrasound screening also indicated abnormalities.However,the possibility of CMV infection could not be ruled out in case 2 because CMV‑DNA of newborn urine was not detected.
In recent years, high‑throughput sequencing technology has developed rapidly, which not only plays an important role in prenatal diagnosis, but also gradually increases the research on pathogens in various clinical samples.Among them, CNV‑seq, as a first‑line prenatal diagnostic technology, is applied in the detection of genetic diseases or genetic diseases, as well as in the analysis of the whole genome genetic information of pathogenic microorganisms in clinical samples.In 2017, EliasHage et al.applied NGS high‑throughput sequencing technology to isolate CMVS from different samples of clinical CMV infection, including blood, urine and diseased tissues of children with organ transplantation and congenital CMV infection, conducted whole‑genome sequencing of CMV and sequence analysis of CMV‑rich targets.The diversity of CMV genome changes and the mutational characteristics of antiviral resistance were revealed[15].At the same time, the high‑throughput sequencing technology for the whole CMV genome is also constantly optimized.Some research teams help to improve the CMV gene profile through the development of high‑quality CMV‑DNA preparation and the optimization of NGS sequencing process[14].In particular, high‑throughput sequencing studies on CMV in the urine of children with congenital CMV infection have found that the use of highly sensitive deep sequencing methods can help to understand the genotyping and clinical genetic diversity of CMV[16].However, the above studies focus on CMV genome sequencing after isolation of CMV virus from clinical samples,and do not involve high‑throughput sequencing technology to study the CMV‑DNA load in the samples.Previous studies have conducted high‑throughput sequencing on peripheral blood of patients with hematopoietic stem cell transplantation, and detected gene variation in CMV‑infected virus strain profiles and low load of CMV‑DNA through targeted enrichment[17].Another study of genome‑wide single nucleotide polymorphism (SNP)genotyping found that multiple CNV regions were associated with HIV‑1 viral load[18].Recently, Huada Corporation has carried out bioinformatics analysis of pathogen detection by high‑throughput sequencing technology CNV‑seq, which uses two Metaphan2 and Kraken metagenomics tools to construct bioinformatics process of pathogen microbial detection[19-20].In this study, the above CNV‑seq technique was used to detect CMV in all 3 cases.Through the analysis of the ratio of detected sequence number to total sequence number in sequencing data, it was found that CMV existed in the amniotic fluid samples of all 3 patients.Their comparison values were 179/2048166(0.000087), 15619/1815815(0.008602) and 13227/2736964(0.004833), respectively.To confirm the results of CMV detection by CNV‑seq, CMV‑DNA nucleic acid quantification was performed using cytomegalovirus nucleic acid detection kit.The results suggested that the CMV viral loads of the 3 patients were 1227.76Copies/mL, 11639.00Copies/mL and 13754.60Copies/ml, respectively.Previous studies have suggested that the amniotic cytomegaloviral load is highly likely to be correlated with fetal prognosis.It is believed that when the CMV‑DNA load is larger than 103Copies/mL, the probability of mother‑to‑child transmission can be predicted, while when the CMV‑DNA load is larger than 105Copies/mL, symptomatic infection can be predicted[21].In this study, no obvious clinical manifestations were found in the follow‑up of the 3 patients and their newborns, which was consistent with the results of the above studies.
As a widely used prenatal gene detection technology, CNV‑seq technology can not only diagnose intrauterine infection of CMV or congenital CMV infection in newborns, but also detect fetal chromosome abnormalities or genetic abnormalities.This is of great clinical significance for prenatal diagnosis of CMV infection.It can not only clarify early diagnosis of CMV infection to guide clinical diagnosis and improve prognosis, but also detect birth defects that may be caused by CMV infection or other genetic factors as early as possible.Based on the above results, we conclude that CNV‑seq may have certain application value in the clinical diagnosis of CMV infection, and can provide the corresponding clinical basis for the construction of a one‑stop prenatal diagnosis platform for CMV infection.However, it is necessary to continue to expand the sample size and conduct prospective randomized controlled studies in multiple prenatal diagnosis centers to provide a basis for the diagnosis of cytomegalovirus infection in pregnancy and better apply in the field of prenatal diagnosis.
 Journal of Hainan Medical College2023年14期
Journal of Hainan Medical College2023年14期
- Journal of Hainan Medical College的其它文章
- Research progress on key genes of vitamin D signaling pathway
- Research progress on the influence of local hemodynamics on carotid atherosclerosis
- Exploration of the molecular mechanism of Qishen decoction in regulating miR-495/FTO pathway mediated macrophage polarization to improve insulin resistance therapy of type 2 diabetes
- Intervention of Xuduan Zhongzi Formula on spermatogenesis epididymal morphological changes in a mice model of oligospermia
- Expression of miR-9-5p and RHOA in aluminum-induced rat cognitive dysfunction
- miR-483-5p regulates osteoclast generation by targeting Timp2
