Bacterial community in response to packaging conditions in farmed gilthead seabream
Apollon Thoms, Spyros J.Konteles, Sotiris Ouzounis, Spyros Pptheodorou,Aliki Tskni, Dimitr Houhoul,*, Theofni Tsironi
a Department of Food Science and Technology, Faculty of Food Sciences, University of West Attica, Athens, 11521, Greece
b Medical Image and Signal Processing Laboratory, Department of Biomedical Engineering, University of West Attica, Athens, 11521, Greece
c Laboratory of Food Process Engineering, Department of Food Science and Human Nutrition, Agricultural University of Athens, Athens, 11855, Greece
Keywords:Fish Preservation Next generation sequencing Modified atmosphere packaging Microbiome
ABSTRACT The objective of this study was to evaluate the effect of modified atmosphere packaging (MAP) on fish skin, gills and intestines bacterial microbiome.
1.Introduction
Aquaculture has expanded significantly during the last 30 years both in production volume and geographical region.Its growth is so intense and rapid, that it is estimated that by the year 2025 more than 50% of world’s fish production will be farmed (Ottinger et al., 2016).The duration of fish transportation from the aquaculture facilities to the fish processing unit is often longer than one day, so fish should be properly pre-packed immediately after harvesting and transported in flake ice to maintain high quality.Significant effort has been exerted with the aim to improve fish quality and extend shelf life at any stage of the supply chain, including improvements in fish diet (Alexi et al., 2017; Kotzamanis et al., 2020; Su´arez et al., 2010), harvesting protocols(Zampacavallo et al., 2015; Bagni et al., 2007; Ntzimani et al., 2021),post-harvest processing (Bosco, 2010; ´Alvarez et al., 2009; Cakli et al.,2007), packaging (Tsironi et al., 2018; Bouletis et al., 2017) as well as storage conditions (Mercier et al., 2017; Margeirsson et al., 2012; Tsironi, Gogou, et al., 2008 and b; Tsironi et al., 2011).
Spoilage of fish stored under refrigerated conditions is attributed mainly to microbial activity (Gram & Huss, 1996).In the case of aerobically packed, chilled Mediterranean fish, such as gilthead seabream,European sea bass and red sea bream,Pseudomonasspp.have been reported as the dominant spoilage factor (Neumeyer et al., 1997; Tsironi et al., 2020).Shelf life estimation of fresh fish has been performed based on the correlation of microbial load and the sensory parameters of fish,considering a limit of acceptability of 107cfu/g for total viable count orPseudomonasspp.(ICMSF, 1986; Papaharisis et al., 2019; Tsironi,Ntzimani, et al., 2019).Pseudomonasspp.and total viable count levels of 106–107cfu/g have been reported at the sensory rejection time in aerobically packed, cultured gilthead seabream, sea bass and turbot in relevant studies (Koutsoumanis & Nychas, 2000; Rodrıguez et al., 2006;Tsironi et al., 2015).
As regards packaging, one of the techniques that have been widely applied to delay the deterioration of fish quality is atmosphere modification in the package headspace.The gas composition in the package affects significantly the microbial activity, the main cause of fish quality deterioration during storage (Ahmed et al., 2017; Bouletis et al., 2017).The preservative effect of Modified Atmosphere Packaging (MAP) on gilthead seabream flesh has been reported in the literature (Drosinos &Nychas, 1996; Gim´enez et al., 2002; Parlapani et al., 2014; Garrido et al., 2016), indicating significant extension of shelf life, depending on storage conditions.The diversification of the dominant microbiota in MA-packaged fish depends on the geographical origination, water temperature and storage conditions (Tsironi et al., 2020; Tsironi &Taoukis, 2018).Gram-positive bacteria, such as lactobacilli, have been reported as resistant to high CO2concentration in the package headspace, playing a significant role in the spoilage process of MAP finfish species from warm waters, such as the Mediterranean basis (DeWitt &Oliveira, 2016; Sivertsvik et al., 2002; Stenstr¨om, 1985).Lactic acid bacteria,Brochothrix thermosphactaand H2S-producing bacteria have been reported as the dominant spoilage bacteria in MAP gilthead seabream, using high CO2concentrations (Drosinos et al., 1997; Tsironi et al., 2011, 2008b and; Parlapani et al., 2014).
Recent studies, based on molecular analysis methods such as 16S rRNA gene sequencing, reported that the synthesis of spoilage microbiota of fish is strongly dependent on storage temperature and packaging atmosphere (Parlapani et al., 2015).Species such asCarnobacterium maltaromaticum,Carnobacterium divergensandVagococcus fluvialisdominated spoilage of MAP gilthead seabream fillets under 60% CO2, 10% O2, 30% N2at 5◦C, whilePseudomonas veroniidominated at 0◦C.16S rRNA gene sequencing that have been used to characterize several microbial communities has been introduced as one of the most powerful tools for microbiological analyses (Jagadeesan et al., 2019).The introduction of Next Generation Sequencing (NGS)represents another important, fundamental technological advance in the biological sciences since the development of the polymerase chain reaction (PCR) in the mid-1980s.High performance and speed of NGS on 16S rRNA gene significantly reduces the cost and facilitates the research of metagenomics, providing a more complete assessment compared to a culture-based analysis.16S rRNA hypervariable regions exhibit different degrees of sequence diversity, and not every hypervariable region is able to distinguish among all bacteria with equal specificity.Few applications on NGS have been reported recently for the determination of microbial flora in food products, i.e., dairy products (Ribani et al., 2018),fish (Silbande et al., 2018; Tsironi, Anjos, et al., 2019 and b) and shrimp(Yang Chen, Lee, Chen, Lin, & Hor, 2014).
Microbiota associated with the skin, gills and gut has been investigated as a function of fish age, environmental conditions, diet and water quality in different fish species.Skin microbiota has been reported as more diverse and variable in community composition than gut microbiota (Guivier et al., 2019; Kuang et al., 2020).During the chilled preservation of whole fish, microbial growth is observed at the outer contaminated tissues of fish, i.e.skin and gills.Growth of gut bacteria is another cause of the production of metabolic compounds, which affects significantly the odor and overall quality of whole fish.The objective of this study was to evaluate the effect of modified atmosphere packaging(MAP) on fish skin, gills and intestines bacterial microbiome.
2.Materials and methods
2.1. Provision and handling of fish samples
Fifty (50) whole, gilthead seabream (Sparus aurata) (weight 380–420 g) were provided by Selonda Aquaculture S.A.(Attica, Greece)and used for the packaging experiment.All fish were fed with commercial feed, came from the same batch and harvested from the same net cage.According to the aquaculture’s practices, fish was fasted one day before slaughter, which was carried out by immersion in ice-cold water(ice shock).Then, fish was placed in polystyrene boxes with appropriate quantity of flake ice and delivered to the laboratory within 24 h after slaughter.Fish body temperature was measured upon receipt of the samples (0οC).
2.2. Packaging and storage of gilthead seabream
Fish samples were randomly divided in two groups.One group (25 specimens) was stored in polystyrene boxes with flaked ice (ice/fish 0.5:1 w/w) and stored aerobically in a high-precision low-temperature incubator (Sanyo MIR 153, Sanyo Electric, Ora-Gun, Gunma, Japan)under controlled isothermal conditions (0 ±0.2◦C) (AIR).Flake ice was replenished in the boxes during storage.For the other group of samples(25 specimens), fish was individually packed in HDPE pouches under modified atmosphere (60% CO2, 30% N2, 10% O2) (C200, MULTIVAC Sepp Haggenmüller GmbH & Co.KG., Wolfertschwenden, Germany)with gas-to-product volume ratio equal to 3:1.The pouches were packed in polystyrene boxes along with flaked ice and stored in a high-precision low-temperature incubator (Sanyo MIR 153, Sanyo Electric, Ora-Gun,Gunma, Japan) under controlled isothermal conditions (0 ± 0.2◦C)(MAP).
2.3. Sampling from skin, gills, and intestines
At days 0 and 10 of isothermal storage at 0◦C, ten (10) whole, fish samples were randomly taken and analyzed regarding the bacterial microbiota of skin, gills, and intestines (60 fish tissue samples were used in total).Skin samples were taken by swabbing several times from eight(8) different spots of the fish from head to tail.Gills swabs were taken from both filaments between the first, second and third arch.As regards the bacterial microbiome of intestines, the samples were prepared as follows: under sterile conditions, the intestines of each fish were removed, and their content was taken by squeezing the lower intestinal tract.The samples also contained remnants of the feed as well as cells from fish mucosa.
2.4. DNA extraction and Next Generation Sequencing
Samples were aseptically homogenized in a laminar flow hood.DNA extraction was performed using the NucleoSpin Tissue kit (Macherey-Nagel, GmbH & Co.KG, Germany) according to the manufacturer’s instructions, with the addition of a Proteinase K overnight incubation step at 65◦C.Extracted DNA was quantified using a spectrophotometer at 260 nm and 280 nm.After DNA extraction, 16S rRNA genes were amplified using domain-level bacterial primers that contain sequencing adapters and unique, sample-specific sequences.Multiple samples barcoded and sequenced simultaneously on a single Ion PGM 318 chip resulted in sufficient number of reads.The number of total reads per sample was between 2.5 ×106and 4.5 ×106.Approximately 50–65% of these reads passed stringency filters and of these, 60–75% mapped to the databases.The kit includes 2 sets of primers that can be used to amplify the corresponding hypervariable regions of the 16S rDNA gene in bacteria (Primer set V2-4-8, Primer set V3-6,7–9).After generating amplicons, the Ion Plus™ Fragment Library Kit was used to ligate barcoded adapters and synthesize libraries.Barcoded libraries from all samples were pooled and templated on the OneTouch2™ system followed by 400 bp sequencing on the Ion PGM.Automated analysis, annotation and taxonomic assignment occurs via the Ion Reporter Software pipeline.Classification of reads is through alignment to either the curated MicroSEQ ID or curated Green genes databases.
2.5. Statistical analyses
Statistical analysis was carried out for skin, gill and intestines microbiota individually, as well as for fish microbiota as unity.Initially,the number of the families within each set of samples was calculated, to quantify the families’ richness in each packaging type with the specnumber function from the R-package vegan (Oksanen, 2014).Then analysis of variance was employed to test if the family richness was statistically significant across the samples.The variance analysis among the 3 tested conditions was performed with the kruskal-Wallis test(Kruskal & Wallis, 1952), which was followed by a pairwise Wilcoxon test (Wilcoxon, 1945).The latter was carried out to examine which packaging type was responsible for the statistical significance amongst the 3 groups (Initial, AIR, MAP).The second step of the analyses was to investigate the diversity of the families for each sample per packaging type.The calculation of the diversity was carried out with diversity function in vegan according to the metrics of Shannon diversity which considers the Families abundance and evenness.Then, in order to test if there is a statistically significant difference among the Families diversity of the 3 tested conditions it was implemented analysis of variance for the Shannon Indexes.Again, for the analysis of variance a kruskal-Wallis test was conducted, which was then followed by a pairwise Wilcoxon test.Kruskal-Wallis and Wilcoxon are non-parametric tests and were selected due to the limited number of available samples, which follow mostly a continuous uniform distribution.The third step of statistical analyses was to search if the microbiome composition was affected by the packaging type.A non-parametric permutational Multivariate Analysis of Variance (perMANOVA) implemented with the function adonis2, based on Bray-Curtis dissimilarity matrix (Oksanen, 2014).This test would unveil the differences in microbiome composition.The null hypothesis of the perMANOVA test is that the tested groups do not differ in spread or position in the multivariate space.The fourth step was the analyses of multivariate homogeneity of group dispersions which was done only for the unified microbiome with the betadisper function(Oksanen, 2014).This analysis was done to examine the distances between the different microbiome Families and their packaging type centroids.Such an analysis can be also interpreted as a measure of multivariate beta diversity among tested groups since the beta diversity can be defined as the variability in Families composition among sampling units for a given group (Anderson et al., 2006).Finally, a Non-metric multidimensional scaling (nMDS) plot (Kruskal, 1964) was conducted in order to represent the original position of communities in multidimensional space based on the packaging using a reduced number of dimensions that can be easily visualized.Finally, a correlation test of multivariate independence (Szekely et al., 2007) was performed between the microbiomes of skin and intestines samples in order to see if there is a correlation between the two microbiome communities.The test was implemented with the dcor.test function of the energy package in R.This test is a non-parametric test that can be used as a measure of multivariate dependence between (random) vectors.
Venn diagrams were generated to illustrate unique and shared families using Venny 2.1.0 (Oliveros, 2007-2015).The relationships between the bacterial community and the sensory scorings have been evaluated using multivariate statistics by the XLSTAT statistical software program (Addinsoft, New York, USA).
2.6. Sensory evaluation
Sensory parameters of fish samples were evaluated using the Quality Index Method (QIM) as described by Lougovois et al.(2003).According to this method, the body, eyes and gills of fish were sensorially evaluated regarding specific characteristics such as odor, color, appearance,(Table 1).The freshness evaluation was performed on a demerit scale,which means that the defects were scored.Thus, the higher the score (2 or 3), the more intense the deterioration of the fish quality while freshness was graded 0.The range of Quality Index of gilthead seabream is from 0 to 16 (fresh to spoiled).Sensory analysis was carried out by a team of 5 trained assessors, on day 0, 4, 8, and 10, under the same conditions (room temperature/light).Each member assessed randomly selected fish from each packaging type (AIR and MAP).
3.Results
3.1. Microbiome composition among the different packaging types
3.1.1.Microbiome composition at phylum level
Microbiome analysis was performed at phylum and family level for the skin, gills, and fish intestines.In Fig.1 the analysis of the microbiome is depicted (Relative abundance %) at phylum level for: (a) skin, (b)gills, and (c) intestines of seabream samples, 24 h after harvesting(Initial state) as well as following 10 days of isothermal storage at 0◦C(AIR) or packed under MAP and stored at 0◦C (MAP), indicating the dominant microorganisms (at phylum level) in the tested fish tissues initially and after 10 days of storage at alternative packaging conditions.
The initial fish skin microbiome consisted mainly ofProteobacteria(80.48%), followed byFirmicutes,BacteroidetesandActinobacteria.After 10 days of aerobic storage in ice flakes (AIR),Proteobacteriawas the dominant phylum on fish skin (92.74%), whileActinobacteriaalong withFirmicutesexhibited reduction.Similarly, on MAP fish, the most prevailing phylum wasProteobacteria, whileFirmicuteswas nearly extinguished (1.51%).
As regards microbiome of gills, it consisted, almost exclusively, of bacteria belonging to the family ofProteobacteria(97.82%), which remained dominant after 10 days of aerobic storage (AIR, 99.87%) or under MAP (97.54%).Similarly to skin and gills, the initial microbiome of fish intestines consisted by 66.6%Proteobacteria, followed byFirmicutesandBacteroidetes.During 10 days of iced storage under aerobicconditions (AIR)Proteobacteriapercentage increased up to 86% whileFirmicutesdecreased to 9.85% andBacteroidetesto 0.63%.In contrast, for fish stored under MAP, theProteobacteriaslightly decreased whileBacteroidetes, significantly increased.
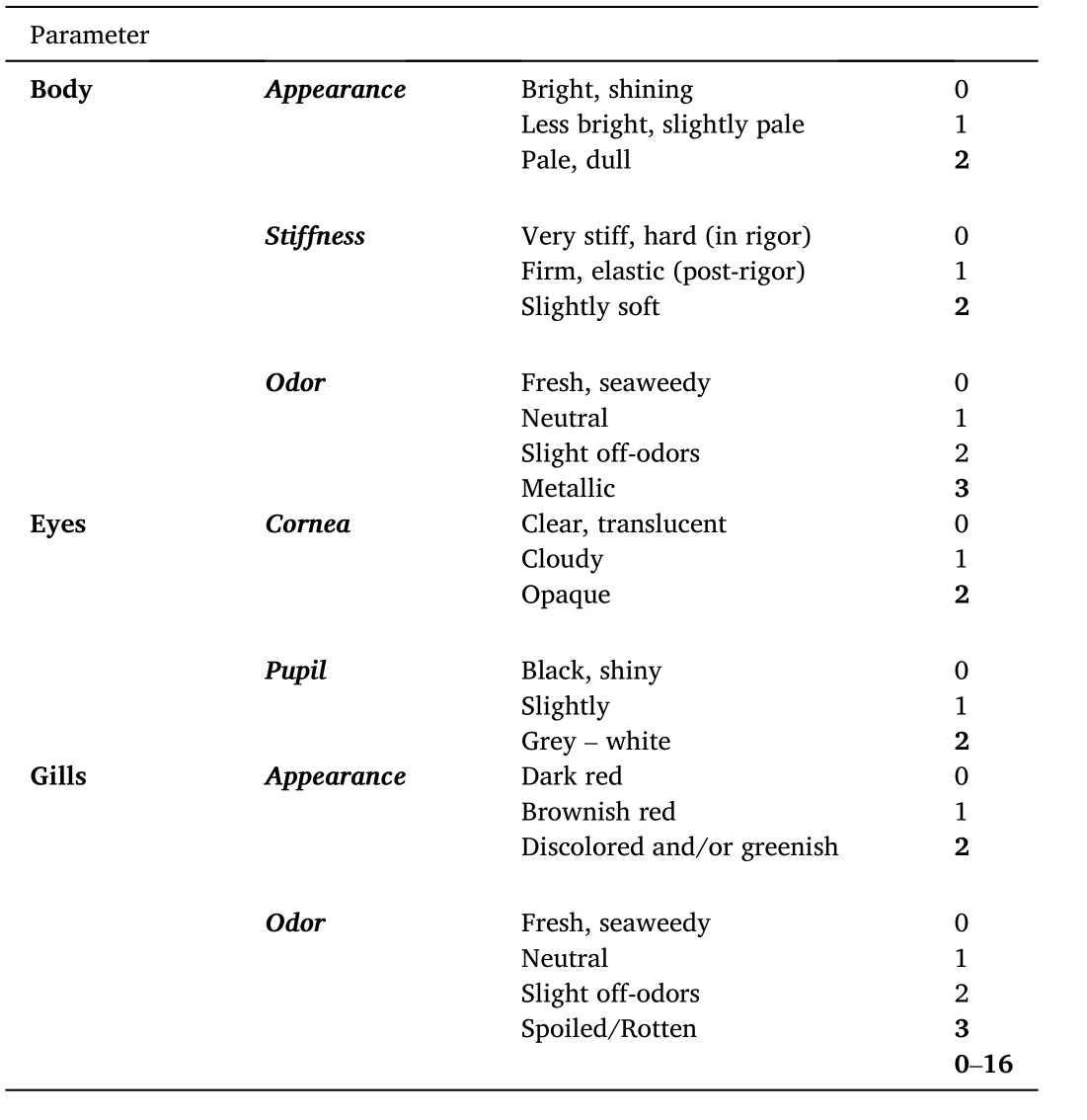
Table 1 Quality Index Method (QIM) for sensory evaluation of whole gilthead seabream(Sparus aurata).
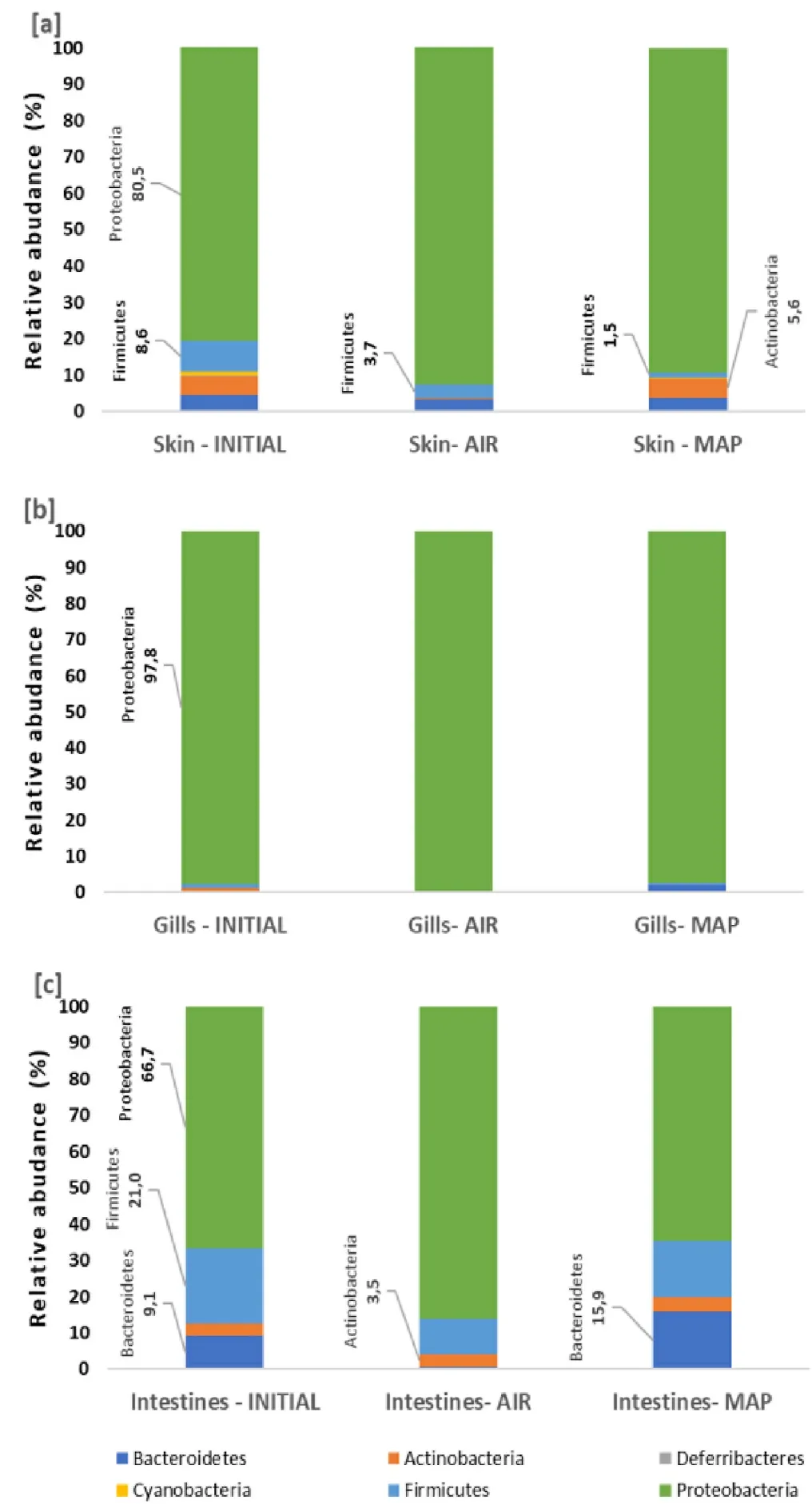
Fig.1.Phylum taxonomic composition of (a) skin, (b) gills and (c) intestines microbiomes of seabreams, 24 h after their catch and storage in ice (INITIAL),as well as after 10 days packed on ice (AIR) or under MAP conditions and on ice (MAP).
3.1.2.Microbiome composition at family level
In Fig.2a the Relative Abundance (%) of the skin microbiome of gilthead seabream 24 h after harvesting and packed in ice-flakes is depicted, indicating the dominant microorganisms (at family level level)in the tested fish tissues initially and after 10 days of storage at alternative packaging conditions.Initially the dominant families were,
Comamonadaceae(53.5%),Enterobacteriaceae(13.01%), andMoraxellaceae(5.89%).After 10 days of aerobic storage on ice (AIR) the microbiota had changed and the dominant families werePseudoalteromonadaceae(45.59%),Shewanellaceae(22.11%) andPsychromonadaceae(17.47%) while on MAP fish the main families wereShewanellaceae
(37.4%),Psychromonadaceae(29.53%) andRhodocyclaceae(22.16%).Regarding the gills microbiome (Fig.2b) at day 0 (initial state), the prevalence of the familyRhodocyclaceaewas evident (94.86%).However, after 10 days of iced storage, the gill microbiome was modified.For gills in aerobically stored fish (AIR), the familiesPsychromonadaceae(56.67%),Rhodocyclaceae(21.72%), andShewanellaceae(16.18%) prevailed.The sum of these three families corresponds to 94.57% of the total microbiome.
On the gills of the fish preserved under MAP, the predominant bacterial families wereShewanellaceae(37.4%),Psychromonadaceae(29.5%) andRhodocyclaceae(22.16%).In this case the sum of these families was 89.04%.
Finally, the microbiome of fish intestines (Figure 2c), 24 h after harvesting, consisted of the familyComamonadaceae(34.2%), followed byAnaplasmataceae(10.6%),Bacillaceae(8.74%) andEnterobacteriaceae(6.2%).In this case, at the end of the storage period (day 10), the intestinal microbiome of fish stored on ice consisted of the familiesPseudoalteromonadaceae(34.1%),Comamonadaceae(21.2%),Enterobacteriaceae(10.1%) andPsychromonadaceae(8.5%) while on the MAP samplesComamonadaceae(31.1%) prevailed, followed byPorphyromonadaceae(11.5%) andClostridiaceae7.3%).
3.2. Microbial diversity amongst skin, gills, and intestines
3.2.1.Statistical analysis of families’richness
From the analysis of variance which was performed in skin gill, and intestines microbiota it was found that there is a statistical significance of the mean families’ richness across the different packaging types for skin, gill, and intestines, where richness is defined as the number of different species in each microbiota tested.More thoroughly, the pairwise comparisons showed that there is a statistically significant difference in the families’ richness not only between the initial seabream microbiome and AIR samples but also between the initial fish and MAP samples.Furthermore, it was found that the AIR and MAP samples had also a statistical significance in the families’ richness.However, when the richness of fish microbiota as unity was investigated among the initial stage and the two packaging types, 10 days of iced storage, it was found that there was a statistical difference between MAP, the initial fish and AIR samples, whereas there was no difference in richness between initial fish and AIR samples (Table 2, Table 3, Fig.3).
3.2.2.Statistical analysis of families’diversity
When the alpha-diversity of the families in skin, gill, intestines, and unified (total) microbiota was compared among the packaging types, it was found that there was a statistically significant difference among the microbiota of skin, gill, and intestines for the tested groups (Table 4,Fig.4).
3.3. Statistical analysis of microbiome composition
The non-parametric permutational Multivariate Analysis of Variance for each tissue type (skin, gill, intestines) was performed with regard to the packaging type that each sample had, revealed that there is a strong statistically significant effect of the packaging type on the microbiome composition.On the other hand, when the same test was done in the unified microbiota there was still a significant difference among the packaging types but the R2is low which means there is a lot of variation in the model.This is due to the fact that we have mixed the microbiomefrom different tissues (Τable 5).For the unified microbiome, an analysis of multivariate homogeneity of group dispersions was also carried out and showed that there is statistical deference in beta diversity of skin,gill, and intestines microbiome among the 3 tested groups.Finally, from the nMDS plots (Fig.5) we can see that the samples of the skin, gill and intestine are clustered in different areas according to the packaging type and have no dispersion since they are from the same cage whereas, the unified microbiota shows a dispersion in the packaging clusters.

Table 2 Number of families for each microbiome type according to fish packaging method.
3.4. Bacterial diversity between packaging types
In order to evaluate the shared versus unique bacterial microbiota by packaging type (AIR or MAP), the respective Venn diagrams were constructed (Fig.6a–c).Fish skin 1 day after harvesting presented higher bacterial richness and unique bacterial composition than skin samples stored for 10 days at 0◦C at aerobic conditions or under MAP (26.5%,12.2% and 2%, respectively.A core group with 16 (32.7%) taxa were common to all three samples (skin initially and after 10 days at 0◦C.34.7% of the observed families were common on fish skin at the end of storage period (10 days) at aerobic conditions and under MAP (Fig.6a).The most abundant species for fish skin initially wasHylemonellaspp.,followed byShigella sonneiandAcinetobacter johnsonii.At the end of storage, the most abundant species in fish skin werePseudoalteromonas spongia,Shewanellaspp.andParabacteroides distasonisfor aerobically packed fish andHylemonellaspp.,Brachymonasspp.andShewanellaspp.for MAP samples.For fish gills after 1 day of harvesting, the dominant species wereZoogloea oryzae,Rhodocyclus tenuisandDechloromonas agitata, while at the end of storage under aerobic conditions the most abundant species werePsychromonas arctica,Zoogloea oryzaeandShewanellaspp.and for MA packagingPsychromonas arctica,Shewanellaspp.,Rhodocyclus tenuisandPophyromonasspp.Finally, for fish intestines initially (after 1 day from harvesting) the most abundant species wereHylemonellaspp.,Neorickettsdiaspp.,Cloacibacteriumspp.andShigella sonnei.At the end of storage (10 days at 0◦C) the dominant species wereHylemonellaspp.,Pseudoalteromonas spongia,Psychromonas arcticaandShigella sonneiwhile for MAP samples the most abundant species wereHylemonellaspp.,Psychromonas arctica,Shigella sonnei,Pseudoalteromonas spongiaandParabacteroides distasonis.
3.5. Sensory evaluation
In Fig.7 the sensory evaluation of fish samples after 10 days of isothermal storage at 0◦C is illustrated, for aerobically stored and MAP fish.Slight sensorial degradation of fish was detected on day 5 of storage in aerobically stored fish, based on off-odors in the belly region as well as decay in body stiffness, when fish was judged as acceptable by the panelists.From day 8 of fish storage at 0◦C, slight quality degradation was detected in MAP fish.Quality loss was related with slight decrease in body stiffness, development of off-odor in the belly region, as well as discoloration color of gills.However, in aerobically stored fish, quality degradation was more pronounced regarding flesh stiffness and offodors, compared to MAP fish.At day 10 of fish storage at 0◦C, quality degradation of the MAP fish was more intense compared to the aerobically stored samples (Fig.7).
Based on the Agglomerative hierarchical clustering as illustrated in Fig.8 at the similarity plot of the sensory scoring and microbiome data of the tested fish samples (AIR and MAP) initially and after 10 days of isothermal storage at 0◦C, the fish packaging types should be grouped into 2 clusters.According to Fig.8, a similarity is observed between the gills sensory and microbiome data parameters.Microbiome data and sensory scoring indicate grouping between skin and intestine, while storage time does not affect significantly the dominant bacterial families.

Table 3 Statistical Analysis of families’ richness of fish microbiota stored under AIR or MAP (p-values of the pairwise Wilcoxon test).
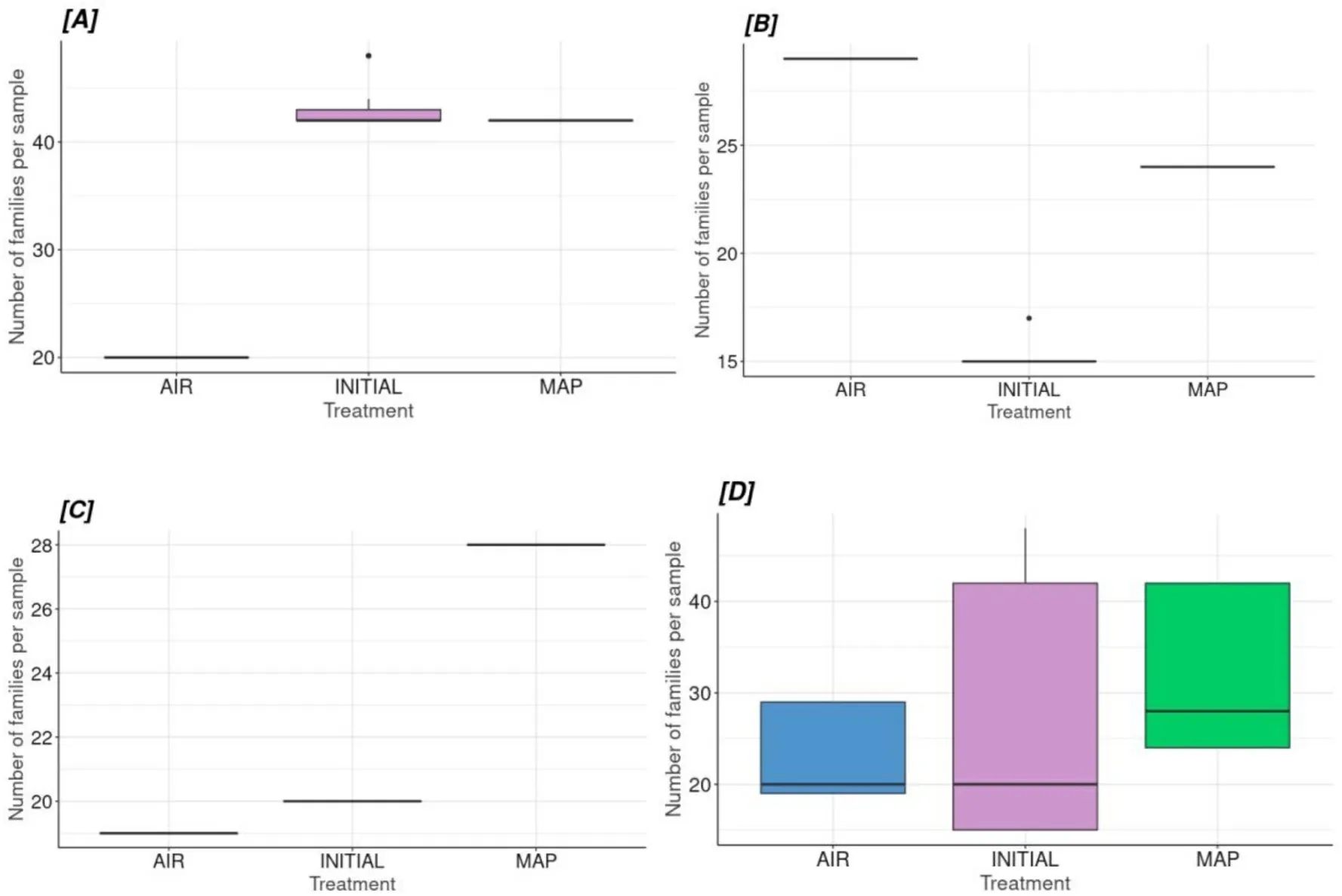
Fig.3.Families’ richness for each tissue (A) Skin, (B) Intestines, (C) Gill, and (D) Unified microbiome according to treatment type.

Table 4 Statistical Analysis of families’ diversity of fish microbiota stored under AIR or MAP (p-values of the pairwise Wilcoxon test).
4.Discussion
Fresh fish is a perishable food product with high nutritional value and significant potential of value addition if its shelf life is extended by the application of appropriate processing and packaging technologies.The aim of this study was to investigate the effect of MAP on the microbial community in fish skin, gills and intestines and sensory scoring using NGS.The use of MAP in whole fresh gilthead seabream may have advantages as regards fish quality retention during transportation and storage, since the bacteriostatic activity of CO2and inhibitory effect of MAP on quality deterioration of perishable seafood products is documented (Tsironi, Ntzimani, et al., 2019; Zhuang et al., 2020).Considering that one of the main factor of fish quality degradation is bacterial activity, the microbiota on fish skin, gills, and intestines was mapped by NGS at the beginning (day 1) and at the end of storage (day 10).
The NGS microbiome analysis of skin, gills, and intestines, initially at phylum level, showed thatProteobacteriawas predominant in fish samples 24 h after their catch and storage on ice.Proteobacteriawas also predominant 10 days after storage at 0◦C in samples that were either stored aerobically (AIR) or under MAP.The presence ofProteobacteriain aquaculture fish is common, since it has been documented for gilthead seabream (Salgueiro et al., 2020), salmon (Lokesh & Kiron, 2016) and Chinook salmon (Steiner et al., 2021).Recently, also, Rosado et al.(2019) reported that the skin and gill microbiome of sea bass and gilthead seabream was dominated byProteobacteriaandBacteroidetes.As regards the microbiome of the intestines, the main phyla of the fresh-catch fish wereProteobacteriaalong withFirmicutes, andActinobacteria, that has, also, been reported by Estruch et al.(2015) for the gastrointestinal microbiome of gilthead seabream.
Fig.4.Families’ alpha-diversity for each tissue (A) Skin, (B) Intestines, (C) Gill and (D) Unified microbiome according to treatment type.
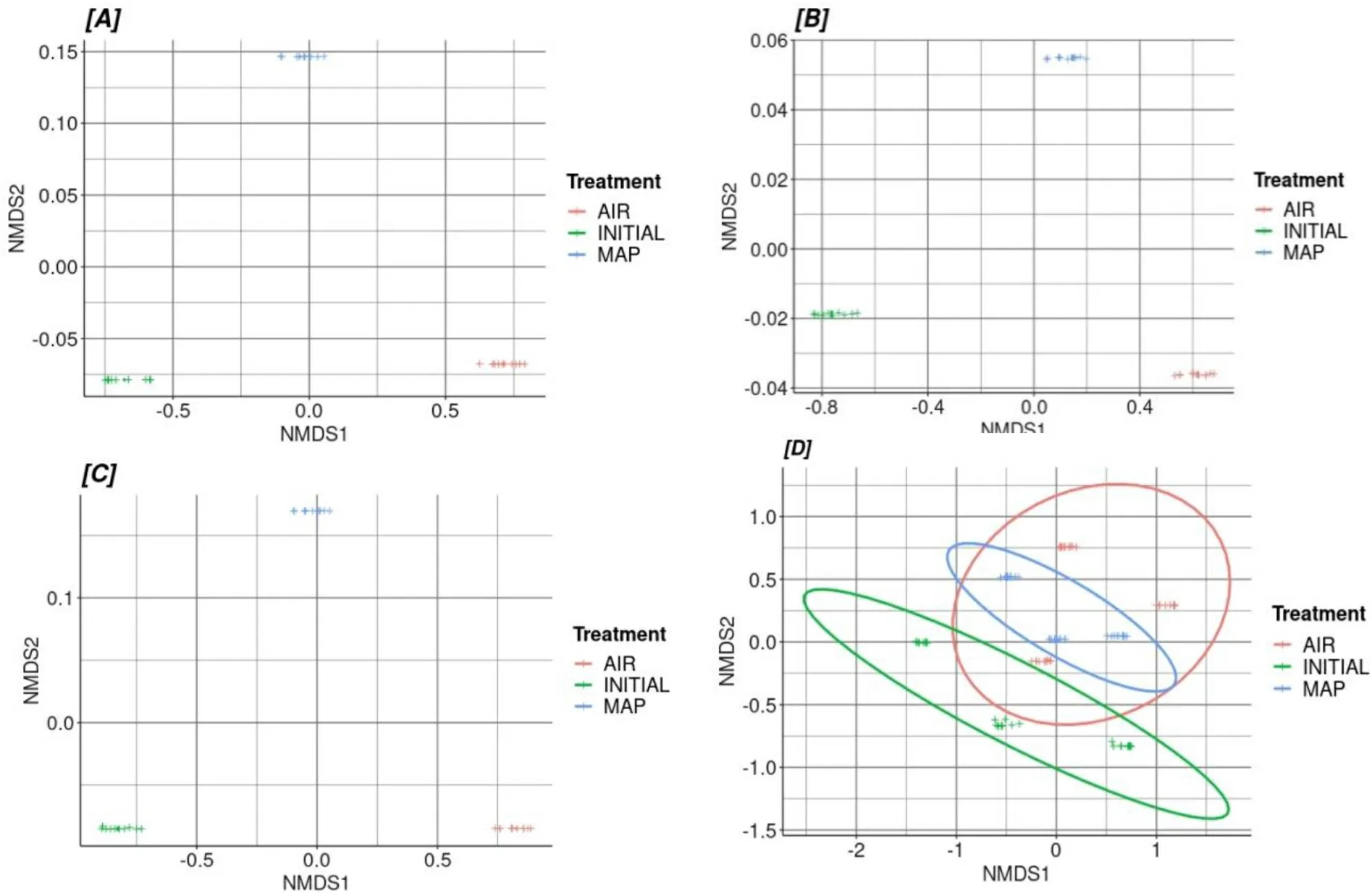
Fig.5.Non-metric multidimensional scaling plots based on a Bray-Curtis resemblance matrix for each tissue (A) Skin, (B) Intestines, (C) Gill and (D) Unified microbiome according to treatment type.

Fig.7.Average score (QIM) for different quality attributes (a: appearance, b:body stiffness, c: odor, d: cornea, e: pupil, f: gills appearance, g: gills odor, h:overall QIM score) used for the sensory evaluation of fish stored aerobically(AIR) or under MAP.
NGS analysis at the family level showed that the fish skin microbiome was initially composed by 49 different families, thought that just three of them were dominant:Comamonadaceae,EnterobacteriaceaeandMoraxellaceae.Similar results have been reported by Salgueiro et al.(2020) where the microbiome of gilthead seabream collected from a land tank was investigated.Regarding gilthead seabream initial intestinal microbiome, it was composed by 29 different families whereComamonadaceae,Enterobacteriaceae,Moraxellaceaealong withPorphyromonadaceaeprevailed.Following statistical analyses, a positive correlation was found between skin and intestine microbiome.This type of correlation has, also, been reported by Chaillou et al.(2015) that analyzed seafood spoilage microbiota and reported that a combination of environmental and fish gut microbiota prevailed.Wong and Rawls(2012) reported that fish intestinal microbiota is affected by the farming environment microbiota and Giatsis et al.(2015) showed that the rearing environment affected the gut microbiota in tilapia larvae.An interaction between the microbiota of sea water, gut microbiome and fish skin microbiome is evident.Briefly, this interaction has been attributed to fish that are fed at the same time and later, almost simultaneously, they empty their intestinal content in the sea.That means for a period of time, depending on sea currents, the bacterial population in the aquaculture water is strongly affected by the intestinal microbiome,and part of it remains on the fish skin.
The initial microbiome of the gilthead seabream gills was dominated(94.9%) by the familyRhodocyclaceae.Bacteria of this family have been detected by Giatsis et al.(2015) in recirculating aquaculture system(RAS) as well as in active suspension (AS) systems.FamilyRhodocyclaceaeis consisted of 12 genera (https://lpsn.dsmz.de/family/rhodocycla ceae) that exhibit versatile metabolism; anoxygenic photoheterotrophic(nitrogen-fixing aerobes) and sulfuroxidizing chemoautotrophic, methylotrophic, and anaerobic with propionic acid as one of the end-products.Genera of this family have been isolated from various environments including sewage treatment plants, ponds, and rivers, and participate in the bioremediation of organic wastes (Oren, 2014).The dominance of this family on the gills could, most likely, be attributed to the high concentration of nitrogen in the rearing aquaculture environment.In aquacultures nitrogen comes from two sources: the feed which is abundantly dispersed in the sea as well as the fish feces.In many cases,the fish density in the cages is rather high, and therefore sea currents are often weak to replenish water in cages, and thus the environment is ideal for the growth of anaerobic or aerobic denitrifying bacteria such as those of the familyRhodocyclaceae(Haro-Moreno et al., 2020).
Several studies have shown the positive effect of MAP application on fish quality as reviewed by Mendes, (2019) and Zhuang et al.(2020).Most of these works dealt with fish that had undergone some type of treatment before being packaged so that their microbiome was modified.The originality of the present study was based on the immediate packaging of whole fish without any pre-processing step and the application of novel NGS analysis for the characterization of the microbiome on three different fish tissues, i.e.skin, gills and intestines.During the 10-days storage of gilthead seabream in air or under MAP, there was a qualitative and quantitative differentiation in the skin, gills and intestines microbiome.In particular, the statistical analyses of family richness showed that AIR and MAP samples differentiated the total number of families among the samples tested.A decrease in richness index was observed– means lower number of families within each packaging – except the case of MAP application in the gill microbiome.Yet, in skin, gill, and intestines, as well as the "Unified" microbiome the number of families was higher in MAP than in AIR fish.Furthermore, the analyses of families diversity (alpha – diversity), showed that both AIR and MA packaging affected the number of families in each microbiome(skin, gill and, intestines) as well as the abundance of each family.The increased diversity of microbiota developed under MAP conditions has been reported by Drosinos and Nychas (1996).In general, during the 10-day cold storage, psychotropic and psychrophilic families of bacteria,were favored and grown, and several families that were predominant in the initial microbiome, were replaced at the end of the fish shelf life.For example, on fish skin, the initially dominant familiesComamonadaceae,EnterobacteriaceaeandMoraxellaceaeafter 10 days of aerobic storage were replaced by the familiesPseudoalteromonadaceae,Psychromonadaceae, andShewanellaceaethat all were psychotropic and/or psychrophilic.Along with cold storage, the use of MAP was also a factor that differentiated fish microbiome.The case of the familyPseudoalteromonadaceaewas distinctive, since after 10 days of iced storage it prevailed (45.5%) on the skin of fish stored in aerobic conditions and was practically absent from the MAP samples.The members of the familyPseudoalteromonadaceaeare aerobic, and thus they did not grow under MAP conditions.An opposite case is that of the familyComamonadaceae, which practically remained unaffected by the application of MAP both on the skin and intestines microbiota.Additionally,in the case of gills microbiome after 10 days of MAP storage at 0◦C, the initially dominant familyRhodocyclaceaethough it was significantly reduced and replaced by the familiesPsychromonadaceae,Shewanellaceae, was not eliminated.It was evident that MAP may modify the fish microbiome but only in the context of the initial microbiome since groups of bacteria may not be inhibited by the atmosphere modification.
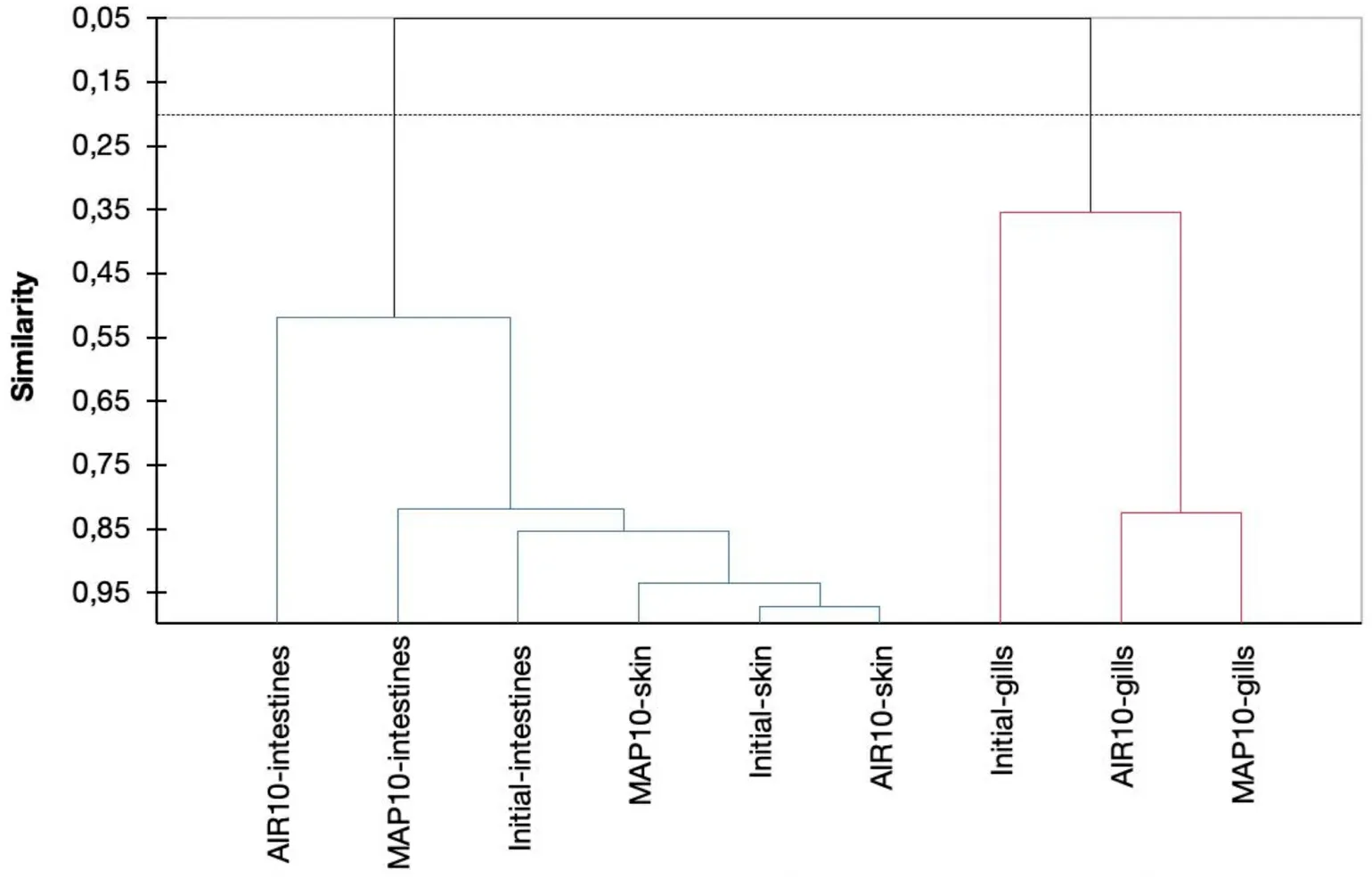
Fig.8.Similarity plot of the sensory scoring and microbiome data of the tested fish samples (AIR and MAP) initially and after 10 days of isothermal storage at 0 ◦C(Agglomerative hierarchical clustering).
The sensory evaluation of whole fish indicated that up to day 8 of storage at 0◦C, MAP samples exhibited higher sensory scorings than the conventional aerobic storage.However, at day 10 of iced storage (11 days after harvesting), both AIR and MAP samples were sensorially rejected.
There is a large amount of reported data on the shelf life of seafood(Zhuang et al., 2020, Odeyemi, Burke, Bolch, & Stanley, 2018) as well as fish, such as gilthead seabream packed under MAP or conventionally in aerobic conditions (Mendes, 2019).Lougovois et al.(2003) reported that whole gilthead seabream stored aerobically in ice was still sensorially acceptable after 14–15 days.Also, Cakli et al., (2007) reported 14 days shelf life for whole, gutted and ungutted sea bass and gilthead seabream stored in ice.In the latter work, fish had been washed once with tap water prior experimentation.It is evident that the shelf life of fish and seafood stored at low temperatures, either under MAP/vacuum or aerobically, depends on a wide range of parameters, including rearing conditions, slaughtering method, handling protocols and potential application of disinfection washing pre-treatments (Ntzimani et al.,2021).However, it is well documented that among the important parameters that affect fish shelf life are their initial microbial load, the type of bacteria that compose the microbiota, the processing as pre-treatment(gutted, fillet, etc.), packaging conditions as well as the temperature fluctuations during their transportation and storage (Tsironi, Lougovois,et al., 2019).
In this work the main factor for gilthead seabream sensory rejection by the assessors was the unpleasant odor, both on the skin and gills,mainly in the samples that were packed under MA.The odor of the MAP samples may be mainly attributed to the core families that were identified at the end of the day 10 of storage, i.e.Comamonadaceae,ShewanellaceaeandPsychromonadaceae.The bacteria of the familyComamonadaceaehad paramount presence in the fish microbiota, both on skin, and intestines, and was not affected by MA.This family includes anaerobic denitrifier and phosphorus-removing bacteria, often isolated from soil, and ponds (Willems, 2014).Yet, it has been reported that low dissolved oxygen water concentration (lower than 1 mg L-1) favors its presence (Sadaie et al., 2007).The distinctive presence of this family in the MAP fish perhaps could be attributed to the feed (nitrogen and phosphorus) that are not consumed by the fish, and potentially low sea current.However, it should be noted that the presence of these bacteria in farming facilities is considered as positive because they may remove the excessive quantities of nitrogen and phosphorus and improve water quality (Li et al., 2017).This was one of the prevailing families of the intestinal microbiome of gilthead seabream, and as it was statistically shown, there was a high correlation between the skin and intestinal microbiome.The familyShewanellaceae– also one of the predominant families both in the AIR and MAP samples – includes psychrotolerant,spoilage bacteria that are putative anaerobic, and more resistant to CO2thanPseudomonasand thus can grow under MAP conditions (Antunes-Rohling et al., 2019), producing H2S as well as trimethylamine and off-flavour dimethylamine compounds (Satomi et al., 2007).As far it concerns the bacteria of the familyPsychromonadaceae, they are inhabitants of sea ice, and aquatic environments.They are facultative anaerobes, but some species may be aerotolerant or aerobes.Bacterial of this family have been part of the spoilage microbiota of Pacific oysters,and Eastern oysters during refrigerated storage (Chen et al., 2019).
The results of the study indicate the linkage between the packaging conditions (aerobic or MAP) and the bacterial microbiome of three different fish tissues, i.e.skin, gills and intestines during iced storage of whole gilthead seabream.Novel application of NGS enabled the accurate identification of microbial taxa, including uncultivable organisms and those present in small numbers, providing a more detailed and efficient mapping of the bacterial community of the tested tissues.The bacterial microbiome was also correlated with the sensory scoring of fish using a typical QIM methodology.Future studies may focus on the investigation of other physical (colour, texture) and chemical fish quality parameters (volatile basic nitrogen, lipid oxidation,K-value or novelomicstools, such as proteomic analysis of the different fish tissues)on different fish species and correlation with the microbiome data.This approach may allow a systematic quality assessment of fish and evaluation of the effect of processing, packaging and storage conditions on the quality and shelf life of perishable food products.
5.Conclusion
The results of the study show the potential of MAP for modifying the initial bacterial microbiome in whole gilthead seabream during iced storage.The bacterial community is fish is greatly dependent on the aquaculture environment and may affect the quality deterioration of fish during subsequent transportation and storage.Appropriate packaging and retention of initial quality of fish until further processing (e.g.gutting, filleting) may extend shelf life by inhibiting microbial load and altering the dominant bacteria, that are related to the development of unpleasant off-odors and degradation of sensorial quality.NGS may be a powerful tool for the accurate identification of microbial taxa in different fish tissues, including also uncultivable organisms and those present at low levels (below the detection limits of the conventional culture-based techniques), providing the systematic and detailed evaluation of the spoilage mechanism of MAP fish products.
CRediT authorship contribution statement
Apollon Thomas:Methodology, Formal analysis, Investigation,Writing – original draft, Visualization.Spyros J.Konteles:Methodology, Formal analysis, Investigation, Writing – original draft, Visualization.Sotiris Ouzounis:Methodology, Formal analysis, Writing –original draft, Visualization.Spyros Papatheodorou:Methodology,Formal analysis, Investigation.Aliki Tsakni:Methodology, Formal analysis, Investigation, Writing – original draft, Visualization.Dimitra Houhoula:Conceptualization, Writing – review & editing, Supervision,Project administration, Funding acquisition.Theofania Tsironi:Conceptualization, Writing – review & editing, Supervision, Project administration.
Acknowledgment
This Project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-CurieGrant agreement 872217.
 Aquaculture and Fisheries2023年4期
Aquaculture and Fisheries2023年4期
- Aquaculture and Fisheries的其它文章
- Editorial: Improving fish from catch to the consumer: Post harvest handling, processing, packaging, transportation and storage
- Biogenic amines in fresh fish and fishery products and emerging control
- The role of digital technologies in supporting and improving fishery and aquaculture across the supply chain – Quo Vadis?
- Integrating omics technologies for improved quality and safety of seafood products
- Seasonal variation in the biochemical composition, condition index, and meat yield of the non-indigenous pearl oyster Pinctada imbricata radiata(Leach, 1814) from the West of the Aegean Sea, Greece
- Effective algorithmic operational framework for fish texture evaluation in industry: Achieving maturity
