Scientists Map Single-Cell Spatial Distribution Atlas of Macaque Cortex
A recent study published inCellsuccessfully mapped the cell-type taxonomy in the macaque cortex and revealed the relationship between cell-type composition and various primate brain regions, by using the self-developed spatial transcriptome sequencing technology Stereo-seq and snRNA-seq technology.These findings provide a molecular and cellular basis for further investigation into neural circuits.
This collaborative study was conducted by a research team of nearly 100 scientists from the Institute of Neuroscience (ION), Center for Excellence in Brain Science and Intelligence Technology of the Chinese Academy of Sciences, BGI Research, Lingang Laboratory,Shanghai Center for Brain Science and Brain-Inspired Technology, Tencent AI Lab, as well as Karolinska Institute and KTH Royal Institute of Technology in Sweden.
Primates have a vast number of neurons that form complex and intricate neural circuits supporting advanced cognition and behavior.Disruptions in these cells and circuits can lead to various brain disorders.Understanding the composition and spatial distribution of cells in the brain, as well as the relationships between them, is a fundamental question in neuroscience,comparable to the periodic table in chemistry, the world map in geographic discoveries, or the DNA base sequence discovered through human genome sequencing.Compared to other species, primates, including macaques as the closest animal model to humans, have higher cognitive and social abilities, as well as larger cortices and more cell types.For example, the macaque brain,with over six billion cells, can be classified into hundreds of cell types based on their molecular, morphological,or physiological features, and their spatial distribution spans hundreds of distinct brain regions.Deciphering the composition and spatial distribution patterns of cell subtypes in the cortex is crucial for understanding the organizational principles of primate brains.
This study utilized a newly developed large fieldof-view spatial transcriptome method called Stereoseq, and independently developed a method to prepare centimeter-scale thin slices of the macaque brain to meet the experimental needs.By combining large-scale singlecell transcriptome analysis, the research team obtained a comprehensive three-dimensional single-cell atlas of the entire cortex of the crab-eating macaque, providing a guide for systematic analysis of cell-type distribution specificity and regional specificity within the cortex, as well as molecular features (Figure 1).
The team found that glutamatergic neurons,GABAergic neurons, and non-neuronal cells exhibited distinct cortical and regional specificity in their distribution throughout the cerebral cortex (Figure 2).Interestingly, there was a significant correlation between the cell-type composition and the hierarchical organization of brain regions in the visual and somatosensory systems.Brain regions at the same hierarchical level tended to have similar cell-type compositions, revealing the relationship between cell composition and brain region structure.Furthermore,through cross-species comparison with publicly available single-cell data from the human and mouse brains,the research team identified glutamatergic neuron cells specific to primates, which are predominantly located in the layer 4 and highly express genes associated with human diseases, includingFOXP2,DCC, andEPHA3.This study generated a comprehensive dataset of singlecell and spatial transcriptomics for the entire macaque cerebral cortex, providing an important data resource for future related research.The data is now publicly available for sharing at https://macaque.digital-brain.cn/spatial-omics (Figure 3).
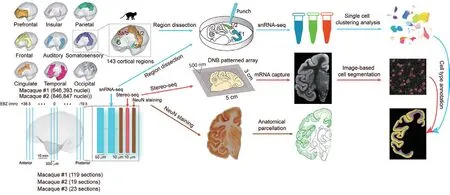
Figure 1.Overall experimental design.We employed the Stereo-seq technique to collect spatial transcriptome data from 161 10-micron-thick coronal sections of the left hemisphere of three macaque monkeys.Additionally, we obtained single-cell transcriptome data from millions of cortical cells using a combination of microdissection and single-nucleus RNA sequencing technology.By integrating the single-cell and spatial transcriptome data, we constructed a three-dimensional single-cell atlas of the entire cortex of the crab-eating monkey.(Image by Cell)
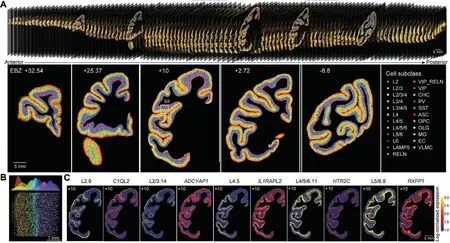
Figure 2.Spatial distribution map of cell types in the macaque cortex.(A) Spatial distribution of hundreds of cell types in different regions of the macaque brain (top) and on five representative slices (bottom).(B) Layer-specific distribution of diverse cell types.(C) Spatial distribution/expression patterns of five cell types and their characteristic genes.(Image by Cell)
Throughout the research process, the team crossed multiple institutions and fields, utilizing China's domes tically developed core technologies and large platforms.The research was carried out in an organized mode with clear objectives, task orientation, complementary division of work, and unity in efforts.In the future, the team will continue to focus on mechanisms and target development of brain diseases, brain cell and structural evolution, and cellular and molecular mechanisms of brain function.The aim is to promote the continuous generation of original and leading achievements in these fields in China.
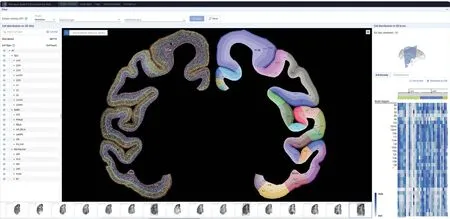
Figure 3.Database of cell types and gene expression in the macaque cortex.
This work entitled “Single-cell spatial transcriptome reveals cell-type organization in macaque cortex” was published online inCellon July 12, 2023.Dr.LI Chengyu, Dr.LIU Zhiyong, Dr.SUN Yidi, Dr.SHEN Zhiming from the Center for Excellence in Brain Science and Intelligence Technology of the Chinese Academy of Sciences, along with Dr.XU Xun, Dr.LIU Longqi,Dr.LI Yuxiang from BGI Research Institute, and Dr.WEI Wu from Lingang Laboratory, as well as Dr.YAO Jianhua, AI Healthcare Chief Scientist of Tencent AI Lab,are the co-corresponding authors of the paper.Dr.CHEN Ao and Dr.LEI Ying from BGI Research, Dr.LIAO Sha,Mr.ZHU Zhiyong (Master’s student), Dr.SUN Yidi, Dr.LI Chao, Dr.MENG Juan (PhD student), Dr.LIANG Zhifeng,Dr.YUAN Nini, and Mr.YANG Hao from the Center for Excellence in Brain Science and Intelligence Technology,along with Dr.BAI Yiqin, a postdoctoral researcher from Lingang Laboratory, Dr.LIU Zhen, a PhD student from Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences, and Mr.WU Zihan, a researcher from Tencent AI Lab, are the co-first authors of the paper.The research was supported by the National Science and Technology Innovation 2030 Major Program, Shanghai Municipal Science and Technology Major Project, and projects from Ministry of Science and Technology, National Natural Science Foundation of China, and Shenzhen Municipal Government.
 Bulletin of the Chinese Academy of Sciences2023年3期
Bulletin of the Chinese Academy of Sciences2023年3期
- Bulletin of the Chinese Academy of Sciences的其它文章
- Could Gut Bacteria Hold a Cure for Sepsis?
- Looking into the Tissue Adjacent to Tumors
- Daya Bay Collaboration Awarded 2023 High Energy and Particle Physics Prize by European Physical Society
- Largest Optical Time-domain Survey Telescope in Northern Hemisphere Goes into Operation
- CAS Paleogeneticist Awarded UNESCO Prize
- “Magnetically Arrested Disk”Revealed by Multiwavelength Observation
